Abstract
Inhalation of nanoparticles has been implicated in respiratory morbidity and mortality. In particular, carbon black nanoparticles are found in many different environmental exposures. Macrophages take up inhaled nanoparticles and respond via release of inflammatory mediators and in some cases cell death. Based on new data, we propose that exposure of macrophages (both a macrophage cell line and primary human alveolar macrophages) to carbon black nanoparticles induces pyroptosis, an inflammasome-dependent form of cell death. Exposure of macrophages to carbon black nanoparticles resulted in inflammasome activation as defined by cleavage of caspase 1 to its active form and downstream IL-1β release. The cell death that occurred with carbon black nanoparticle exposure was identified as pyroptosis by the protective effect of a caspase 1 inhibitor and a pyroptosis inhibitor. These data demonstrate that carbon black nanoparticle exposure activates caspase 1, increases IL-1β release after LPS priming, and induces the proinflammatory cell death, pyroptosis. The identification of pyroptosis as a cellular response to carbon nanoparticle exposure is novel and relates to environmental and health impacts of carbon-based particulates.
Keywords: Caspase, Cell Death, Inflammation, Innate Immunity, Interleukin, Macrophage, Inflammasome, Nanoparticles, Pyroptosis
Introduction
Macrophages are critical regulators of local immune homeostasis. They are highly adaptive components of the innate immune system and respond in diverse ways to pathogens and other potential danger signals (1–3). In the lung, the alveolar macrophage is the first line of defense against environmental exposures. Alveolar macrophages phagocytose particulate matter, release inflammatory cytokines, and interact with other cells and molecules through the expression of surface receptors. One way in which an immune response is generated in alveolar macrophages is through the phagocytosis of deposited particles within the respiratory tract (4).
The nanoparticle industry has expanded substantially in recent years. A variety of engineered carbon nanoparticles is used in consumer products such as car tires, rubber, and printer toner cartridges (5). Nanoparticles are also being used as novel means of drug delivery. Additionally, carbonaceous nanoparticles are present as an environmental contaminant. Combustion processes are a significant source of carbon nanoparticles. Elemental carbon-based nanoparticles with a diameter of less than 100 nm are a major part of diesel exhaust and ambient pollution (6).
Particulate ambient pollution is known to cause adverse health effects in susceptible individuals and aggravates existing respiratory conditions such as asthma and chronic obstructive pulmonary disease (7). Even moderate levels of ambient air particulates are known to induce acute adverse health effects such as mortality in heart and lung diseases and chronic lung morbidity (8). Ultrafine particles are unique in their ability to bypass mucociliary clearance mechanisms and penetrate into deeper regions of the respiratory tract (9–12). Although bulk elemental carbon is considered chemically inert (as in diamond and graphite), seemingly inert substances have been shown to elicit an inflammatory response when exposure occurs with nanoscale particles compared with an equivalent mass dose of larger particles (11–15). Carbon black (CB)2 nanoparticles can cause cytotoxic injury, increase levels of proinflammatory chemokines, and inhibit cell growth (16).
There are several explanations for this increased toxicity, including the increased surface area of nanoparticles (10, 12, 14, 17–22). In a previous study, acute adverse effects of different types of carbonaceous nanoparticles instilled in mice strongly correlated with particle size and surface area (23). A surface area threshold of ∼20 cm2 was defined for acute lung inflammation in mice below which no inflammatory responses were observed (23). CB nanoparticles showed higher surface reactivity compared with a similar dose of larger particles (24). CB nanoparticles have also been shown to induce oxidative stress in alveolar macrophages, and it is believed that this capacity for oxidation may be mediated by particle surface functionality (19, 25–29). A recent study showed that the oxidative potency of CB nanoparticles correlates with their surface area and inflammatory responses (30). A possible mechanism for CB particle-related inflammation involves direct and indirect reactive oxygen species (ROS) generation by particle-cell interactions, which in turn activates redox-sensitive transcription of proinflammatory genes (30). ROS have been implicated as the cause of significant inflammation and, in some cases, cell death (31).
One possible outcome of macrophage exposure to nanoparticles is cell death. Cell death may be categorized according to several characteristics including noninflammatory or proinflammatory and accidental or programmed. Apoptosis, perhaps the best characterized of these mechanisms, is a programmed and noninflammatory process. It is characterized by distinctive DNA cleavage as well as activation of the executioner caspases 3 and 9 (31, 32). In contrast to apoptosis, necrosis is defined as an accidental and proinflammatory form of cell death in which the plasma membrane loses its integrity, allowing rapid fluid influx, leading to cell swelling and lysis (32–34). Pyroptosis is a recently described mechanism of cell death, sharing unique characteristics with both necrosis and apoptosis (32–36). It is defined by its dependence on inflammasome activation and caspase 1 activity. Inflammasomes, which can differ in their subunit composition, have been shown to activate caspase 1, which in the setting of a microbial stimulus activates the proinflammatory cytokines IL-1β and IL-18 (33, 34, 37). Like apoptosis, pyroptosis is a form of programmed cell death. But unlike apoptosis, pyroptosis is characterized by loss of membrane integrity. This is due to caspase 1-dependent insertion of a pore into the membrane, leading to fluid influx, cell swelling, and lysis (38). Pyroptosis ultimately leads to release of cellular contents and inflammation (32–34, 36, 38, 39).
The recent expansion of the nanotechnology industry as well as the continually growing sources of combustion derived pollution warrants investigation into the potential health effects of these nanoparticles. In this study we examined the effect of CB nanoparticles on the inflammasome and pyroptosis. The data show that macrophage exposure to 20 ± 6 nm CB nanoparticles induces caspase 1 activation and IL-1β release and the proinflammatory form of cell death, pyroptosis.
EXPERIMENTAL PROCEDURES
Source of Manufactured Nanomaterial
TiO2 and CB nanoparticles were purchased from Degussa, GmbH (Düsseldorf, Germany). The manufacturer's stated average diameters of titanium dioxide (TiO2) nanoparticles (Degussa P25) and CB nanoparticles (Degussa Printex 90) are 21 and 14 nm, respectively. The nanoparticles were used as received from the manufacturer without modification.
Bulk Characterization of Nanoparticles
Powder x-ray diffraction (XRD) was used to identify crystalline phases of the sample. XRD was performed using a Bruker D-5000 q-q diffractometer with Kevex-sensitive detector (Madison, WI). High resolution transmission electron microscopy (TEM; (JEOL JEM-2100F, Japan) operating at 200 kV was used to image the nanoparticles and measure their diameters to compare the average diameter to the manufacturer's specifications. Samples for TEM analysis were deposited from methanol suspensions onto copper grids. Dynamic light scattering (DLS, Beckman Coulter Delsa Nano C, Brea, CA) was used to measure hydrodynamic diameter of the nanoparticle aggregates in reduced serum media (Opti-MEM, Invitrogen), which was used as a cell culture medium in the cytotoxicity experiments. Inductively coupled plasma optical emission spectroscopy analysis was performed to check for metal impurities in CB nanoparticles. The nanoparticles were digested in concentrated nitric acid at 90 °C before the inductively coupled plasma analysis. The digested solutions were filtered and centrifuged for 30 min at 14,000 rpm to remove nanoparticles and aggregates that were not dissolved. The final solutions were analyzed by inductively coupled plasma optical emission spectroscopy (Varian 720 ES, Walnut Creek, CA).
Surface Characterization of Nanoparticles
Surface area and surface composition of the TiO2 and CB nanoparticles were examined. Surface area measurements of powdered samples were performed on an automated multipoint Brunauer-Emmett-Teller (BET) surface area apparatus (Quantachrome Nova 4200e, Boynton Beach, FL) using nitrogen gas as the adsorbent. Samples were degassed at 100 °C for 24 h under vacuum before the analysis. The surface area of TiO2 and CB nanoparticles was calculated using the seven-point BET method. X-ray photoelectron spectroscopy was used to probe the surface chemical composition characteristics of the powdered samples (Ultra-Axis DLD, Kratos, Manchester, UK). The system has been described before (40).
Demonstration of Intracellular CB Nanoparticles (TEM)
Samples were fixed overnight with 2.5% glutaraldehyde in 0.1 m cacodylate buffer. Post-fixation was carried out for 1 h at room temperature with a buffered 1% osmium tetroxide solution reduced with 1.5% potassium ferrocyanide. Samples were en bloc-stained using 2.5% uranyl acetate. Cells were then rinsed and dehydrated. Infiltration of Spurr's epoxy resin and acetone was carried out over several days to 100% resin and cured overnight in a 70 °C oven. Sections of 100-nm thickness were cut using a Leica EM UC6 ultramicrotome. Grids were then counterstained with 5% uranyl acetate for 12 min and Reynold's lead citrate for 5 min. Samples were imaged using a JEOL 1230 transmission electron microscope.
Cell Culture
RAW264.7 cells were obtained from ATCC (#TIB-71) and maintained in DMEM with 10% fetal bovine serum and gentamycin, 40 μg/ml. Cells were subcultured every 2–3 days. Experiments were run in 6-well Costar tissue culture plates, 96-well assay plates, or coverslip chamber slides.
Human Alveolar Macrophages
To obtain normal human alveolar macrophages, subjects were recruited who were nonsmokers with no underlying medical conditions and on no medications other than possible birth control. After informed consent was obtained, subjects underwent standard flexible bronchoscopy. Bronchoalveolar lavage was performed by instilling 20 ml of normal saline into a tertiary bronchus up to five times in three different lung segments. The first collection of five was discarded for possible contamination from upper airway secretions or by lidocaine, which is used to locally anesthetize the subject during the procedure. The remaining lavage was transported to the laboratory where fluid was filtered through sterile gauze and centrifuged at 200 × g for 5 min to pellet cellular material. Cells were washed twice in PBS and finally re-suspended in RPMI plus Glutamax for cell culture. Cyto-prep slides were also made with the cells and stained with Wright stain. Slides were microscopically examined to ensure that greater than 95% of the cells were macrophages (41–43).
The cells were then placed in culture and exposed to CB nanoparticles. All procedures and protocols described in this communication were approved by the University of Iowa Institutional Review Board. Written informed consent was obtained, and all clinical investigation was conducted according to the principles expressed in the Declaration of Helsinki.
Whole Cell Protein Isolation
Whole cell protein was obtained by lysing the cells on ice for 20 min in 200 μl of lysis buffer (0.05 m Tris, pH 7.4, 0.15 m NaCl, 1% Nonidet P-40, with added protease and phosphatase inhibitors: 1 protease minitab (Roche Biochemicals)/10 ml and 100 μl of 100× phosphatase inhibitor mixture (Calbiochem)/10 ml. The lysates were sonicated for 20 s, kept at 4 °C for 30 min, and spun at 15,000 × g for 10 min, and the supernatant was saved. Protein determinations were made using the Bradford Protein assay from Bio-Rad. Cell lysates were stored at −70 °C until use.
Cell Supernatant Protein Isolation
To isolate proteins from cell supernatants, macrophages were cultured in Opti-MEM® from Invitrogen to allow for reduced serum culture. Cell supernatant protein was obtained by concentrating the supernatants in Amicon p10 filter tubes, spun at 3000 × g for 30 min. Protein determinations were made using the Bradford Protein assay from Bio-Rad. Concentrated supernatants were stored at −70 °C until use.
Western Analysis
Western analysis for the presence of active caspase 1 was performed on whole cell proteins and concentrated supernatants from RAW cell experiments. 30 μg of protein was mixed 1:1 with 2− sample buffer (20% glycerol, 4% SDS, 10% β-mercaptoethanol, 0.05% bromphenol blue, and 1.25 m Tris, pH 6.8; all chemicals were from Sigma), heated to 95 °C for 5 min, loaded onto a 10% SDS-PAGE gel, and run at 100 V for 90 min. Cell proteins were transferred to PVDF (Bio-Rad) by semidry transfer (Bio-Rad). Equal loading of the protein groups on the blots was evaluated using Ponceau S, a staining solution designed for staining proteins on PVDF membranes or by stripping and reprobing with antibodies to β-actin or GAPDH. The PVDF was dried and then incubated with the primary antibody overnight in 5% milk. The blots were washed ×4 with TBS and incubated for 1 h with horseradish peroxidase-conjugated anti-rabbit or mouse IgG antibody. Immunoreactive bands were developed using a chemiluminescent substrate (ECL Plus, Amersham Biosciences). An autoradiograph was obtained with exposure times of 10 s to 2 min.
IL-1β Release
For these studies RAW cells were cultured in standard medium for 24 h with and without LPS (10 ng/ml). After the culture period, the supernatants were harvested and stored at −70 °C until assayed. The amount of IL-1β in the supernatant was measured by ELISA (R & D Systems, Minneapolis, MN).
Cell Survival Analysis
For analysis of cell survival, macrophages were cultured in 96-well tissue culture plates. After incubations with nanoparticles, plasma membrane integrity was assayed by two methods (LDH release and PrestoBlue assay). Triplicate cultures were performed on all experiments. LDH released into the supernatant was monitored using CytoTox-ONETM Homogeneous Membrane Integrity Assay, which measures LDH release via a coupled fluorescent assay (Promega). PrestoBlue Cell Viability Assay was done following the manufacturer's protocol.
Whole Cell DNA Isolation/DNA Analysis
For DNA analysis, cells were cultured in 100-mm tissue culture plates. After incubations with particles, whole cells were harvested in 500 μl of PBS, and DNA was isolated (DNeasy Blood and Tissue kit, Qiagen, Valencia, CA). DNA concentration was measured, and equal concentrations were loaded and run on a SYBR Green gel (E-Gel 2% with SYBR Safe, Invitrogen). The gel was visualized with ultraviolet light, and samples were examined for laddering.
Quantitative RT-PCR
Total RNA was isolated using the RNAqueous-4PCR kit (Ambion Inc., Austin, TX) following the manufacturer's instructions. RNA quality and quantity were assessed with Experion automated electrophoresis system (Bio-Rad) using the Experion RNA StdSens Analysis kit according to the manufacturer's protocol. RNA quality was considered adequate for use if the 28 S/18 S ratio was >1.2 and the RNA Quality Indicator (RQI) was >7. Total RNA (300 ng) was reverse-transcribed to cDNA using a iScript cDNA Synthesis kit (Bio-Rad) following the manufacturer's instructions. PCR reactions were performed as previously described (44). Specificity of the amplification was confirmed using melting curve analysis. Data were collected and recorded by CFX Manager Software and expressed as a function of threshold cycle (CT). The relative quantity of the gene of interest was then normalized to relative quantity of HPRT (ΔΔCT). The sample mRNA abundance was calculated by the formula 2−(ΔΔCT).
Specific primer sets used are as follows (5′ to 3′): IL-1β forward (5′-CTCCAGGGACAGGATATGGA-3′) and reverse (5′-TTCTGCTTGAGAG GTGCTGA-3′); IL-18 forward (5′-ACAGCTTCGGGAAGAGGAAAGGAA-3′) and reverse (5′-TGTCTTCTACTGGTTCAGCAGCCA-3′); TNFα forward (5′-AGGACACCATGAGCACTGAAAGCA-3′) and reverse (5′-TTGAGGGTTTGCTACAACATGGGC-3′).
RESULTS
Nanoparticle Characterization
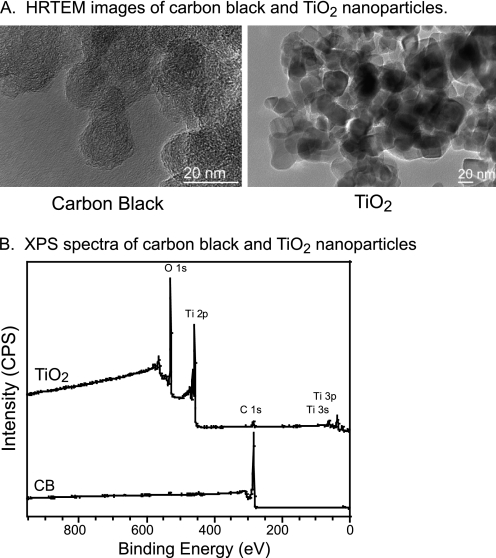
The results of particle characterization are summarized in Table 1. Primary particle diameters obtained from the TEM images (Fig. 1A) indicate that TiO2 nanoparticles have a diameter of 23 ± 3 nm, and CB nanoparticles have a diameter of 20 ± 6 nm (Table 1). For the CB nanoparticles, this result is close to the manufacturer's specified diameter of 14 nm. The aggregate size distributions of TiO2 and CB nanoparticles were measured using DLS in Opti-MEM media. Both types of nanoparticles form aggregates in the cell culture media. TiO2 nanoparticles have bimodal size distribution with peaks at 146 ± 60 and 985 ± 450 nm with a larger size distribution dominating over a smaller size distribution. CB nanoparticles have aggregate sizes of 175 ± 80 nm (Table 1). The XRD analysis of TiO2 nanoparticles indicates that there are anatase and rutile crystalline phases present in the sample (45). A diffraction pattern for CB nanoparticles is similar to the literature reports for CB (46) and indicates crystalline phases of graphitic domains. X-ray photoelectron spectroscopy analysis of TiO2 nanoparticles indicates 33, 61, and 6% of titanium, oxygen, and carbon, respectively, showing close to stoichiometric titanium:oxygen ratio and residual hydrocarbon and hydroxyl groups present on the surface of the sample (Fig. 1B). High resolution x-ray photoelectron spectroscopy analysis of CB nanoparticles shows the presence of carbon at 285.0 eV with traces of oxygen at 530.0 eV (Fig. 1B). Inductively coupled plasma optical emission spectroscopy analysis of CB nanoparticles digested in concentrated nitric acid revealed small amounts of metal impurities such as calcium (0.02%) and potassium (0.04%) present in the sample. The BET-specific surface areas of TiO2 and CB nanoparticles calculated using the 7-point BET method are 41 ± 1 and 279 ± 6 m2/g, respectively (Table 1). CB is considered to be a microporous material. The external surface area of CB nanoparticles calculated using the Halsey method (47) is 176 ± 9 m2/g.
TABLE 1.
Summary of physicochemical characterization data of the nanoparticles
| Physiochemical characteristics | Detection method | TiO2 | Carbon black |
|---|---|---|---|
| Primary particle size | TEM | 23 ± 3 nm | 20 ± 6 nm |
| Phase | XRD | Anatase/rutile | Graphitic domains |
| BET surface area | BET | 41 ± 1 m2/g | 279 ± 6 m2/g |
| Aggregate size in Opti-MEM media | DLS | 146 ± 60 nm | 175 ± 80 nm |
| 985 ± 450 nm |
FIGURE 1.
Nanoparticle characterization. A, shown is high resolution TEM images of CB and TiO2 nanoparticles. The scale bar in both images is 20 nm. Both nanoparticles form aggregates when in solution. Particles were sonicated for 1 h and vortexed vigorously before cell exposure. Primary particle diameters were obtained from the images. B, surface composition of TiO2 and CB nanoparticles was measured using x-ray photoelectron spectroscopy.
CB Nanoparticle Exposure Induces Macrophage Cell Death (RAW264.7 Cells)
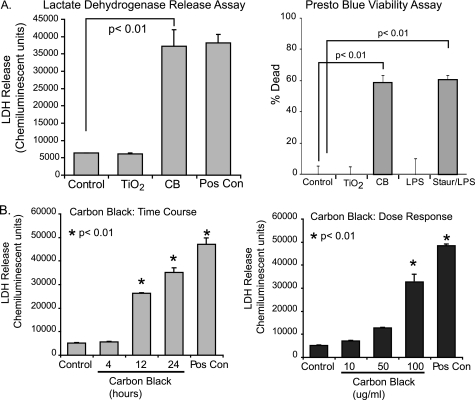
To evaluate the effect of nanoparticles on macrophage viability, RAW264.7 cells were seeded into 96-well tissue culture plates and cultured for 24 h with TiO2 (30 μg/cm2) or CB nanoparticles (30 μg/cm2). At the end of the culture period, one group was treated with ATP as a positive control for cell death (31, 38, 45–47). Samples were assayed for LDH release. Fig. 2A shows that CB nanoparticles and not TiO2 nanoparticles induce LDH release from macrophages. To confirm cell death with an alternative assay, a PrestoBlue cell viability reagent was used. PrestoBlue analyzes cell death by determining the level of reducing activity associated with living cells. RAW264.7 cells were seeded into 96-well tissue culture plates and cultured for 24 h in the same conditions. As with the LDH release assay, the PrestoBlue analysis showed cell killing with exposure to CB nanoparticles and not TiO2 nanoparticles.
FIGURE 2.
CB nanoparticles induce macrophage cell death. A, RAW264.7 cells were exposed to CB (30 μg/cm2) or TiO2 (30 μg/cm2) nanoparticles for 24 h. One group was treated with ATP as a positive control for LDH release. At the end of the culture period, LDH analysis was performed to determine cell viability. The experiment was repeated using identical conditions. A PrestoBlue cell viability assay was performed as indicated by the manufacturer. Significance was determined using nonpaired Student's t test. Staur, staurosporine. B, RAW264.7 cells were exposed to CB nanoparticles (30 μg/cm2) for 4, 12, and 24 h. ATP was used as a positive control for LDH release. At the end of the culture period, an LDH assay was performed to determine cell viability. Additionally, RAW264.7 cells were exposed to CB nanoparticles at 3, 15, or 30 μg/cm2 for 24 h. An LDH assay was performed. Significance was determined using nonpaired Student's t test.
To further characterize the cell death caused by CB nanoparticle exposure, both time and dose-response experiments were performed. Fig. 2B demonstrates an increase in LDH release by 12 h that continues increasing through 24 h of particle exposure. Optimal effects were seen at a dose of 30 μg/cm2. As a composite, these data show that CB nanoparticles, and not TiO2 nanoparticles, decrease plasma membrane integrity leading to cell death.
Nanoparticle-induced Plasma Membrane Disruption Is Characterized by an Increase in Cell Size
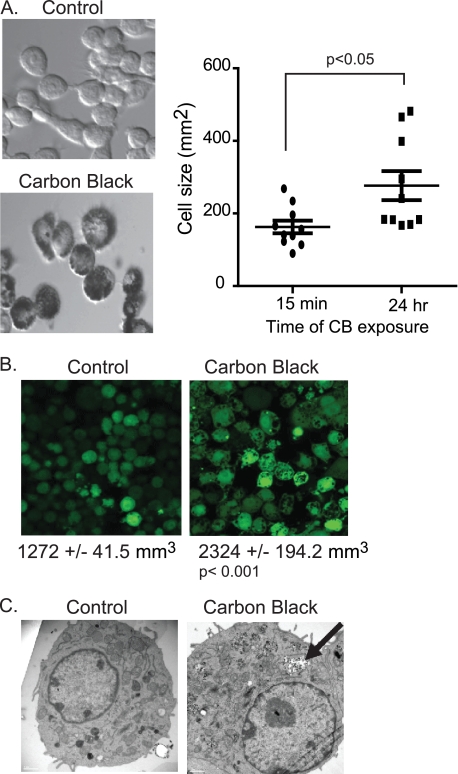
The LDH and PrestoBlue assays suggest that exposure to CB nanoparticles disrupts the plasma membrane. To determine whether this reduced plasma membrane integrity affected cell size, cell size was quantified after exposure to CB nanoparticles. In the first experiment (Fig. 3A), cells were exposed to CB nanoparticles, and images were obtained immediately (15 min) and again after 24 h. Cell size was determined by drawing circles around representative cells from 10 fields and calculating area using ImageJ software. Fig. 3A shows an increase in cell size after exposure to CB nanoparticles (mean of 162.7 ± 17.57 mm2 for control cells compared with 276.9 ± 39.99 mm2 for CB nanoparticle exposed macrophages). To analyze volume changes, macrophages were loaded with a fluorescent tracer, cell tracker green 5-chloromethylfluorescein diacetate (Invitrogen). Fig. 3B shows an increase in volume with CB nanoparticle exposure. To confirm that the macrophages internalize the particles, transmission electron microscopy was used to analyze particle exposed macrophages. Fig. 3C demonstrates that CB nanoparticles are taken up by macrophages and appear to localize both in the cytosol and in membrane-bound vesicles.
FIGURE 3.
CB nanoparticles cause an increase in macrophage cell size. A, bright field images of CB nanoparticle-exposed cells were obtained 15 min after exposure and again after 24 h. Circles were drawn around representative cells from 10 fields and then used to calculate cellular area. B, fluorescent images of cells were obtained before CB nanoparticle exposure and again 24 h after exposure. Cells were loaded with cell tracker green 5-chloromethylfluorescein diacetate. Average cellular volume was calculated from the level of fluorescence per cell. C, transmission electron microscopy images of control and CB nanoparticle-exposed cells.
CB Nanoparticles Activate the Inflammasome (Caspase 1 and IL-1β Release)
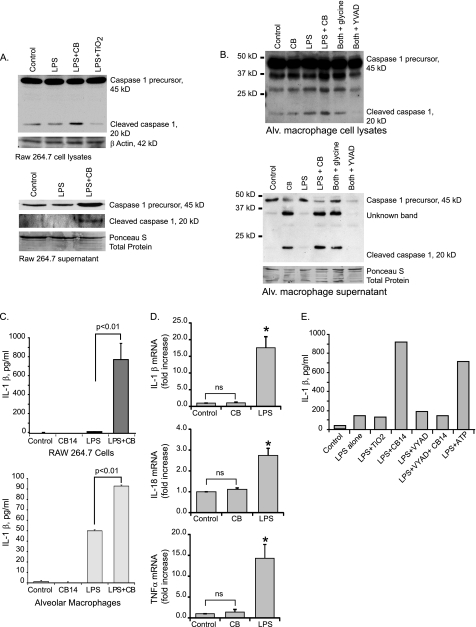
Both necrosis and pyroptosis are characterized by LDH release and loss of plasma membrane integrity. We asked if CB nanoparticle exposure activates the inflammasome (central to the pyroptosis form of cell death). The inflammasome two primary activation markers, caspase 1 activity and IL-1β release, were measured. Cells were primed for 3 h with LPS (10 ng/ml). After priming, media were replaced, and fresh LPS was added along with CB and TiO2 nanoparticles for an additional 6 h as previously described (48). Western analysis was performed using whole cell lysates as well as concentrated supernatants. Fig. 4A shows that in RAW264.7 cells, significantly more of the 20-kDa cleaved caspase 1 was present in both the lysates and supernatants of cells exposed to CB nanoparticles as opposed to those exposed to TiO2 nanoparticles. Recent studies have defined the release of active caspase 1 as a valid measure of inflammasome activity (49–53).
FIGURE 4.
CB nanoparticles activate the inflammasome. A, activation of caspase 1 in RAW264.7 cells is shown. 3 × 106 RAW264.7 cells per well were seeded into culture plates. Cells were primed for 3 h with LPS (10 ng/ml). After 3 h medium was aspirated off cells and replaced with fresh medium. LPS was re-added, and CB (30 μg/cm2) or TiO2 (30 μg/cm2) nanoparticles were added for an additional 6 h. After the culture period, whole cell lysates were harvested in Western lysis buffer. Supernatants were concentrated, and proteins were analyzed. Western analysis for caspase 1 was performed on cellular lysates and concentrated supernatants. B, 3 × 106 human alveolar (Alv.) macrophages per well were seeded into culture plates. Cells were primed and cultured as detailed above. Western analysis for caspase 1 was performed on cellular lysates and concentrated supernatants. β-Actin and Ponceau S stain demonstrate equal loading. C, 1 × 106 RAW264.7 cells per well were seeded into culture plates. Cells were cultured as above, except for an extended second incubation time (16–24 h). Supernatants were harvested, and IL-1β levels were measured by ELISA. Significance was determined using nonpaired Student's t test. D, expression of IL-1β, IL-18, and TNFα mRNA in human alveolar macrophages exposed to LPS or CB nanoparticles is shown. 3 × 106 human alveolar macrophages per well were seeded into culture plates. Cells were incubated with CB nanoparticles (30 μg/cm2) or LPS (10 ng/ml) for 4 h. RNA was isolated, and quantitative real-time PCR was performed. *, p < 0.01 by Student's t test (compared with control). ns, not significant. E, 1 × 106 RAW264.7 cells per well were seeded into culture plates. Cells were primed for 3 h with LPS (10 ng/ml). For the second incubation period, LPS was re-added, and CB nanoparticles (30 μg/cm2), TiO2 nanoparticles (30 μg/cm2), YVAD (100 μm), and ATP were added for an additional 24 h. Supernatants were harvested, and IL-1β levels were measured by ELISA.
To determine whether caspase 1 activation also occurred in a relevant human primary cell, human alveolar macrophages were cultured in the same conditions. Cell lysates and supernatants were harvested, and Western analysis was performed for active caspase 1. Fig. 4B shows that in human alveolar macrophages, CB nanoparticle exposure caused activation of caspase 1 as demonstrated by the presence of the 20-kDa cleaved caspase 1 in both cell lysates and supernatants. To confirm the caspase 1 activation, IL-1β release was measured using an ELISA. Fig. 4C demonstrates that with LPS priming, both RAW264.7 cells and human alveolar macrophages increase IL-1β release with CB nanoparticle exposure (p < 0.01).
Fig. 4C shows that CB alone does not induce IL-1β protein release from macrophages. To further examine the role of LPS priming in the CB augmentation of IL-1β release, we examined LPS- or CB-exposed cells for IL-1β, IL-18, and TNFα mRNAs. We found that LPS, but not CB, induced transcript up-regulation (Fig. 4D). This supports our conclusion that CB alone activates the inflammasome leading to cell death (pyroptosis), whereas CB plus a microbial stimulus leads to both pyroptosis and IL-1β release.
To confirm that the increase in IL-1β release with CB nanoparticles was due to caspase 1 activation, an experiment was performed using the caspase 1 inhibitor, YVAD. Fig. 4E shows that the increase in IL-1β release with CB nanoparticles is blocked in the YVAD-exposed cells. As a composite, these data suggest that CB nanoparticle exposure in macrophages activates the inflammasome as shown by caspase 1 activation and IL-1β release.
CB Nanoparticle-induced Cell Death Is Not Due to Apoptosis
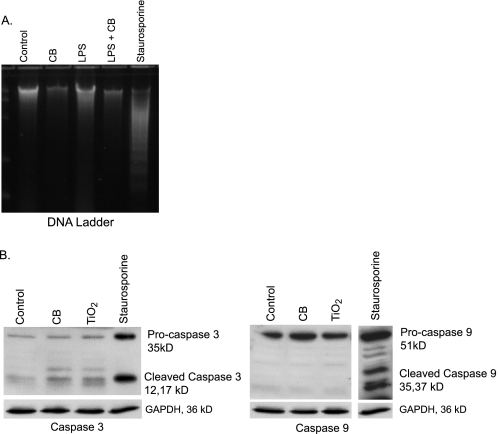
Because the CB nanoparticle-induced cell death is characterized by loss of plasma membrane integrity, it is unlikely that it is an apoptotic process. However, to confirm that apoptosis was not involved, we examined the effect of CB nanoparticles on apoptosis. Macrophages were exposed to CB or TiO2 nanoparticles for 16–24 h. Staurosporine was used as a positive control for apoptosis. After the incubation period, DNA was isolated from whole cell lysates. The DNA concentration was measured, and equal amounts were run on a SYBR Green gel and visualized using ultraviolet light. Fig. 5A shows that although the cells exposed to staurosporine show distinctive DNA laddering characteristic of apoptosis, none of the CB nanoparticle-exposed cells show DNA laddering. In a second set of experiments, macrophages from CB nanoparticle-exposed cells were lysed, proteins were isolated, and Western analysis was performed for activation of two caspases linked to apoptosis, caspase 3 and caspase 9 (31). Fig. 5B shows that caspases 3 and 9 were activated in cells exposed to staurosporine but not in those exposed to CB nanoparticles. Taken in combination with the LDH and PrestoBlue assays, this demonstrates that CB nanoparticles induce non-apoptotic cell death.
FIGURE 5.
Nanoparticle-induced cell death is not apoptosis. A, 5 × 106 RAW264.7 cells were seeded into 100-mm culture dishes and incubated with LPS (10 ng/ml), CB nanoparticles (30 μg/cm2), or both for 24 h. One group was exposed to staurosporine (1 μm) as a positive control for apoptosis. After the culture period, DNA was isolated. DNA concentration was measured, and equal concentrations were loaded and run on a SYBR Green gel. The gel was visualized with ultraviolet light, and samples were examined for laddering. B, 3 × 106 RAW264.7 cells/well were seeded into culture plates. Cells were exposed to CB nanoparticles (30 μg/cm2), TiO2 nanoparticles (30 μg/cm2), or staurosporine (1 μm) for 24 h. Proteins were isolated, and Western analysis for the apoptosis-associated caspases 3 and 9 was performed.
Inhibition of Caspase 1 Blocks CB Nanoparticle-induced Cell Death in Macrophages
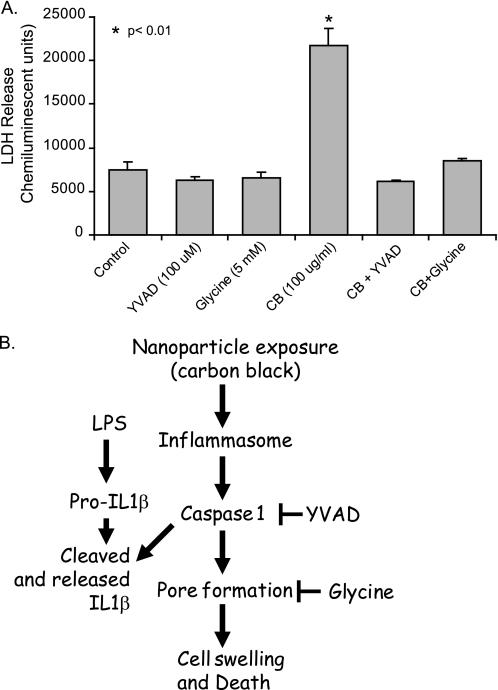
In light of the observation that CB nanoparticles activate the inflammasome and induce cell death in macrophages, we next sought to characterize the specific mechanism of cell death. The cell death modality pyroptosis is characterized by caspase 1 activation and the subsequent opening of cell membrane pores, resulting in an influx of extracellular fluid and eventual cell lysis (38). To confirm that the observed non-apoptotic cell death was pyroptosis, we examined the effects of a caspase 1 inhibitor (YVAD) and a pyroptosis inhibitor (glycine) on RAW264.7 cells exposed to CB nanoparticles. RAW264.7 cells were seeded into 96-well tissue culture plates and cultured for 24 h with CB nanoparticles (30 μg/cm2), CB nanoparticles in the presence of YVAD, or CB nanoparticles in the presence of glycine. At the end of the culture period, all samples were assayed for LDH release. Fig. 6A shows that, as previously demonstrated, CB nanoparticle-exposed cells showed decreased membrane integrity as evidenced by LDH release. Both YVAD and glycine attenuated the effects of CB nanoparticle-induced LDH release. Cells exposed to CB nanoparticles in the presence of either of these inhibitors released near control levels of LDH.
FIGURE 6.
Blocking caspase 1 activation protects macrophages from CB nanoparticles toxicity. A, RAW264.7 cells were exposed to CB nanoparticles (30 μg/cm2), CB nanoparticles in combination with the pyroptosis inhibitor glycine (5 mm), CB nanoparticles in combination with the caspase 1 inhibitor YVAD (100 μm), or TiO2 nanoparticles (30 μg/cm2) for 24 h. At the end of the culture period, LDH analysis was performed to determine cell viability. B, the diagram summarizes the CB nanoparticle-induced pathway to inflammasome activation and pyroptosis, as supported by this study.
DISCUSSION
In this study we found that CB nanoparticles induced cell death in macrophages and that this occurred in the absence of any detectable transition metals. These data show that after phagocytosis of the CB nanoparticles, macrophages increased in size; that is, the opposite of the cellular condensation associated with apoptosis. Macrophage exposure to CB nanoparticles led to inflammasome activation, as characterized by caspase 1 activation and IL-1β release. The identification of the cell death undergone by exposed cells as pyroptosis was confirmed by the inhibiting effects of both a caspase 1 inhibitor and a pyroptosis inhibitor on CB nanoparticle-induced cell death (Fig. 6B).
A daily dose of CB in the working environment can be as large as 120 μg/kg person, assuming a threshold limiting value for respirable carbon black of 2.5 mg/m3 (54). Additionally, an 8-h average amount of elemental carbon detected on a heavily traveled roadway in Harlem was 6.2 μg/m3 (55), which corresponds to the alveoli burden of 7 μg, assuming a respiratory volume of 0.7 m3/h and 0.2 alveolar deposition fraction for 20-nm particles based on a human deposition model (56). In the current study 30 μg/cm2 of CB nanoparticles were applied to cell culture wells. Although it is difficult to compare these doses with the mass/mass and mass/volume concentrations discussed above, it is within the mass range that is observed in occupational and environmental settings but does represent a substantial dose for respirable carbon black.
Pyroptosis, a proinflammatory form of cell death, proceeds through the activation of the inflammasome, leading to cleavage of caspase 1 into its active form. Once activated, caspase 1 cleaves the proinflammatory cytokines IL-1β and IL-18 into their active forms, allowing for their release into the extracellular environment. Although indicative of caspase 1 activation, the release of these inflammatory cytokines is not required for caspase 1 activation or pyroptosis. The production of pro-IL-1β and pro-IL-18 is mediated by toll-like receptors. The release of active IL-1β and IL-18 associated with pyroptosis in previous reports (36, 39, 58–65) has all included toll-like receptor stimulatory effects of microbial antigens. Thus, although priming cells with LPS before nanoparticle exposure allows for an additional confirmation of caspase 1 activation, inflammasome activation can occur separately from LPS priming. This is supported by previous reports showing that caspase 1-induced cell death may proceed independently of IL-1β and IL-18 secretion when a microbial stimulus is not present (38, 66).
Pyroptosis is characterized by a loss of membrane integrity (32–34, 38). Although the exact mechanism of this remains unknown, it has been demonstrated that membrane pore formation occurs, leading to cell swelling and necrosis-like lysis. Our data supports pyroptosis after CB nanoparticle exposure with evidence of cell swelling, caspase 1 activation, and cell death.
There have been a number of inflammasomes characterized with different subunits. Although this study does not specify which inflammasome CB nanoparticles activate, it has been shown that particulate matter including silica, asbestos, monosodium urate crystals, cholesterol crystals (37, 67, 68), and aluminum adjuvants (69) induce a caspase 1-dependent inflammatory response mediated by the NALP3 inflammasome (31, 37, 70–72). As such, these data suggest that CB nanoparticles activate the NALP3 inflammasome as well, although ongoing studies will further characterize the CB nanoparticle inflammasome. To the best of our knowledge this is the first instance in which nanoparticles have been implicated in inducing pyroptosis (31).
Several mechanisms of inflammasome activation have been proposed, including the generation of ROS (7,13, 18, 73–76, 78), potassium efflux (50), cathepsin B (57), and phagosomal destabilization (28, 48, 72, 77). Disintegration of the cellular membrane by CB nanoparticles can cause the production of ROS. Aam and Fonnum (28) showed that low doses of CB nanoparticles activate rat alveolar macrophages to produce ROS. They suggested that the ERK MAPK pathway participates in intracellular signaling leading to the ROS generation. Hornung et al. (48) demonstrated that the NALP3 inflammasome activation induced by silica crystals and aluminum salts could be replicated via sterile lysosomal damage, implicating intracellular pH or cathepsin B activity in inflammasome activation. Even nanoparticles made from materials that are considered inert in bulk form have been found to induce pulmonary inflammation when exposure occurs with nanoscale particles. Although this study did not find TiO2 nanoparticles to be inflammasome activating in macrophages, Yazdi et al. (51) showed that in human keratinocytes nano-TiO2 activates the NALP3 inflammasome and induces IL-1β. Any or all of the potential mechanisms discussed may apply to the CB nanoparticle mechanism of inflammasome activation. Further investigation into the ROS generated by alveolar macrophages in response to CB nanoparticle exposure is warranted.
The present study shows that macrophage exposure to CB nanoparticles activates the inflammasome leading to pyroptosis. CB merits further investigation into its mechanisms of inflammation modulation (increased IL-1β release) and pyroptosis. As a primary component in ambient pollution and diesel exhaust and a component of toners in printers used in office buildings worldwide, CB nanoparticles are a critical target for study. A better understanding of their mechanism of inflammasome activation may allow us to appropriately regulate potential health hazards.
This work was supported, in whole or in part, by National Institutes of Health Grants R01 HL079901 and R01HL96625 (to M. M. M.) and R01HL96625 (to V. H. G.). This publication was made possible by National Center for Research Resources, National Institutes of Health Grant UL1RR024979.
- CB
- carbon black
- NALP3
- NACHT domain, leucine-rich-repeat (LRR) domain, and pyrin domain (PYD)-containing protein 3
- ROS
- reactive oxygen species
- YVAD
- benzyloxycarbonyl-Tyr-Val-Ala-Asp(OMe)-fluoromethyl ketone
- TEM
- transmission electron microscopy
- BET
- Brunauer-Emmett-Teller
- DLS
- dynamic light scattering
- XRD
- x-ray diffraction.
REFERENCES
- 1. Opitz B., van Laak V., Eitel J., Suttorp N. (2010) Am. J. Respir. Crit. Care Med. 181, 1294–1309 [DOI] [PubMed] [Google Scholar]
- 2. Harada R. N., Repine J. E. (1985) Chest 87, 247–252 [DOI] [PubMed] [Google Scholar]
- 3. Twigg H. L., 3rd (2004) Semin. Respir. Crit. Care Med. 25, 21–31 [DOI] [PubMed] [Google Scholar]
- 4. Sibille Y., Reynolds H. Y. (1990) Am. Rev. Respir. Dis. 141, 471–501 [DOI] [PubMed] [Google Scholar]
- 5. Loeffler J., Hedderich R., Fiedeler U., Malsch I., Túquerres G., Koskinen J., Linden M., Lojkowski W., Moritz T., Zins M., Bernabeu E., Larena A. (2010) in Nanoroad SME European Project, European Union, Brussels, Belgium [Google Scholar]
- 6. Möller W., Brown D. M., Kreyling W. G., Stone V. (2005) Part Fibre Toxicol. 2, 7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Hussain S., Thomassen L. C., Ferecatu I., Borot M. C., Andreau K., Martens J. A., Fleury J., Baeza-Squiban A., Marano F., Boland S. (2010) Part. Fibre Toxicol. 7, 10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Lundborg M., Johard U., Låstbom L., Gerde P., Camner P. (2001) Environ Res. 86, 244–253 [DOI] [PubMed] [Google Scholar]
- 9. Lippmann M., Yeates D. B., Albert R. E. (1980) Br. J. Ind. Med. 37, 337–362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Oberdörster G. (1994) Ann. Occup. Hyg. 38, 601–615, 421–402 [DOI] [PubMed] [Google Scholar]
- 11. Ferin J., Oberdörster G., Penney D. P. (1992) Am. J. Respir. Cell Mol. Biol. 6, 535–542 [DOI] [PubMed] [Google Scholar]
- 12. Oberdörster G., Ferin J., Lehnert B. E. (1994) Environ. Health Perspect. 102, Suppl. 5, 173–179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Beck-Speier I., Dayal N., Karg E., Maier K. L., Schumann G., Schulz H., Semmler M., Takenaka S., Stettmaier K., Bors W., Ghio A., Samet J. M., Heyder J. (2005) Free Radic. Biol. Med. 38, 1080–1092 [DOI] [PubMed] [Google Scholar]
- 14. Stone V., Shaw J., Brown D. M., Macnee W., Faux S. P., Donaldson K. (1998) Toxicol. In Vitro 12, 649–659 [DOI] [PubMed] [Google Scholar]
- 15. Renwick L. C., Brown D., Clouter A., Donaldson K. (2004) Occup. Environ. Med. 61, 442–447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Yamawaki H., Iwai N. (2006) Circ. J. 70, 129–140 [DOI] [PubMed] [Google Scholar]
- 17. Beck-Speier I., Dayal N., Karg E., Maier K. L., Roth C., Ziesenis A., Heyder J. (2001) Environ. Health Perspect. 109, Suppl. 4, 613–618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hussain S., Boland S., Baeza-Squiban A., Hamel R., Thomassen L. C., Martens J. A., Billon-Galland M. A., Fleury-Feith J., Moisan F., Pairon J. C., Marano F. (2009) Toxicology 260, 142–149 [DOI] [PubMed] [Google Scholar]
- 19. Koike E., Kobayashi T. (2006) Chemosphere 65, 946–951 [DOI] [PubMed] [Google Scholar]
- 20. Mauderly J. L., Snipes M. B., Barr E. B., Belinsky S. A., Bond J. A., Brooks A. L., Chang I. Y., Cheng Y. S., Gillett N. A., Griffith W. C., Henderson R. F., Mitchell C. E., Nikula K. J., Thomassen D. G. (1994) Research Report (Health Effects Institute) pp. 1–75; discussion pp. 77–97 [PubMed] [Google Scholar]
- 21. Moss O. R., Wong V. A. (2006) Inhal. Toxicol. 18, 711–716 [DOI] [PubMed] [Google Scholar]
- 22. Oberdörster G., Oberdörster E., Oberdörster J. (2005) Environ. Health Perspect. 113, 823–839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Stoeger T., Reinhard C., Takenaka S., Schroeppel A., Karg E., Ritter B., Heyder J., Schulz H. (2006) Environ. Health Perspect. 114, 328–333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Brown D. M., Dickson C., Duncan P., Al-Attili F., Stone V. (2010) Nanotechnology 21, 215104/215101–215104/215109 [DOI] [PubMed] [Google Scholar]
- 25. Barlow P. G., Clouter-Baker A., Donaldson K., Maccallum J., Stone V. (2005) Part Fibre Toxicol. 2, 11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ma J. Y., Ma J. K. (2002) J. Environ. Sci. Health C Environ Carcinog Ecotoxicol. Rev. 20, 117–147 [DOI] [PubMed] [Google Scholar]
- 27. Ito T., Ikeda M., Yamasaki H., Sagai M., Tomita T. (2000) Environ Toxicol. Pharmacol. 9, 1–8 [DOI] [PubMed] [Google Scholar]
- 28. Aam B. B., Fonnum F. (2007) Arch. Toxicol. 81, 441–446 [DOI] [PubMed] [Google Scholar]
- 29. Dick C. A., Brown D. M., Donaldson K., Stone V. (2003) Inhal. Toxicol. 15, 39–52 [DOI] [PubMed] [Google Scholar]
- 30. Stoeger T., Takenaka S., Frankenberger B., Ritter B., Karg E., Maier K., Schulz H., Schmid O. (2009) Environ. Health Perspect. 117, 54–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Martinon F., Mayor A., Tschopp J. (2009) Annu. Rev. Immunol. 27, 229–265 [DOI] [PubMed] [Google Scholar]
- 32. Fink S. L., Cookson B. T. (2005) Infect. Immun. 73, 1907–1916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kepp O., Galluzzi L., Zitvogel L., Kroemer G. (2010) Eur. J. Immunol. 40, 627–630 [DOI] [PubMed] [Google Scholar]
- 34. Bergsbaken T., Fink S. L., Cookson B. T. (2009) Nat. Rev. Microbiol. 7, 99–109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Schroder K., Tschopp J. (2010) Cell 140, 821–832 [DOI] [PubMed] [Google Scholar]
- 36. Suzuki T., Franchi L., Toma C., Ashida H., Ogawa M., Yoshikawa Y., Mimuro H., Inohara N., Sasakawa C., Nuñez G. (2007) PLoS Pathog. 3, e111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Cassel S. L., Joly S., Sutterwala F. S. (2009) Semin. Immunol. 21, 194–198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Fink S. L., Cookson B. T. (2006) Cell. Microbiol. 8, 1812–1825 [DOI] [PubMed] [Google Scholar]
- 39. Miao E. A., Leaf I. A., Treuting P. M., Mao D. P., Dors M., Sarkar A., Warren S. E., Wewers M. D., Aderem A. (2010) Nat. Immunol. 11, 1136–1142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Baltrusaitis J., Usher C. R., Grassian V. H. (2007) Phys. Chem. Chem. Phys. 9, 3011–3024 [DOI] [PubMed] [Google Scholar]
- 41. Monick M. M., Powers L. S., Barrett C. W., Hinde S., Ashare A., Groskreutz D. J., Nyunoya T., Coleman M., Spitz D. R., Hunninghake G. W. (2008) J. Immunol. 180, 7485–7496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Monick M. M., Powers L. S., Gross T. J., Flaherty D. M., Barrett C. W., Hunninghake G. W. (2006) J. Immunol. 177, 1636–1645 [DOI] [PubMed] [Google Scholar]
- 43. Monick M. M., Powers L. S., Walters K., Lovan N., Zhang M., Gerke A., Hansdottir S., Hunninghake G. W. (2010) J. Immunol. 185, 5425–5435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Hansdottir S., Monick M. M. (2011) Vitam. Horm. 86, 217–237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Brough D., Rothwell N. J. (2007) J. Cell Sci. 120, 772–781 [DOI] [PubMed] [Google Scholar]
- 46. Perregaux D., Gabel C. A. (1994) J. Biol. Chem. 269, 15195–15203 [PubMed] [Google Scholar]
- 47. Walev I., Reske K., Palmer M., Valeva A., Bhakdi S. (1995) EMBO J. 14, 1607–1614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Hornung V., Bauernfeind F., Halle A., Samstad E. O., Kono H., Rock K. L., Fitzgerald K. A., Latz E. (2008) Nat. Immunol. 9, 847–856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Dostert C., Pétrilli V., Van Bruggen R., Steele C., Mossman B. T., Tschopp J. (2008) Science 320, 674–677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Pétrilli V., Papin S., Dostert C., Mayor A., Martinon F., Tschopp J. (2007) Cell death Differ. 14, 1583–1589 [DOI] [PubMed] [Google Scholar]
- 51. Yazdi A. S., Guarda G., Riteau N., Drexler S. K., Tardivel A., Couillin I., Tschopp J. (2010) Proc. Natl. Acad. Sci. U.S.A. 107, 19449–19454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Pazár B., Ea H. K., Narayan S., Kolly L., Bagnoud N., Chobaz V., Roger T., Lioté F., So A., Busso N. (2011) J. Immunol. 186, 2495–2502 [DOI] [PubMed] [Google Scholar]
- 53. Guarda G., Zenger M., Yazdi A. S., Schroder K., Ferrero I., Menu P., Tardivel A., Mattmann C., Tschopp J. (2011) J. Immunol. 186, 2529–2534 [DOI] [PubMed] [Google Scholar]
- 54. Vesterdal L. K., Folkmann J. K., Jacobsen N. R., Sheykhzade M., Wallin H., Loft S., Møller P. (2010) Part. Fibre Toxicol. 7, 33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Kinney P. L., Aggarwal M., Northridge M. E., Janssen N. A., Shepard P. (2000) Environ. Health Perspect. 108, 213–218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Cassee F. R., Frijer J. I., Subramaniam R., Asgharian B., Miller F. J., van Bree L., Rombout P. J. (1999) Development of a Model for Human and Rat Airway Particle Deposition: Implications for Risk Assessment, National Institute of Public Health and the Environment, Bilthoven, The Netherlands [Google Scholar]
- 57. Davis M. J., Swanson J. A. (2010) J. Leukoc. Biol. 88, 813–822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Sauer J. D., Witte C. E., Zemansky J., Hanson B., Lauer P., Portnoy D. A. (2010) Cell Host Microbe. 7, 412–419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Silveira T. N., Zamboni D. S. (2010) Infect. Immun. 78, 1403–1413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Ngai S., Batty S., Liao K. C., Mogridge J. (2010) Febs J 277, 119–127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Whitfield N. N., Byrne B. G., Swanson M. S. (2010) Infect. Immun. 78, 423–432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Rupper A. C., Cardelli J. A. (2008) Infect. Immun. 76, 2304–2315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Fink S. L., Bergsbaken T., Cookson B. T. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 4312–4317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Bergsbaken T., Cookson B. T. (2007) PLoS Pathog. 3, e161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Fink S. L., Cookson B. T. (2007) Cell. Microbiol. 9, 2562–2570 [DOI] [PubMed] [Google Scholar]
- 66. Monack D. M., Navarre W. W., Falkow S. (2001) Microbes Infect. 3, 1201–1212 [DOI] [PubMed] [Google Scholar]
- 67. Duewell P., Kono H., Rayner K. J., Sirois C. M., Vladimer G., Bauernfeind F. G., Abela G. S., Franchi L., Nuñez G., Schnurr M., Espevik T., Lien E., Fitzgerald K. A., Rock K. L., Moore K. J., Wright S. D., Hornung V., Latz E. (2010) Nature 464, 1357–1361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Cassel S. L., Eisenbarth S. C., Iyer S. S., Sadler J. J., Colegio O. R., Tephly L. A., Carter A. B., Rothman P. B., Flavell R. A., Sutterwala F. S. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 9035–9040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Eisenbarth S. C., Colegio O. R., O'Connor W., Sutterwala F. S., Flavell R. A. (2008) Nature 453, 1122–1126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Martinon F., Gaide O., Pétrilli V., Mayor A., Tschopp J. (2007) Semin. Immunopathol. 29, 213–229 [DOI] [PubMed] [Google Scholar]
- 71. Martinon F., Pétrilli V., Mayor A., Tardivel A., Tschopp J. (2006) Nature 440, 237–241 [DOI] [PubMed] [Google Scholar]
- 72. Chen G. Y., Nuñez G. (2010) Nat. Rev. Immunol. 10, 826–837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Zhou R., Yazdi A. S., Menu P., Tschopp J. (2011) Nature 469, 221–225 [DOI] [PubMed] [Google Scholar]
- 74. Tschopp J., Schroder K. (2010) Nat. Rev. Immunol. 10, 210–215 [DOI] [PubMed] [Google Scholar]
- 75. Slane B. G., Aykin-Burns N., Smith B. J., Kalen A. L., Goswami P. C., Domann F. E., Spitz D. R. (2006) Cancer Res. 66, 7615–7620 [DOI] [PubMed] [Google Scholar]
- 76. Hu Y., Mao K., Zeng Y., Chen S., Tao Z., Yang C., Sun S., Wu X., Meng G., Sun B. (2010) J. Immunol. 185, 7699–7705 [DOI] [PubMed] [Google Scholar]
- 77. Cassel S. L., Sutterwala F. S. (2010) Eur. J. Immunol. 40, 607–611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Val S., Hussain S., Boland S., Hamel R., Baeza-Squiban A., Marano F. (2009) Inhal. Toxicol. 21, Suppl. 1, 115–122 [DOI] [PubMed] [Google Scholar]