Abstract
The mitochondrial uncoupling proteins 2 and 3 (UCP2 and -3) are known to curtail oxidative stress and participate in a wide array of cellular functions, including insulin secretion and the regulation of satiety. However, the molecular control mechanism(s) governing these proteins remains elusive. Here we reveal that UCP2 and UCP3 contain reactive cysteine residues that can be conjugated to glutathione. We further demonstrate that this modification controls UCP2 and UCP3 function. Both reactive oxygen species and glutathionylation were found to activate and deactivate UCP3-dependent increases in non-phosphorylating respiration. We identified both Cys25 and Cys259 as the major glutathionylation sites on UCP3. Additional experiments in thymocytes from wild-type and UCP2 null mice demonstrated that glutathionylation similarly diminishes non-phosphorylating respiration. Our results illustrate that UCP2- and UCP3-mediated state 4 respiration is controlled by reversible glutathionylation. Altogether, these findings advance our understanding of the roles UCP2 and UCP3 play in modulating metabolic efficiency, cell signaling, and oxidative stress processes.
Keywords: Mitochondria, Mitochondrial Metabolism, Oxidative Stress, Post-translational Modification, Reactive Oxygen Species (ROS), UCP, Glutathionylation, Proton Leak
Introduction
UCP2 and -3 belong to the mitochondrial anion carrier family and are ∼73% homologous to each other, and they are both ∼58% homologous to the highly thermogenic uncoupling protein, UCP1. Whereas UCP3 is expressed in skeletal muscle and brown adipose tissue (BAT)3 and to some extent in the heart, UCP2 is found in a wide variety of tissues (1). UCP2 and UCP3 have been shown to diminish oxidative stress by lowering the mitochondrial membrane potential (2, 3). Acute increases in reactive oxygen species (ROS) production increase proton conductance through both UCP2 and UCP3, providing a negative feedback loop to limit further mitochondrial ROS formation (4, 5). Furthermore, UCP2 and UCP3 have also been implicated in many physiologic functions, suggesting that the common underlying mechanism may be ROS-mediated cell signaling.
Previous work has reported that UCP3 protects against insulin resistance and obesity (6–9), whereas UCP2 has been implicated a broad range of functions, including hormone secretion from the pancreas, immune cell function, and feeding behavior (10–13). For UCP2, most of these functions are linked to ROS level buffering (14–16). Indeed, UCP2 null mice on various genetic backgrounds exhibit oxidative stress in many tissue types and a decrease in the circulating glutathione (GSH)/glutathione disulfide ratio (2, 12). Increased expression of UCP2 in cancer cells is associated with the acquisition of drug-resistant phenotypes, a phenomenon related to the ROS-quenching function of UCP2 (17, 18). The absence of UCP3 in skeletal muscle increases oxidative damage and perturbs skeletal muscle metabolism (19). Increased UCP3 expression in muscle augments fatty acid metabolism and also curtails ROS production during fatty acid oxidation (20, 21). Remarkably, although they have been linked to a plethora of cellular processes, the molecular control of UCP2 and UCP3 has remained elusive.
Reactive cysteine residues of cellular proteins are known sites of regulation by conjugation to GSH, a process referred to as glutathionylation. This is especially relevant for mitochondria because a large fraction of the mitochondrial proteome contains exposed thiols that can be covalently modified by GSH. We were intrigued by the fact that UCP2 and UCP3 proteins contain several cysteine residues located in predicted membrane-spanning domains and in a loop region in the mitochondrial matrix (22). Here, we provide the first evidence that implicates reversible glutathionylation in the regulation of UCP2 and UCP3 function. We also show that glutathionylation and ROS-induced deglutathionylation work in tandem to turn UCP2 and UCP3 off and on, respectively. This novel mechanism for UCP2 and UCP3 control provides new insight into the putative role of these proteins for cellular ROS buffering. These results also improved our understanding of the function of these proteins in various physiological processes.
MATERIALS AND METHODS
Thymocyte Isolation
Thymocytes were isolated from UCP2 null (knock-out (KO)) and wild-type (WT) mice as described previously (23). The thymus was immediately removed and placed in ice-cold glucose-free Dulbecco's modified Eagle's medium (DMEM) containing 5 mm glutamine, 1 mm pyruvate, 2% FBS and buffered with HEPES, pH 7.4 (Glc-free DMEM) and then pressed through a metal tea strainer. Cells were washed twice with Glc-free DMEM and counted for oxygen consumption rate determinations.
Primary Cell Culture and Treatment
Primary myoblasts were isolated from WT and UCP3 null (KO) mice as described (24). Primary myoblasts were maintained on Matrigel-coated 60-mm2 plates in DMEM containing 5 mm dextrose, 1 mm pyruvate, 4 mm glutamine, 10% (v/v) fetal bovine serum (FBS), and 1% antimycotics antibiotics (A.A.). For experiments, primary myoblasts were grown to 70% and then differentiated for up to 7 days in DMEM containing 5 mm dextrose, 1 mm pyruvate, 4 mm glutamine, 2% FBS, and 1% A. A. Cells were then exposed to the superoxide-producing quinone menadione (0–20 μm), butathionine sulfoximine (BSO; 0–100 μm), or diamide (0–500 μm). Differentiated myotubes were then treated accordingly for experimentation.
Isolation of Mitochondria from Mouse Skeletal Muscle and BAT
Muscle mitochondria were isolated as described previously (25). Skeletal muscle from forelimbs, hind limbs, and the pectoral region were pooled, cleaned, and placed in basic medium (140 mm KCl, 20 mm HEPES, 5 mm MgCl2, 1 mm EGTA, pH 7.0). Following mincing, tissue was placed in homogenizing medium (basic medium supplemented with 1 mm ATP and 1% BSA (w/v)) and one unit of protease (subtilisin A) and homogenized using a glass/Teflon Potter-Elvehjem tissue grinder. Mitochondria were then isolated as described (25). BAT mitochondria were isolated using a standard protocol, as described (26). Interscapular BAT was dissected from 2–3 mice/genotype, cleaned, and pooled in a 250 mm sucrose solution. The tissue was minced and then homogenized by hand using a glass/Teflon Potter-Elvehjem tissue grinder in a 250 mm sucrose solution containing 0.2% BSA. Mitochondria were isolated as described (26). Protein concentration was determined by Bradford assay using BSA as the standard.
Mitochondrial preparations were kept on ice, and all experiments were conducted within 4 h following completion of the isolation procedure. Experiments in skeletal muscle mitochondria were conducted in incubation medium containing 120 mm KCl, 1 mm EGTA, 5 mm KH2PO4, 5 mm MgCl2, and 5 mm HEPES, pH 7.4, supplemented with 0.3% defatted BSA. Experiments in BAT mitochondria were conducted in incubation medium containing 125 mm sucrose, 20 mm Tris-HCl, 2 mm MgCl2, 4 mm KH2PO4, 1 mm EDTA, pH 7.2. An aliquot of mitochondria from BAT and skeletal muscle was frozen at −20 °C for immunoblot analysis.
Clark-type Oxygen Electrode Measurements
Oxygen consumption was measured in isolated mitochondria (0.5 mg/ml) or thymocytes at 37 °C using a Clark-type oxygen electrode (Hansatech, Norfolk, UK) and incubated in the appropriate incubation medium assumed to contain 406 nmol of oxygen/ml at 37 °C. Mitochondria were preincubated in the absence or presence of 2 μm diamide for skeletal muscle mitochondria and 100 μm diamide for BAT mitochondria with 1 μg/ml of oligomycin for 10 min at 37 °C before oxygen consumption measurements. 10 mm pyruvate and 5 mm malate were the substrates.
For determinations in thymocytes, 20 × 106 cells were incubated in DMEM containing 5 mm glucose, 4 mm glutamine, and 1 mm pyruvate for 15 min at 37 °C containing oligomycin (1 μg/ml) and in the presence or absence of diamide (100 μm). Following incubations, cells were then placed in the Clark-type electrode chamber for oxygen consumption measurements. An aliquot of thymocytes was kept for immunoblot analysis of UCP2 levels.
Mitochondrial Membrane Potential
Membrane potential (Δψm) was determined fluorimetrically, using safranin O dye (5 μm; excitation 485 nm, emission 580 nm) in isolated mouse mitochondria (0.5 mg/ml) incubated in the requisite incubation medium at 37 °C (27). Prior to assessments, mitochondria were preincubated for 10 min in diamide (0–200 μm) and then treated with 1 μg/ml oligomycin to induce state 4 respiration conditions. The substrates used were 5 mm pyruvate and 3 mm malate for BAT and 5 mm succinate for skeletal muscle.
In Situ Determination of Mitochondrial Bioenergetics
The XF24 extracellular flux analyzer (Seahorse Bioscience, North Billerica, MA) was employed to determine the mitochondrial bioenergetic characteristics of primary myotubes exposed to menadione, diamide, and/or BSO. Primary myoblasts were seeded at 50,000 cells/ml in Matrigel-coated 24-well Seahorse XF24 culture plates. Upon differentiation and reaching confluence, the cells were exposed to menadione (0–20 μm) or BSO (0–100 μm) for 24 h and then incubated for 30 min at 37 °C in HCO3-free DMEM containing 5–10 mm glucose, 4 mm l-glutamine, and 1 mm pyruvate. Measurement of oxygen consumption rates (OCRs) was performed for three measurement intervals to assess basal metabolic rate (one measurement interval consists of a 2-min mixing, 2-min incubation, and 2-min measurement step). State 4 respiration, maximal metabolic rate, and extramitochondrial O2 consumption were ascertained by injecting oligomycin (1 μg/ml), carbonyl cyanide p-trifluoromethoxyphenylhydrazone (1 μm), and antimycin A (1 μm). The impact of glutathionylation on state 4 respiration was determined by exposing cells to 100 μm diamide for 15 min prior to the injection of oligomcycin. OCR was normalized to total cellular protein/well using the Bradford assay.
Biotinylated Iodoacetamide (BIAM) and Biotinylated Glutathione Ethyl Ester (BioGEE) Labeling of UCP3 in Vitro
Reactive cysteines and glutathionylation sites were tested by BIAM (Invitrogen) and BioGEE (Invitrogen) labeling (28, 29). Briefly, partially purified UCP3 was diluted to 4 mg/ml in sucrose buffer (250 mm sucrose, Tris-HCl, pH 7) containing 100 μm BIAM or BioGEE and incubated for 30 min at room temperature in the dark under constant agitation. For control experiments, the samples were preincubated for 30 min at room temperature in 10 mm dithiothreitol (DTT) or 5 mm N-ethylmaleimide (NEM). Samples were then passed through a PD-10 Sephadex G25 desalting column (GE Healthcare) and eluted by centrifugation at 1000 rpm for 1 min (30). BIAM and BioGEE-labeled proteins were then pulled out of solution with streptavidin-coated beads overnight at 4 °C. Beads were then collected, and the supernatant fraction was placed in an ice-cold minitube containing Laemmli buffer. Beads were separated from the BIAM and BioGEE-labeled proteins using sucrose media containing 5 m urea. Upon removal of the deproteinated beads, the sample was placed in an ice-cold minitube containing Laemmli buffer constituting the pull-down fraction. Samples were electrophoresed under non-reducing conditions, and then UCP3 was detected by immunoblot using anti-UCP3 antibody (Abcam).
Glutaredoxin (GRx) Glutathionylation of UCP3
The glutathionylation of UCP3 was tested with glutaredoxin 1 (GRx1; Sigma). For GRx1, experiments were performed as described previously (31) with some modifications. Purified UCP3 was incubated in sucrose buffer for 15 min at 37 °C under constant agitation and in the dark with 0.5 mm GSH in the absence or presence of 10 units of GRx1. The samples were then incubated in 200 μm BioGEE for 30 min at room temperature under constant agitation and in the dark. The reaction was quenched with 5 mm NEM and passed through a PD-10 Sephadex G25 desalting column. The amount of UCP3 was accessed by immunoblot using anti-UCP3 antibody (Abcam).
UCP2 Glutathionylation in Kidney Lysate
Kidneys from WT and UCP2 null (KO) mice were excised, flash-frozen, and stored at −80 °C. On the day of experiments, kidneys were placed in 10 ml of Trizma (Tris base)-buffered sucrose solution (125 mm sucrose, 25 mm Trizma, 2 mm MgCl2, 1 mm EGTA, pH 7.2) and homogenized on ice using a glass/Teflon Potter-Elvehjem tissue grinder. The homogenate was then centrifuged at 150 × g for 5 min at 4 °C to remove any particulates and whole cells, and the supernatant was then tested for protein content. Protein was diluted to 4 mg/ml in sucrose buffer and incubated for 30 min in the dark at room temperature under constant agitation with 100 μm BioGEE. Reactions were quenched by the addition of NEM. 15 μg of total protein was then loaded in a 6–15% linear gradient SDS-gel and then electrophoresed under standard conditions. UCP2 biotinylated with BioGEE was detected by probing blots for 1 h at room temperature in the dark using an anti-biotin HRP-conjugated antibody (1:4000 dilution in 5% nonfat skim milk; Abcam). The electrophoretic mobility of UCP2 in the biotin blots was confirmed by probing for UCP2 on the same gel with anti-UCP2 antibody. The assay was optimized by loading various amounts of protein (5–60 μg) and by varying antibody dilution (1:500 to 1:4000) and incubation time (30 min to 24 h).
UCP3 Glutathionylation in Isolated Mitochondria
To test whether UCP3 was glutathionylated, we employed mitochondria isolated from the skeletal muscle of WT and UCP3 null (KO) mice. Mitochondria were diluted to 4 mg/ml in sucrose buffer and incubated in BioGEE. Reactions were stopped by the addition of NEM. Protein was then loaded in a 6–15% linear gradient SDS-gel and then electrophoresed under standard conditions. Detection of biotinylation was performed using anti-biotin HRP-conjugated antibody (1:4000 dilution in 5% nonfat skim milk; Abcam). The electrophoretic mobility of UCP3 in the biotin blots was confirmed by probing for these proteins on the same gel with anti-UCP3 (1:1000 dilution, 24-h incubation at 4 °C; Abcam) antibody.
Site-directed Mutagenesis
CMV plasmid constructs containing the full-length human UCP3 were purchased from Origene Technologies (Rockville, MD). For the site-directed mutagenesis, suitable primers (from Sigma; 5′-gcaggcacagcagccgcttttgctgacctcgt-3′ and 3′-cgtccgtgtcgtcggcgaaaacgactggagca-5′ for UCP3C25A, 5′-ttggagccggcttcgctgccacagtggtgg-3′ and 3′-aacctcggccgaagcgacggtgtcaccacc-5′ for UCP3C230A, and 5′-tacttcagccccctcgacgctatgataaagatggtggc-3′ and 3′-atgaagtcgggggagctgcgatactatttctaccaccg-5′ for UCP3C259A, respectively) were used to change codons from cysteine to alanine. Primers were designed using the QuikChange II mutagenesis primer design application (Agilent Technologies). Mutants were generated using the QuikChange II site-directed mutagenesis kit (Agilent Technologies). Plasmids carrying the appropriate point mutation were confirmed using BLAST and Expasy.
Transient transfections were carried out with Lipofectamine 2000 according to the manufacturer's instructions. Briefly, HEK293 cells were seeded at 90% confluence in Matrigel-coated 60-mm dishes in antibiotic/antimycotic-free low glucose DMEM containing 10% FBS. Cells were treated with plasmid-Lipofectamine complex in antibiotic/antimycotic-free DMEM containing 10% FBS and incubated for ∼36 h in the cell incubator. The cell monolayer was washed once with PBS and then incubated for 1 h at 37 °C in serum and antimycotic/antibiotic-free DMEM containing 1 mm BioGEE. The cell monolayer was then quickly washed twice, and the cells were lysed on ice with precipitation buffer containing 25 mm NEM. BioGEE-labeled proteins were isolated as described above. UCP3 was detected by immunoblot as described above.
Regulation of Glutathionylation by ROS
Primary WT myotubes were treated for 1 h with serum-free LG DMEM containing 1 mm BioGEE. Monolayers were then washed once with serum-free DMEM and then treated with serum-free DMEM containing 0–100 μm H2O2 for 15 min. Cells were then immediately washed with PBS and lysed with precipitation buffer containing 25 mm NEM. BioGEE-labeled proteins were extracted as described above, and the presence of UCP3 was tested by immunoblot. Protein content was tested by a BCA assay.
Cells exposed to BioGEE and H2O2 as described above were also tested for ROS levels. Immediately following H2O2 exposure, WT cells were incubated for 30 min at 37 °C in differentiating media containing 20 μm dichlorofluorescein diacetate. Fluorescence was determined as described (17).
Complex I Activity
Primary WT and UCP3 null (KO) myotubes were treated with 100 μm diamide for 30 min and then isolated by trypsinization. Mitochondria were isolated by sonication and differential centrifugation as described (32) with some modifications. 50 mm NEM was included in the mitochondrial isolation buffer. Mitochondria were permeabilized with 1% (w/v) digitonin on ice for 30 min, and the specific activity of complex I was then determined using 1.5 mm NADH and 0.5 mg/ml mitochondrial protein. NADH consumption was monitored as described (32). Mitochondrial protein concentration was determined using the Bradford assay.
Detection of Protein-Glutathione Adducts
Primary myotubes from WT mice were treated with diamide (0–500 μm) for 1 h in serum-free media and then lysed in a radioimmune precipitation buffer containing 50 mm NEM (to quench diamide reaction). Protein-glutathione (PSSG) adducts were detected using anti-PSSG antibody (Virogen). PSSG adducts were detected as described (33).
Determination of GSH Levels
Primary myotubes were exposed to BSO (0–100 μm) for 24 h and then trypsinized and washed with PBS, and then total GSH levels were tested using a GSH detection kit provided by Sigma.
Diamide and Menadione Toxicity
WT and KO primary myotubes were seeded, grown, and differentiated in Matrigel-coated 96-well plates. Upon full differentiation, cells were treated for 24 h with differentiating medium containing 0–40 μm menadione or for 30 min with serum-free DMEM containing 0–500 μm diamide. Following the exposure, cell monolayers were washed, probed with propidium iodide, and then tested for amount of cell death as described (32).
Immunoblots
Immunoblots were carried out as described previously (32). The membranes were blocked and probed for 3–24 h at 4 °C with primary antibodies directed against UCP3 (1:1000 dilution for cell lysate, 1:500 dilution of detection following BioGEE pull-down from cell lysate, 1:2500 dilution for purified UCP3; Abcam), UCP2 (1:2000, N-19; Santa Cruz Biotechnology, Inc.), UCP1 (1:10,000; Sigma), protein-glutathione adducts (1:500; Virogen), troponin T (1:1000; Abcam), and SDH (1:2000; Santa Cruz Biotechnology, Inc.). SDH and troponin T served as the loading standards. Membranes were then incubated for 1 h at room temperature with the requisite horseradish peroxidase-conjugated secondary antibodies (anti-rabbit, anti-mouse, or anti-goat 1:2000; Santa Cruz Biotechnology, Inc.). Blots were incubated in enhanced chemiluminescent substrate for visualization (ECL kit, Thermo Scientific).
Statistical Analysis
Unless otherwise stated, all data are expressed as mean ± S.D. Statistical analysis was performed using Statview software (SAS Institute Inc.) except for Student's t tests, which were performed in Microsoft Excel. Comparison of control with treated groups was performed with Student's t test, and for comparison of multiple treatments in the same group with a control, one-way ANOVA with a post hoc Tukey's test was used.
RESULTS
UCP3 Is Covalently Modified by Glutathione
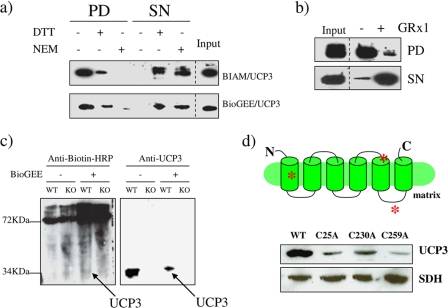
As a first approach, we determined if partially purified UCP3 protein could be covalently modified with the two agents BIAM and BioGEE in vitro. BIAM is routinely used to test the presence of reactive cysteines on proteins, whereas BioGEE, a cell-permeable glutathione molecule attached to biotin, is used to test if proteins can be glutathionylated. We found that UCP3 was readily recovered following BIAM treatment and streptavidin pull-down (Fig. 1a). By maintaining the cysteines in a reduced state or by blocking the cysteines with NEM, the yield of UCP3 in the pull-down was significantly diminished, confirming the presence of reactive cysteines in this protein (Fig. 1a). Similar observations were made with BioGEE (Fig. 1a). The level of recoverable UCP3 following BioGEE treatment was decreased substantially by the reducing and blocking agents DTT and NEM, revealing the requirement of cysteines for the conjugation of GSH to UCP3 (Fig. 1a). Glutathionylation of proteins has been shown to be mediated by GRx, a thiol-rich enzyme involved in antioxidative defense (34). Incubation of UCP3 with GRx1 and GSH prior to BioGEE treatment and pull-down with streptavidin significantly diminished the recovery of UCP3 (Fig. 1b). In contrast, incubations performed in the absence of GRx1 increased UCP3 recovery following BioGEE treatment and streptavidin pull-down. These data indicate that the glutathionylation of UCP3 is enzymatically mediated in vitro by the GSH exchanger, GRx. We next tested whether BioGEE could covalently modify UCP3 in mitochondria from skeletal muscle. Mitochondria were treated with BioGEE and then tested for biotin content by immunoblot using anti-biotin antibodies. Immunoblotting for BioGEE-modified mitochondrial proteins revealed bands at ∼34 kDa, which corresponds to the electrophoretic mobility of UCP3 (Fig. 1c). Moreover, these immunoreactive bands were only detected in mitochondria from WT mice. Furthermore, a slight shift in the electrophoretic mobility of UCP3 in the BioGEE-treated WT mitochondria was observed. This is most likely due to the covalent modification of this protein with BioGEE (Fig. 1c). Detection of BioGEE-modified proteins with anti-biotin antibodies also revealed a number of other immunoreactive bands at a range of molecular weights indicating that a number of mitochondrial proteins can be modified by GSH in skeletal muscle mitochondria. This is not surprising because the mitochondrial proteome is known to be very susceptible to glutathionylation (35). To confirm that UCP3 is glutathionylated and to identify the site(s) of glutathionylation, we generated UCP3 genes carrying mutations for various cysteines and expressed the mutant genes in HEK293 cells. Following a 1-h exposure of cells carrying either wild type or mutant UCP3 genes to BioGEE, cells were lysed, and the BioGEE proteins were enriched with streptavidin beads for immunoblot analysis. Mutation of Cys25 significantly decreased UCP3 enrichment (Fig. 1d). Changing Cys230 to Ala also decreased UCP3 enrichment but not to the same degree as the Cys25 mutation. When Cys259 (which is located in the last loop region of UCP3 contacting the matrix) was mutated, a much greater decrease in UCP3 enrichment was recorded (Fig. 1d). Hence, Cys259 appears to be the major site for GSH conjugation in UCP3, although contributions from other sites, such as Cys25, cannot be ruled out.
FIGURE 1.
a, UCP3 contains reactive cysteine groups and is covalently modified by GSH. Reactive and modifiable cysteines were detected in vitro using purified UCP3 as described under “Materials and Methods.” Purified UCP3 protein was treated with BIAM or BioGEE and pulled out of solution with streptavidin beads. UCP3 levels were detected by immunoblot in the BIAM or BioGEE pull-down (PD) and supernatant from pull-down (SN) fractions. Treatment with 10 mm DTT or 5 mm NEM prior to BIAM or BioGEE exposure served as a control. b, the glutathionylation of UCP3 in vitro is enzymatically mediated by GRx1. Following exposure to GSH or GSH + GRx1, purified UCP3 was incubated with BioGEE and then pulled out of solution with streptavidin beads. Presence of UCP3 was tested by immunoblot in the BioGEE pull-down and supernatant from pull-down. In all panels, input corresponds to the total protein used in the reaction. c, UCP3 is modified by BioGEE in isolated skeletal muscle mitochondria. Skeletal muscle mitochondria isolated from WT and UCP3 null (KO) mice were treated with or without BioGEE and then subjected to immunodetection for proteins modified by biotin using anti-biotin antibodies. UCP3 was subsequently detected on the same gels using anti-UCP3 antibody as described under “Materials and Methods.” d, analysis of the site of glutathionylation in UCP3. Diagram depicts the putative locations of Cys25, Cys230, and Cys259 (red asterisks). UCP3 genes carrying point mutations for either cysteine were overexpressed in HEK293 cells. Cells were then lysed, treated with BioGEE, and then tested for the amount of UCP3 following elution with streptavidin beads. UCP3 was subsequently detected on the same gels using anti-UCP3 antibody as described under “Materials and Methods.”
Glutathionylation Inhibits UCP3-mediated State 4 Respiration
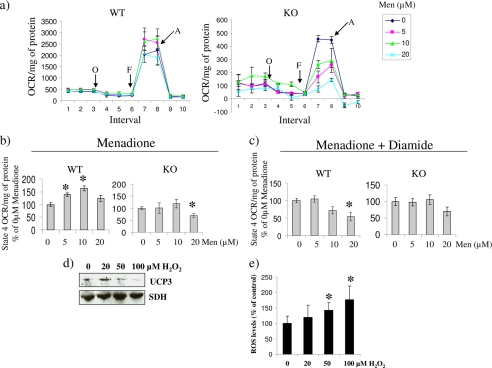
Because the in vitro determinations above indicated that UCP3 can be conjugated to GSH, we next tested if glutathionylation of UCP3 impacts mitochondrial metabolism. Diamide, a strong catalyst for GSH conjugation to proteins, was employed to induce glutathionylation in mitochondria isolated from skeletal muscle and brown adipose tissue. We performed the experiment using a range of diamide concentrations (0–50 μm for skeletal muscle and 0–200 μm for BAT mitochondria). An acute 10-min exposure to diamide (2 and 10 μm) increased the mitochondrial membrane potential under oligomycin-induced state 4 (non-phosphorylating) conditions in muscle mitochondria from WT but not UCP3 null mice (Fig. 2a). Higher concentrations of diamide (20–50 μm) induced a steady decline in membrane polarization in both WT and UCP3 null mitochondria, indicating that these higher concentrations had more promiscuous effects on mitochondrial metabolism. The steady decline in membrane potential is probably due to induction of mitochondrial permeability transition (MPT). Oxygen consumption was similarly affected by the induction of glutathionylation. Clark-type electrode experiments revealed that non-phosphorylating O2 consumption in WT mitochondria was decreased by an acute 10-min exposure to 2 μm diamide, an effect not observed in UCP3 null mitochondria (Fig. 2b). The cysteine residues shared between UCP1 and UCP3 are highly conserved. Hence, we next tested the effect of glutathionylation on UCP1 function. Diamide concentrations ranging from 50 to 100 μm induced significant increases in membrane polarization in BAT mitochondria from WT mice (Fig. 2c). In contrast, BAT mitochondria from the UCP3 null mice were far less responsive to the diamide treatments. We further tested the effects of diamide on BAT mitochondria bioenergetics by assessing O2 consumption using a Clark-type electrode. Acute treatment with 100 μm diamide diminished state 4 O2 consumption in the WT but not UCP3 null mitochondria from BAT, confirming the observations made with safranin O (Fig. 2d). It is also intriguing to note that high amounts of diamide did not collapse membrane potential as they did in skeletal muscle mitochondria, highlighting the metabolic differences between skeletal muscle and BAT mitochondria. Hence, although UCP3 is controlled by glutathionylation, our data indicate that UCP1 may be regulated by different mechanisms. The control of UCP3 by glutathionylation was confirmed in situ by measuring O2 consumption in intact WT and KO primary myotubes. Myotubes from WT mice displayed diminished state 4 respiration following a 15-min treatment with 100 μm diamide (Fig. 2e). These effects were absent in myotubes from UCP3 null mice.
FIGURE 2.
a and c, impact of diamide on the membrane potential of mitochondria isolated from skeletal muscle and BAT. Skeletal muscle and BAT mitochondria from WT and UCP3 null (KO) mice were treated acutely with different concentrations of diamide for 10 min and tested for mitochondrial inner membrane polarization using safranin O. Reactions were performed under oligomycin-induced state 4 conditions. *, p < 0.05; **, p < 0.02, n = 5–7, Student's t test. The single and double asterisks correspond to significance when compared with the WT control (0 μm diamide). Error bars in a are S.E. b and d, oligomycin-induced state 4 oxygen consumption in skeletal muscle mitochondria isolated from WT and UCP3 null (KO) mice. Mitochondria were treated with or without 2 μm diamide (skeletal muscle) and 100 μm diamide (BAT) with oligomycin and then tested for state 4 respiration. *, p < 0.05, n = 4–6, Student's t test. e, diamide-induced glutathionylation inhibits state 4 respiration in myotubes from WT but not UCP3 null (KO) mice. Shown is an in situ assessment of the impact of diamide on oligomycin-induced state 4 respiration using the Seahorse extracellular flux analyzer. Following treatment of WT and UCP3 null (KO) myotubes for 15 min with or without 100 μm diamide, the state 4 OCR (nmol/min) was tested. State 4 respiration was initiated following diamide treatment. p < 0.05, n = 4, Student's t test. The asterisk corresponds to significance when compared with no diamide treatment at each interval. f, In situ analysis of the diamide-mediated control of UCP3 carrying mutations at Cys25, Cys230, and Cys259. Analyses were conducted using the Seahorse extracellular flux analyzer. Confluent HEK293 cells transiently transfected with WT or mutant (C25A, C230A, or C259A) UCP3 genes were acutely treated for 15 min with or without 100 μm diamide, and then oligomycin-induced state 4 OCR was assessed. Basal OCR was measured prior to diamide treatment, and data were expressed as a percentage of basal OCR. p < 0.05, n = 4, Student's t test. The asterisk corresponds to significance when compared with no diamide treatment. Error bars, S.D.
To determine which Cys residue(s) played a functional role in controlling UCP3 following glutathionylation, we transiently transfected HEK293 cells with UCP3 genes carrying Cys to Ala mutations followed by the assessments of state 4 respiration in situ. Mutation of either Cys25 or Cys259 blunted the diamide-induced inhibition of UCP3 (Fig. 2f). In contrast, diamide treatment inhibited UCP3-mediated state 4 respiration in cells carrying the Cys230 mutant gene. Hence, Cys25 and Cys259 appear to be functionally important for the regulation of UCP3 by glutathionylation.
Diamide Toxicity
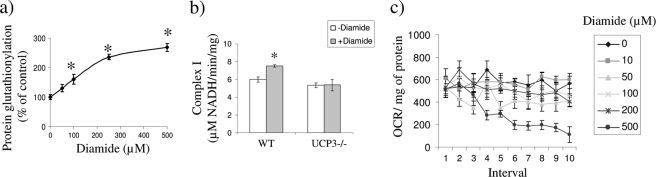
Diamide is used frequently to induce the glutathionylation of proteins and has been used routinely to test the effects of glutathionylation on bioenergetics (33). However, diamide is also known to induce opening of the mitochondrial permeability pore and induce cell death. Hence, to confirm that acute diamide treatment was not having any adverse effects on cell metabolism or viability, we conducted a range of toxicological and functional assessments. These assessments also allowed us to determine if the effects of diamide on state 4 respiration were due to the inhibition/activation of other mitochondrial processes. It is important to note that for a majority of our determinations, we exposed cells to diamide for no more than 1 h to avoid cell toxicity. As shown in supplemental Fig. S3, acute diamide exposure did not induce any significant increases in cell death (except in the case of 500 μm in KO cells). Acute treatment of WT myotubes with 100 μm diamide induced a 50% increase in cellular glutathionylation, confirming that this compound catalyzes the modification of proteins with GSH (Fig. 3a). Complex I activity was also tested because it is a key component of the respiratory chain and is known to be inhibited by glutathionylation. Complex I was not inhibited by the acute exposure to 100 μm diamide (Fig. 3b). This observation was confirmed by testing the impact of different concentrations of diamide on basal O2 consumption in situ. Concentrations of ≤200 μm did not affect basal O2 consumption for at least 1 h in the WT myotubes (Fig. 3c). Hence, these correlative data indicate that acute exposure to diamide concentrations below 500 μm does not have adverse effects on cellular respiration, metabolism, or cell viability.
FIGURE 3.
Assessment of the impact of diamide on mitochondrial bioenergetics and enzyme function. a, diamide treatment increases protein-glutathione adduct formation. WT primary myotubes were exposed to varying concentrations of diamide (0–500 μm) for 1 h in serum-free media. Protein-glutathione adducts were then detected by immunoblot using anti-PSSG adduct antibody (Virogen) and quantified. n = 3; *, p < 0.05, one-way ANOVA with Tukey's post hoc test. b, impact of diamide on complex I activity. WT and UCP3 null (KO) primary myotubes were exposed to 0 or 100 μm diamide for 30 min in serum-free media. Mitochondria were subsequently isolated and permeabilized for complex I activity analysis. n = 4. c, impact of diamide treatment on basal OCR. WT primary myotubes were treated acutely with diamide following assessment of basal respiration. The effect of diamide on basal respiration was tested over a period of 1 h. Basal OCR was determined in WT primary myotubes before and after exposure to 0–500 μm diamide. n = 3. Error bars, S.D.
Depletion of GSH Prevents UCP3 Deactivation by Glutathionylation
The inhibitory role of glutathionylation on UCP3-dependent state 4 respiration was investigated further in GSH-depleted cells. GSH was depleted using BSO, a compound that inhibits GSH synthesis. It has been reported previously that a 24-h exposure to BSO is required to deplete mitochondrial GSH (35). Exposure to 25–100 μm BSO for 24 h resulted in a steady decline in the cellular GSH levels (supplemental Fig. S1). Treatment with BSO also did not affect the basal or oligomycin-induced state 4 respiration in the WT primary myotubes (supplemental Fig. S1). Further analyses with cells treated acutely with diamide revealed that depletion of cellular GSH with BSO prevented the diamide-induced decrease in state 4 respiration in the WT cells (supplemental Fig. S2). These results confirm that GSH is required to inhibit UCP3 function. In contrast, WT cells untreated with BSO displayed a robust decrease in state 4 respiration following the acute diamide treatment (supplemental Fig. S2). By contrast, treatment of the UCP3 null cells with diamide or a combination of diamide and BSO had no effect on state 4 respiration. Hence, these observations from intact cells further corroborate the role of glutathionylation in the control of UCP3.
ROS and Glutathione Work to Control UCP3 Activation
Brand and colleagues (4) have shown that ROS activates proton leak through UCP3 in isolated mitochondria. Thus, we tested the impact of chronic ROS exposure on state 4 respiration in situ using intact primary myotubes exposed to menadione, a superoxide-generating quinone (Fig. 4a). In contrast to WT cells, UCP3 null myotubes were more sensitive to menadione treatment, confirming that UCP3 plays a part in minimizing oxidative stress. Indeed, sharp decreases in oligomycin-induced state 4 respiration and maximal respiration (stimulated by carbonyl cyanide p-trifluoromethoxyphenylhydrazone) were observed in the UCP3 null cells treated with menadione. A gradual increase in oligomycin-induced state 4 respiration was observed in primary WT myotubes treated with 5–10 μm menadione (Fig. 4b). This response was absent in the UCP3 null myotubes. Interestingly, a higher concentration of menadione (20 μm) led to a decrease in state 4 respiration in WT myotubes, suggesting that only small amounts of superoxide or related ROS, such as H2O2, are capable of activating UCP3 (Fig. 4b). In addition, diamide treatment blunted the menadione-induced increase in state 4 oxygen consumption rate in the WT cells, indicating that glutathionylation can prevent ROS-induced activation of UCP3 (Fig. 4c). Concentrations of menadione ranging from 0 to 20 μm did not induce increases in cell death in both the WT and the KO cells (supplemental Fig. S3). Furthermore, we have previously used this range of menadione to induce increases in cellular ROS without inducing cell death (17, 32). The regulation of UCP3 by glutathionylation and ROS was confirmed in WT primary myotubes using BioGEE and H2O2. Cells incubated within the cell-permeable BioGEE were treated acutely with 0, 20, 50, and 100 μm H2O2 for 15 min, and then BioGEE-labeled proteins were isolated and tested by immunoblot. In control cells (untreated with H2O2), UCP3 was readily detected following the pull-down of BioGEE-labeled proteins (Fig. 4d). Although 20 μm H2O2 increased the BioGEEylation of UCP3, higher H2O2 partially or completely reversed the modification of UCP3 with this compound. Furthermore, the decrease in UCP3 glutathionylation with BioGEE coincided with a significant increase in cellular ROS levels (Fig. 4e).
FIGURE 4.
a, in situ determination of the mitochondrial bioenergetic parameters of WT and UCP3 null (KO) primary myotubes exposed to varying amounts of menadione for 24 h. Analyses were conducted using the Seahorse extracellular flux analyzer. Following the determination of basal respiration various bioenergetic parameters were tested. State 4 respiration, maximal respiration, and extramitochondrial O2 consumption were tested using oligomycin (O; 1 μg/ml), carbonyl cyanide p-trifluoromethoxyphenylhydrazone (F; 1 μm), and antimycin A (A; 1 μm), respectively. b, state 4 respiration in WT or UCP3 null (KO) myotubes treated with menadione. State 4 respiration was induced with oligomycin. Data were expressed as a percentage of 0 μm menadione. *, p < 0.05, n = 4, one-way ANOVA with Tukey's post hoc test. The asterisk corresponds to comparison with control mean. c, impact of diamide treatment on state 4 respiration in menadione-exposed cells. Oligomycin-induced state 4 respiration was measured following a 15-min exposure to 100 μm diamide. *, p < 0.05, n = 4, one-way ANOVA with Tukey's post hoc test. The asterisk corresponds to comparison with control mean. d, ROS deglutathionylates UCP3. Primary WT myotubes were exposed to BioGEE and then incubated in H2O2 (0–100 μm). Cells were then lysed, and proteins modified by BioGEE were isolated with streptavidin beads. The presence of both UCP3 and SDH was tested by immunoblot following streptavidin pull-down. e, ROS levels in BioGEE-treated primary WT myotubes exposed to H2O2. Following treatment with BioGEE, cells were exposed to H2O2 (0–100 μm) for 15 min and then tested for ROS levels with dichlorofluorescein diacetate (20 μm, 30-min exposure). Data were normalized to protein level and expressed as a percentage of control. Absolute values were calculated by subtracting background fluorescence. *, p < 0.05, n = 6, one-way ANOVA with Tukey's post hoc test. Error bars, S.D.
Several groups have provided evidence that 4-hydroxynonenal (4-HNE), a ROS by-product, activates UCP3 (36, 37). Chronic exposure of WT myotubes to 10 μm 4-HNE had no effect on state 4 respiration (supplemental Fig. S4). However, 10 μm 4-HNE completely disrupted both basal and state 4 O2 consumption rates in the UCP3 null myotubes, indicating that lack of UCP3 sensitized cells to the toxic effects of 4-HNE (supplemental Fig. S4). In comparison with chronic treatments, acute incubations had relatively little effect on basal or state 4 oxygen consumption rates in WT and UCP3 null myotubes (supplemental Fig. S4). These results do point to a role, possibly indirect, of UCP3 in nullifying 4-HNE toxicity but do not support the previously proposed role of 4-HNE specifically in activating UCP3.
Covalent Addition of Glutathione Inhibits UCP2 Function
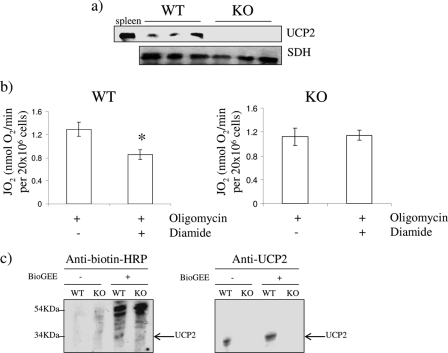
UCP2 shares a high degree of amino acid sequence homology (∼73%) to UCP3 and is also well documented to play a role in controlling mitochondrial ROS emission. Thus, we tested whether glutathionylation also modulates UCP2 function. For these measurements, we used mouse thymocytes, which are well known to express UCP2 (Fig. 5a) (38). Acute treatment with 100 μm diamide decreased oligomycin-induced state 4 respiration in the WT thymocytes (Fig. 5b). In contrast, diamide had no effect on state 4 respiration in thymocytes from UCP2 null mice. Glutathionylation of UCP2 was confirmed by exposure of kidney lysate to BioGEE. Biotin detection with anti-biotin HRP antibodies revealed the presence of a band at ∼33 kDa in the WT kidney extract, corresponding to the molecular mass of UCP2 (Fig. 5c). Also, biotinylation altered the electrophoretic mobility of UCP2, confirming its modification by BioGEE. This band was absent in the kidney extract from UCP2 null mice. Hence, like UCP3, increases in non-phosphorylating respiration associated with UCP2 is also regulated by glutathionylation.
FIGURE 5.
a, immunodetection of UCP2 in thymocytes isolated from WT and UCP2 null (KO) mice. Spleen lysate was used to confirm the electrophoretic mobility of UCP2. SDH was used as the loading control. UCP2 was detected with anti-UCP2 antibodies. b, impact of diamide on oligomycin-induced state 4 respiration in thymocytes from WT and UCP2 null (KO) mice. Cells were treated acutely for 15 min with oligomycin in the presence or absence of 100 μm diamide, and then oxygen consumption was tested. *, p < 0.05, n = 4, Student's t test. c, anti-biotin detection of glutathionylated UCP2. Kidney lysate from WT and UCP2 null (KO) mice was treated with or without BioGEE and then immunodetected with proteins modified by BioGEE using anti-biotin antibodies. Detection of UCP2 on the same gels was used to confirm the electrophoretic mobility of UCP2. Control reactions were performed in the absence of BioGEE. Error bars, S.D.
DISCUSSION
UCP2 and UCP3 are known to participate in a wide variety of physiological processes and are thought to buffer mitochondrial ROS production through a proton leak mechanism. However, the manner in which these proteins are controlled at the molecular level has remained elusive. In this study, we provide the first evidence that UCP2 and UCP3 are controlled by glutathionylation. We also found that this modification is reversed by ROS leading to an increase in state 4 respiration. In vitro studies using purified UCP3 revealed that UCP3 contains reactive cysteine residues that can be readily modified by glutathione. These observations were confirmed and extended using skeletal muscle mitochondria from WT and UCP3 null mice and kidneys from UCP2 null mice.
For a cysteine residue in a protein to be glutathionylated, it must be 1) accessible and 2) reactive. Recent work by others has indicated that allosteric activators or covalent modification of UCP2 or UCP3 is required to induce proton leak to control mitochondrial ROS production. Nadtochiy et al. (39) demonstrated that UCP2-mediated proton leak is induced by nitoalkene-induced covalent modification of cysteine residues, the first recorded covalent modification of UCP2. Brand and colleagues (4, 5, 36) have also showed that ROS and ROS by-products activate UCP1, UCP2, and UCP3. In this case, although the authors do not provide any evidence for the site of ROS or ROS by-product action, they speculated that the UCPs are activated by modification of specific amino acids. In this study, we identified Cys25 and Cys259 as major sites for the control of UCP3 by glutathionylation. Mutation of either of these cysteines resulted in a sharp decrease in the BioGEE-mediated isolation of UCP3. Furthermore, in situ analysis of cellular state 4 respiration revealed that mutation of either Cys25 or Cys259 desensitized UCP3 to the inhibitory effects of diamide-induced glutathionylation. Cys259 sits in a predicted loop region contacting the matrix, making it a perfect candidate for reversible glutathionylation. Intriguingly, Cys25 is located in a predicted transmembrane-spanning portion of the protein. It is difficult to know whether Cys25 is a structural residue or required for the transport function of UCP3 because the three-dimensional structure has not been solved. Also, Figtree et al. (40) recently observed that the sarcolemmal Na+-K+ pump is controlled by the reversible glutathionylation of a cysteinyl group located on a transmembrane-spanning subunit of the protein. Hence, our evidence also indicates that even “inaccessible” cysteines can be key sites for GSH conjugation.
In previous studies, Brand and colleagues (4) showed that ROS and ROS by-products induce proton leak through UCP2 and UCP3 to acutely regulate cellular ROS levels. Indeed, in a prior report, Nègre-Salvayre (2) demonstrated that UCP2 was a negative regulator of mitochondrial hydrogen peroxide production. Furthermore, Nadtochiy et al. (41) showed that ischemic preconditioning activates proton leak through UCP2 and ANT, thereby controlling ROS emission. Here, we substantially extend these observations by showing that 1) UCP3 is activated by ROS in situ, 2) glutathionylation acutely reverses the ROS-mediated activation of UCP3, and 3) glutathionylation can subsequently be reversed by acute treatment with ROS. We found no in situ evidence indicating that 4-HNE, a ROS by-product, performs the same function. Hence, we contend that ROS are responsible for the activation of UCP2- and UCP3-mediated increases in state 4 respiration. Although the ROS-mediated deglutathionylation of UCP2 was not investigated herein, based on similarities in amino acid sequence and the role of UCP2 in preventing ROS toxicity, we hypothesize that UCP2 is activated by ROS in a similar fashion. The treatment of intact cells with 100 μm diamide decreased oligomycin-induced state 4 respiration in a UCP2- and UCP3-dependent fashion. Following our correlative toxicological analysis of the impact of diamide on basal respiration, complex I activity, cell viability, and mitochondrial membrane bioenergetics, we conclude that, based on the genotype specific effect, acute diamide treatment exerts its effects through UCP3 (and UCP2). Effects were dose-dependent because exposure of cells to high concentrations impeded basal respiration and increased cell death when UCP3 was absent. However, despite these observations and because effects of diamide on UCP3 were small but significant, we cannot exclude the possibility that other proteins affecting mitochondrial bioenergetics are involved. For instance, the ANT-mediated proton leak may be affected by diamide-induced glutathionylation. Indeed, ANT can be modulated via cysteine covalent modification and is also reported as a glutathionylation target (42). Hence, the possible role of glutathionylation in the control of proton leak through ANT should also be explored.
It is intriguing that ROS was responsible for the deglutathionylation of UCP3 because ROS is mostly associated with glutathionylation. Indeed, we observed that acute H2O2 exposure increased cellular ROS levels, and this coincided with UCP3 deglutathionylation. In addition, chronic exposure to very low amounts of menadione (5 and 10 μm) was required to activate UCP3. A few proteins have been shown to be deglutathionylated when ROS levels increase; these include monomeric actin and SDH (43, 44). Hence, although ROS is most often associated with increased glutathionylation, it is now more apparent that under certain circumstances, ROS also induce the deglutathionylation of proteins. Furthermore, we provide evidence indicating that ROS-induced deglutathionylation requires an enzymatic system. Indeed, in vitro determinations with purified UCP3 tagged with BioGEE showed that H2O2, even at millimolar concentrations, cannot deglutathionylate UCP3 spontaneously (data not shown). However, as shown in Fig. 4d, ROS-mediated deglutathionylation of UCP3 occurred readily in cells exposed acutely to micromolar amounts of H2O2. Hence, we contend that the ROS species required to mediate this process is actually H2O2 and not superoxide because 1) H2O2 is a well known signaling molecule, 2) superoxide is rapidly dismutated in cells to H2O2, and 3) acute H2O2 exposure deglutathionylated UCP3. Moreover, although menadione does generate superoxide, chronic exposure would increase H2O2 levels as well.
In this study, GRx1 was shown to glutathionylate UCP3 in vitro. The physiological relevance of this finding requires further testing. Both GRx1 and GRx2 mediate glutathionylation reactions (45, 46). GRx proteins are often thought to mediate deglutathionylation reactions; however, more evidence is accumulating indicating that GRx actively glutathionylates target proteins as well. Mitochondria harbor two GRx isoforms with GRx1 in the intermembrane space and GRx2 in the matrix (46). Although less is known about GRx2, this isoform has a moderately high concentration in the matrix and has been shown to glutathionylate mitochondrial proteins (45). Post-translational controls of protein function in vivo require that modifications occur dynamically. In the case of UCP3 and UCP2, we demonstrate that reversible glutathionylation of these proteins is mediated by GRx and ROS.
The results presented herein describe a novel mechanism for the control of UCPs and validate the original observation that UCPs buffer cellular ROS levels following activation by ROS. In the present study, our findings further reveal that UCP1 function is not modulated by diamide-induced glutathionylation. UCP1 does contain cysteine residues located at the same sites as UCP2 and UCP3 (22). However, UCP1 only shares ∼58% homology with UCP2 and UCP3, indicating that the present functional differences may be due to structural differences between the proteins (22). Likely to be of additional relevance is the fact that the redox environment of BAT mitochondria is unique, as evidenced by a very low GSH/GSSG ratio in comparison with skeletal muscle mitochondria, which may influence glutathionylation events (47).4 In addition, skeletal muscle mitochondria harbor both GRx1 and GRx2, whereas BAT mitochondria only contain the former, indicating that glutathionylation may not be as important in regulating BAT mitochondrial function.4 However, this should not be surprising because the function of UCP1 is quite divergent from UCP2 and UCP3. Indeed, UCP1 serves a thermogenic purpose, and studies have demonstrated that UCP1 does not control ROS levels (48). Moreover, there is evidence that UCP1 can be phosphorylated (49). Hence, the control mechanisms governing UCP1 are apparently very different from those governing UCP2 and UCP3. The role of redox state and glutathionylation in the control of metabolic programs in skeletal muscle and BAT requires further exploration.
GSH is the major regulator of cellular redox status due to its high concentration, reductive potential, and involvement in quenching oxidative stress (50). Covalent modification of proteins with GSH is also emerging as a key post-translational modification required for protein regulation in response to changes in redox environment. We show that UCP2 and UCP3 contain reactive cysteines that can be targeted for modification by GSH. Although this is not the first reported covalent modification of UCP2 and UCP3, our findings provide the first evidence of on-off “switch” mechanisms responding to GSH and ROS. Hence, we propose that deglutathionylation/glutathionylation serves as the on-off switch, respectively, for UCP2 and UCP3 (Fig. 6). In our hypothetical model, UCP2/3 is glutathionylated and in an inactive state when mitochondrial ROS levels are maintained at tolerable levels and cell redox state is normal. Small increases in ROS lead to UCP2/3 deglutathionylation, inducing increased state 4 respiration. More research is needed to elucidate the physiological relevance of UCP2/3 activation by deglutathionylation. Our ongoing research findings and the previously published work of others support the idea that this switch mechanism impacts a large number of physiological processes ranging from tumor cytotoxicity to mitigation of oxidative stress, the control of pancreatic insulin and glucagon secretion, ghrelin-induced feeding behavior, and additional signaling in the brain, all processes involving UCP2 and UCP3 (1, 14, 51).
FIGURE 6.
Hypothetical model for the control of UCP2 and UCP3 by glutathionylation and deglutathionylation. Increases in ROS production prompt the deglutathionylation and activation of net proton conductance activity of UCP2 and UCP3. Upon restoration of tolerable ROS levels, UCP2 and UCP3 are reglutathionylated by GRx, effectively deactivating the proton conductance process. Orange UCP2 or UCP3, inactive; green UCP2 or UCP3, active. IMS, intermembrane space; MIM, mitochondrial inner membrane; SH, thiol residue; SSG, protein-glutathione conjugate. Δψm, mitochondrial membrane potential.
Supplementary Material
Acknowledgments
We thank Mahmoud Salkhordeh and Jian Xuan for technical assistance in dissections and primary myoblast purification. We also thank Martin Gerrits and Mahmoud Salkhordeh for previously generating recombinant UCP3. Finally, we thank Dr. Ilona Skerjanc and Tammy Porter (University of Ottawa) for experimental aid and consultation with the site-directed mutagenesis.
This work was supported in part by a grant from the Canadian Institutes of Health Research (CIHR) Institute of Nutrition, Metabolism, and Diabetes (to M. E. H.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S3.
R. J. Mailloux and M.-E. Harper, unpublished data.
- BAT
- brown adipose tissue
- ROS
- reactive oxygen species
- BSO
- butathionine sulfoximine
- OCR
- oxygen consumption rate
- BIAM
- biotinylated iodoacetamide
- BioGEE
- biotinylated glutathione ethyl ester
- GRx
- glutaredoxin
- NEM
- N-ethylmaleimide
- PSSG
- protein-glutathione
- ANOVA
- analysis of variance
- MPT
- mitochondrial permeability transition
- 4-HNE
- 4-hydroxynonenal.
REFERENCES
- 1. Azzu V., Brand M. D. (2010) Trends Biochem. Sci. 35, 298–307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Nègre-Salvayre A., Hirtz C., Carrera G., Cazenave R., Troly M., Salvayre R., Pénicaud L., Casteilla L. (1997) FASEB J. 11, 809–815 [PubMed] [Google Scholar]
- 3. Gustafsson H., Söderdahl T., Jönsson G., Bratteng J. O., Forsby A. (2004) J. Neurosci. Res. 77, 285–291 [DOI] [PubMed] [Google Scholar]
- 4. Echtay K. S., Roussel D., St-Pierre J., Jekabsons M. B., Cadenas S., Stuart J. A., Harper J. A., Roebuck S. J., Morrison A., Pickering S., Clapham J. C., Brand M. D. (2002) Nature 415, 96–99 [DOI] [PubMed] [Google Scholar]
- 5. Echtay K. S., Murphy M. P., Smith R. A., Talbot D. A., Brand M. D. (2002) J. Biol. Chem. 277, 47129–47135 [DOI] [PubMed] [Google Scholar]
- 6. Harper M. E., Green K., Brand M. D. (2008) Annu. Rev. Nutr. 28, 13–33 [DOI] [PubMed] [Google Scholar]
- 7. Choi C. S., Fillmore J. J., Kim J. K., Liu Z. X., Kim S., Collier E. F., Kulkarni A., Distefano A., Hwang Y. J., Kahn M., Chen Y., Yu C., Moore I. K., Reznick R. M., Higashimori T., Shulman G. I. (2007) J. Clin. Invest. 117, 1995–2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Costford S. R., Crawford S. A., Dent R., McPherson R., Harper M. E. (2009) Diabetologia 52, 2405–2415 [DOI] [PubMed] [Google Scholar]
- 9. Costford S. R., Chaudhry S. N., Crawford S. A., Salkhordeh M., Harper M. E. (2008) Am. J. Physiol. Endocrinol. Metab. 295, E1018–E1024 [DOI] [PubMed] [Google Scholar]
- 10. Alves-Guerra M. C., Rousset S., Pecqueur C., Mallat Z., Blanc J., Tedgui A., Bouillaud F., Cassard-Doulcier A. M., Ricquier D., Miroux B. (2003) J. Biol. Chem. 278, 42307–42312 [DOI] [PubMed] [Google Scholar]
- 11. Parton L. E., Ye C. P., Coppari R., Enriori P. J., Choi B., Zhang C. Y., Xu C., Vianna C. R., Balthasar N., Lee C. E., Elmquist J. K., Cowley M. A., Lowell B. B. (2007) Nature 449, 228–232 [DOI] [PubMed] [Google Scholar]
- 12. Pi J., Bai Y., Daniel K. W., Liu D., Lyght O., Edelstein D., Brownlee M., Corkey B. E., Collins S. (2009) Endocrinology 150, 3040–3048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Derdak Z., Mark N. M., Beldi G., Robson S. C., Wands J. R., Baffy G. (2008) Cancer Res. 68, 2813–2819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Andrews Z. B., Liu Z. W., Walllingford N., Erion D. M., Borok E., Friedman J. M., Tschöp M. H., Shanabrough M., Cline G., Shulman G. I., Coppola A., Gao X. B., Horvath T. L., Diano S. (2008) Nature 454, 846–851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Pecqueur C., Alves-Guerra M. C., Gelly C., Levi-Meyrueis C., Couplan E., Collins S., Ricquier D., Bouillaud F., Miroux B. (2001) J. Biol. Chem. 276, 8705–8712 [DOI] [PubMed] [Google Scholar]
- 16. Diao J., Allister E. M., Koshkin V., Lee S. C., Bhattacharjee A., Tang C., Giacca A., Chan C. B., Wheeler M. B. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 12057–12062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Mailloux R. J., Adjeitey C. N., Harper M. E. (2010) PLoS One 5, e13289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Harper M. E., Antoniou A., Villalobos-Menuey E., Russo A., Trauger R., Vendemelio M., George A., Bartholomew R., Carlo D., Shaikh A., Kupperman J., Newell E. W., Bespalov I. A., Wallace S. S., Liu Y., Rogers J. R., Gibbs G. L., Leahy J. L., Camley R. E., Melamede R., Newell M. K. (2002) FASEB J. 16, 1550–1557 [DOI] [PubMed] [Google Scholar]
- 19. Vidal-Puig A. J., Grujic D., Zhang C. Y., Hagen T., Boss O., Ido Y., Szczepanik A., Wade J., Mootha V., Cortright R., Muoio D. M., Lowell B. B. (2000) J. Biol. Chem. 275, 16258–16266 [DOI] [PubMed] [Google Scholar]
- 20. Bezaire V., Spriet L. L., Campbell S., Sabet N., Gerrits M., Bonen A., Harper M. E. (2005) FASEB J. 19, 977–979 [DOI] [PubMed] [Google Scholar]
- 21. Anderson E. J., Yamazaki H., Neufer P. D. (2007) J. Biol. Chem. 282, 31257–31266 [DOI] [PubMed] [Google Scholar]
- 22. Boss O., Samec S., Paoloni-Giacobino A., Rossier C., Dulloo A., Seydoux J., Muzzin P., Giacobino J. P. (1997) FEBS. Lett. 408, 39–42 [DOI] [PubMed] [Google Scholar]
- 23. Krauss S., Buttgereit F., Brand M. D. (1999) Biochim. Biophys. Acta 1412, 129–138 [DOI] [PubMed] [Google Scholar]
- 24. Rando T. A., Blau H. M. (1994) J. Cell Biol. 125, 1275–1287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Seifert E. L., Bézaire V., Estey C., Harper M. E. (2008) J. Biol. Chem. 283, 25124–25131 [DOI] [PubMed] [Google Scholar]
- 26. Shabalina I. G., Jacobsson A., Cannon B., Nedergaard J. (2004) J. Biol. Chem. 279, 38236–38248 [DOI] [PubMed] [Google Scholar]
- 27. Silva J. P., Shabalina I. G., Dufour E., Petrovic N., Backlund E. C., Hultenby K., Wibom R., Nedergaard J., Cannon B., Larsson N. G. (2005) EMBO J. 24, 4061–4070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Reynaert N. L., van der Vliet A., Guala A. S., McGovern T., Hristova M., Pantano C., Heintz N. H., Heim J., Ho Y. S., Matthews D. E., Wouters E. F., Janssen-Heininger Y. M. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 13086–13091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Marino S. M., Li Y., Fomenko D. E., Agisheva N., Cerny R. L., Gladyshev V. N. (2010) Biochemistry 49, 7709–7721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Clavreul N., Adachi T., Pimental D. R., Ido Y., Schöneich C., Cohen R. A. (2006) FASEB J. 20, 518–520 [DOI] [PubMed] [Google Scholar]
- 31. Applegate M. A., Humphries K. M., Szweda L. I. (2008) Biochemistry 47, 473–478 [DOI] [PubMed] [Google Scholar]
- 32. Mailloux R. J., Harper M. E. (2010) FASEB J. 24, 2495–2506 [DOI] [PubMed] [Google Scholar]
- 33. Hill B. G., Higdon A. N., Dranka B. P., Darley-Usmar V. M. (2010) Biochim. Biophys. Acta 1797, 285–295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Gallogly M. M., Starke D. W., Mieyal J. J. (2009) Antioxid. Redox Signal. 11, 1059–1081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Hurd T. R., Costa N. J., Dahm C. C., Beer S. M., Brown S. E., Filipovska A., Murphy M. P. (2005) Antioxid. Redox Signal. 7, 999–1010 [DOI] [PubMed] [Google Scholar]
- 36. Echtay K. S., Esteves T. C., Pakay J. L., Jekabsons M. B., Lambert A. J., Portero-Otín M., Pamplona R., Vidal-Puig A. J., Wang S., Roebuck S. J., Brand M. D. (2003) EMBO J. 22, 4103–4110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Aguirre E., Cadenas S. (2010) Biochim. Biophys. Acta 1797, 1716–1726 [DOI] [PubMed] [Google Scholar]
- 38. Krauss S., Zhang C. Y., Lowell B. B. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 118–122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Nadtochiy S. M., Baker P. R., Freeman B. A., Brookes P. S. (2009) Cardiovasc. Res. 82, 333–340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Figtree G. A., Liu C. C., Bibert S., Hamilton E. J., Garcia A., White C. N., Chia K. K., Cornelius F., Geering K., Rasmussen H. H. (2009) Circ. Res. 105, 185–193 [DOI] [PubMed] [Google Scholar]
- 41. Nadtochiy S. M., Tompkins A. J., Brookes P. S. (2006) Biochem. J. 395, 611–618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Queiroga C. S., Almeida A. S., Martel C., Brenner C., Alves P. M., Vieira H. L. (2010) J. Biol. Chem. 285, 17077–17088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Wang J., Boja E. S., Tan W., Tekle E., Fales H. M., English S., Mieyal J. J., Chock P. B. (2001) J. Biol. Chem. 276, 47763–47766 [DOI] [PubMed] [Google Scholar]
- 44. Chen Y. R., Chen C. L., Pfeiffer D. R., Zweier J. L. (2007) J. Biol. Chem. 282, 32640–32654 [DOI] [PubMed] [Google Scholar]
- 45. Beer S. M., Taylor E. R., Brown S. E., Dahm C. C., Costa N. J., Runswick M. J., Murphy M. P. (2004) J. Biol. Chem. 279, 47939–47951 [DOI] [PubMed] [Google Scholar]
- 46. Gallogly M. M., Mieyal J. J. (2007) Curr. Opin. Pharmacol. 7, 381–391 [DOI] [PubMed] [Google Scholar]
- 47. Barja de Quiroga G., López-Torres M., Pérez-Campo R., Abelenda M., Paz Nava M., Puerta M. L. (1991) Biochem. J. 277, 289–292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Cannon B., Shabalina I. G., Kramarova T. V., Petrovic N., Nedergaard J. (2006) Biochim. Biophys. Acta 1757, 449–458 [DOI] [PubMed] [Google Scholar]
- 49. Carroll A. M., Porter R. K., Morrice N. A. (2008) Biochim. Biophys. Acta 1777, 1060–1065 [DOI] [PubMed] [Google Scholar]
- 50. Vesce S., Jekabsons M. B., Johnson-Cadwell L. I., Nicholls D. G. (2005) J. Biol. Chem. 280, 38720–38728 [DOI] [PubMed] [Google Scholar]
- 51. Bézaire V., Seifert E. L., Harper M. E. (2007) FASEB J. 21, 312–324 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.