Abstract
The Golgi apparatus is a network of polarized cisternae localized to the perinuclear region in mammalian cells. It undergoes extensive vesiculation at the onset of mitosis and its reassembly requires factors that are in part segregated via the mitotic spindle. Here we show that unlike typical Golgi markers, the Golgi-protein p115 partitioned with the spindle poles throughout mitosis. An armadillo-fold in its N terminus mediated a novel interaction between p115 and γ-tubulin and functioned in its centrosomal targeting. Both the N- and C-terminal regions of p115 were required to maintain Golgi structure. Strikingly, p115 was essential for mitotic spindle function and the resolution of the cytokinetic bridge because its depletion resulted in spindle collapse, chromosome missegregation, and failed cytokinesis. We demonstrate that p115 plays a critical role in mitosis progression, implicating it as the only known golgin to regulate both mitosis and apoptosis.
Keywords: Golgi, Membrane, Membrane Biogenesis, Membrane Proteins, Membrane Trafficking
Introduction
The mammalian Golgi apparatus is a polarized organelle consisting of a series of stacked, highly specialized cisternae located at the pericentriolar region of interphase eukaryotic cells. At the onset of mitosis, the Golgi ribbon disassembles into vesicles and tubules that distribute stochastically within the cytoplasm to ensure equal partitioning into the daughter cells (1) and reassembly of a fully functional Golgi apparatus upon mitotic exit. Numerous components involved in Golgi biogenesis have been identified (2–4), however, the details of the mechanism whereby the Golgi apparatus regains its complex organization are not well understood.
The microtubule cytoskeleton is closely associated with the Golgi apparatus in interphase cells (5) and it provides structural integrity to its stacks. Conventionally, the centrosome has been known as the major microtubule organizing center in higher eukaryotes. In animal cells, the centrosome consists of two daughter centrioles surrounded by a proteinaceous pericentriolar material. The pericentriolar material promotes the majority of microtubule polymerization in the cell via a γ-tubulin-containing complex, the key component responsible for microtubule nucleation (6). In addition, microtubules can be nucleated at secondary, non-centrosomal microtubule organizing centers such as the Golgi apparatus by γ-tubulin-dependent (7, 8) or γ-tubulin-independent (9) mechanisms. Although the centrosomal microtubules are mainly involved in Golgi positioning, the Golgi apparatus-derived microtubules provide structure to the organelle (10).
In addition, interactions between golgins (i.e. GM130, giantin, and p115) and the GRASP family of Golgi stacking proteins (i.e. GRASP65 and GRASP55) have been shown to play critical roles in maintaining the structure and dynamic nature of the Golgi apparatus (11–15). Depletion of the Golgi-associated protein p115 leads to disruption of the organelle, implicating it as an important component of Golgi structure (16, 17). p115 is a 962-residue peripheral membrane protein present in the intermediate compartment and cis-Golgi vesicles that functions in endoplasmic reticulum to Golgi trafficking (13, 18, 19). p115 exists as a homodimer; each molecule contains a poorly characterized N-terminal globular head and an extended C-terminal tail region (20) consisting of an acidic domain and four sequential coiled coils (21, 22). It has been proposed that the cis-Golgi cisternae are generated by heterotypic fusion of mitotic vesicles mediated by the interaction between the acidic C terminus of p115 and the Golgi-associated proteins giantin and GM130 (23, 24). More recent studies have also implicated the interaction between the first coiled-coil (CC1) of p115 and SNAREs in the maintenance of Golgi structure (16, 17). During apoptosis, the acidic C terminus of p115 is cleaved, translocates to the nucleus, and plays a pro-apoptotic role (25–27). Recently, the N terminus of p115 was shown to interact with the βCOP subunit of the COPI (Coat protein I) coat (28) and the COG2 subunit of the conserved oligomeric Golgi (COG) complex (29), suggesting an additional membrane recycling model whereby both the p115 N and C termini may be required for COPI vesicle tethering and interphase Golgi integrity.
Recent studies have suggested a functional association between Golgi components and the mitotic apparatus. GM130 and GRASP65 were proposed to regulate the morphology and function of the interphase centrosome and the mitotic spindle (30, 31). In addition, Golgi proteins as well as post-Golgi membrane trafficking have been implicated in cytokinesis completion (32–35).
Here we demonstrate that p115 associates with γ-tubulin throughout the cell cycle and that p115 is required for the structure and function of the late mitotic spindle as well as for the resolution of the cytokinetic bridge. We speculate that p115 knockdown results in a spindle defect due to unstable spindle poles under conditions of chromosome tension and in failed cytokinesis due to impaired Golgi reassembly and post-Golgi trafficking.
EXPERIMENTAL PROCEDURES
Reagents, Transfection, and Synchronization
HeLa, NRK, COS7, U2OS, and 293T cells were grown in DMEM supplemented with 10% FCS, glutamine, and penicillin/streptomycin, at 37 °C in 5% CO2. The HeLa GalNAcT2-GFP cell line was a gift from Dr. Adam Linstedt (Carnegie Melon University, Pittsburg, PA); the HeLa α-tubulin-YFP and the HeLa α-tubulin-YFP/H2B-RFP cell lines were provided by Dr. Don Cleveland (University of California at San Diego, La Jolla, CA). The pSG5-FLAG-human p115 construct was provided by Dr. Yukio Ikehara (Fukuoaka University School of Medicine, Fukuoaka, Japan) and was used as a template to generate all p115 constructs used in this study. A bovine p115 plasmid was provided by Dr. Gerry Waters. siRNA sequences against the human p115 (17, 36) were obtained from Ambion (Applied Biosystems) and transfected using Oliogofectamine (Invitrogen). Controls included either mock transfection or treatment with a predesigned non-silencing siRNA from Ambion (Applied Biosystems). Both mock and non-silencing siRNA treatments gave identical results and were used interchangeably. Plasmids were transfected using Effectene (Qiagen). HeLa or NRK cells were synchronized in S-phase by a 14-h treatment with 2.5 μg/ml of aphidicolin (Calbiochem). Following release, cells were allowed to progress through mitosis and either fixed for indirect immunofluorescence or collected for electron microscopy or total lysate preparation.
GST Pull-down, Immunoprecipitation, and Sucrose Gradient Sedimentation
GST-p115 constructs were transformed into Escherichia coli DH5α cells and induced with 1 mm isopropyl β-d-1-thiogalactopyranoside. Equimolar amounts (20 pmol) of the glutathione-Sepharose 4B beads containing the GST fusion p115 constructs were incubated with 500 μg of postnuclear lysate containing 20 mm Tris-HCl, pH 7.4, 150 mm NaCl, 1% Nonidet P-40, 1 mm EDTA. For immunoprecipitation, HeLa cells were transfected with cDNA encoding the various 3XFLAG-p115 constructs. At 18 h post-transfection, the postnuclear supernatant was prepared as above and pre-cleared with preimmune IgG coupled to Affi-Gel® 10 (Bio-Rad). The pre-cleared lysates were incubated with anti-γ-tubulin antibody after which Protein G-coupled beads (GE Healthcare) were added to the lysates for 3 h. Interphase 293T postnuclear supernatant was prepared in lysis buffer without detergent. Cells were broken using a ball bearing homogenizer. 200 μg of lysate were loaded on top of a continuous 5–40% sucrose gradient and spun at 35,000 × g in a Beckman SW55Ti rotor for 20 h. Protein standards were run in parallel. 1-ml fractions were collected from the top of the gradient, precipitated using trichloroacetic acid (TCA), and resolved by SDS-PAGE.
Immunofluorescence and Electron Microscopy
Cells grown on coverslips were fixed in 3% paraformaldehyde and processed for immunofluorescence microscopy as described previously (25). Confocal images were acquired by capturing Z-series with a 0.25-μm step size on a Leica TCS SP2 AOBS confocal microscope (Leica, Dearfield, IL) using a ×63 oil immersion objective (1.4 NA). Laser lines at 405, 488, 546, and 633 nm were provided by 20-mW Diode, 100 mW Ar, 1.5 mW HeNe, and 10 mW HeNe lasers. The Z-series were projected using the maximum intensity method (ImageJ; rsb.info.nih.gov/ij/). Alternatively, images were acquired using a Nikon PlanApo ×60 A/1.40 oil objective on a Nikon Eclipse TE300 inverted microscope equipped with a Nikon Digital Sight DS-2MBW CCD camera. For electron microscopy HeLa cells were either grown on polylysine-coated glass coverslips or on 60-mm plastic dishes and transfected with either predesigned non-silencing siRNA or p115 siRNA for 96 h. The glass coverslips were fixed with 2.5% glutaraldehyde and postfixed with 1% osmium tetroxide followed by 1% uranyl acetate. The cells on plastic dishes were synchronized with 2.5 μg/ml of aphidicolin as described above. Mitotic cells in cytokinesis were collected 10 h after release from the aphidicolin block, pelleted, and fixed as above. The cell pellets were processed for EM as described previously (37).
Microinjection
HeLa cells stably expressing α-tubulin-YFP were microinjected in prometaphase with preimmune IgG (5 μg/μl) or p115 IgG (5 μg/μl) mixed with the injection marker Alexa Fluor 568 hydrazide (Invitrogen), on a Nikon TE2000-inverted microscope equipped with a cooled charge-coupled device (CCD) camera (Orca-ER; Hamamatsu Photonics, Japan) and a ×40 PlanApo (NA 0.95) objective. Injection pressure was provided by a Pneumatic PicoPump (PV820) (World Precision Instruments, Sarasota, FL). Cells were fixed with 4% paraformaldehyde at 45 or 240 min postinjection and stained with Hoechst 33342 (Invitrogen).
Time Lapse Microscopy
HeLa cells stably co-expressing α-tubulin-YFP and H2B-RFP were mock or p115 siRNA treated. Between 72 and 96 h, cells were imaged with a temperature-controlled DeltaVision RT deconvolution system (Applied Precision Inc.) on an Olympus IX70 microscope, using the softWoRx Suite software (Applied Precision, Inc.). For each field, a Z-series was acquired on a UApo/340 ×40/1.35 oil immersion objective, at 5-min intervals for a duration of 8 h, at 37 °C. Z-series were projected using softWoRx Suite. Time lapse movies were generated using ImageJ and are shown at a display rate of 10 frames/s.
RESULTS
p115 Contains an N-terminal Armadillo-fold That Mediates Its Association with γ-Tubulin
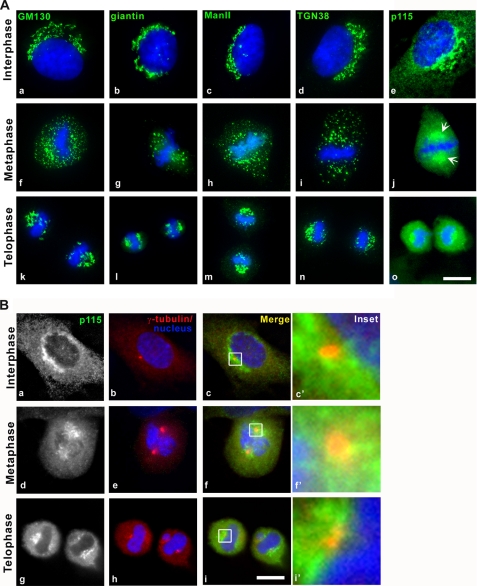
Although several studies suggest that Golgi matrix components are inherited in an orderly fashion, via the mitotic spindle and/or spindle poles (38–40), other reports propose that the Golgi matrix undergoes extensive disassembly in mitosis and that the Golgi apparatus reassembles de novo (41). To distinguish between these proposals, interphase (Fig. 1A, a–e) and mitotic (Fig. 1A, f–o) cells were stained with antibodies against several cis-, medial-, and trans-Golgi marker proteins. During metaphase, GM130, giantin, ManII, and TGN38 displayed the well characterized dispersed-punctate staining pattern typical of the Golgi redistribution into mitotic clusters (Fig. 1A, f–i). In contrast, p115 maintained a spindle pole localization throughout mitosis in several different cell types (Fig. 1A, j, arrows, and supplemental Fig. S1A, arrows). As previously described p115 also displayed significant cytoplasmic staining throughout the cell cycle (Fig. 1A, e, j, and o). During telophase, the distribution of all tested Golgi proteins coincided in the pericentrosomal region (Fig. 1A, k–o). We verified the localization of p115 to the centrosomal area by co-staining with γ-tubulin, a bona fide centrosomal marker (Fig. 1B), and show that p115 segregates with the spindle poles and partially with centrosomes in mitotic cells from two different cell lines (Fig. 1B, d–i, and supplemental Fig. S1A). GM130, which was previously suggested to have a role in spindle function (30), was not present in the spindle pole region or on mitotic spindles, but rather in mitotic remnants stochastically distributed in the cytoplasm (Fig. 1A, f).
FIGURE 1.
p115 segregates with the spindle poles throughout mitosis, whereas most Golgi-associated proteins distribute stochastically. A, indirect immunofluorescence of NRK cells with antibodies against Golgi markers (green) in interphase (a–e), metaphase (f–j), and telophase (k–o) cells. Arrows, p115 enrichment in spindle poles. B, indirect immunofluorescence with antibodies against p115 (green) and γ-tubulin (red). Blue, Hoechst 33342. Images were acquired at equivalent exposures on an inverted microscope, deconvolved, and projected using the IPLab software. Scale bars, 10 μm.
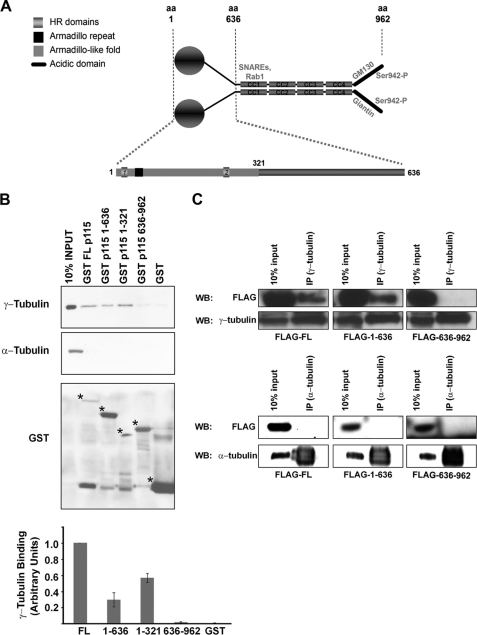
To determine how p115 localized to mitotic spindle poles, we analyzed the human and mouse p115 protein sequence for putative domains that might function in intracellular targeting (www.ebi.ac.uk/interpro/). The globular N-terminal region was predicted to possess an armadillo repeat (residues 60–97) as well as an armadillo-like helical-fold (residues 1–321) (Fig. 2A). The existence and nature of this domain has been confirmed by x-ray crystallography studies (42, 43). Armadillo repeats have been implicated in interactions between cell adhesion proteins and the cytoskeleton (44). Interestingly, an armadillo-fold is absent from other mammalian golgins including GM130, GRASP55 and -65, golgin84, and the cis-Golgi associated protein GMAP-210. We hypothesized that the observed mitotic localization of p115 to spindle poles was mediated by interactions between its armadillo-like helical region and a centrosomal γ-tubulin-containing complex. To test this idea, we performed in vitro binding studies in which purified recombinant full-length p115, as well as truncated forms of the molecule (residues 1–636, 1–321, and 636–962) were fused to GST and incubated with postnuclear cytosolic extracts from interphase (Fig. 2B) or mitotic (supplemental Fig. S1B) cells. Neither control GST alone nor the C terminus of p115 (residues 636–962), which includes the four coiled-coil domains (Fig. 2A), associated with γ-tubulin (Fig. 2B and supplemental Fig. S1, B and C). In contrast, full-length p115, the N-terminal 1–636 construct as well as the armadillo-like helical-fold (1–321) pulled down γ-tubulin efficiently (Fig. 2B and supplemental Fig. S1, B and C), although the binding of the N-terminal region and the 1–321 construct was less efficient than that of the full-length molecule (Fig. 2B and supplemental Fig. S1B). Importantly, the binding profiles were not cell cycle- or GTP-dependent (supplemental Fig. S1B).4 The observed interaction is specific because neither the full-length nor the N-terminal region of p115 showed association to either α-tubulin (Fig. 2B) or β-tubulin,4 which are major components of microtubules and mitotic spindles. The γ-tubulin interaction was saturable by either full-length p115 or the N-terminal 1–636 construct (supplemental Fig. S1C). To determine whether p115 associates with γ-tubulin or a γ-tubulin-containing complex in vivo, 293T cells were transfected with constructs encoding FLAG-tagged versions of full-length, N- or C-terminal domains of p115, and immunoprecipitated with a γ-tubulin antibody (Fig. 2C). In agreement with our in vitro pull-down data ∼5% of both full-length and the N-terminal region of p115 co-immunoprecipitated with endogenous γ-tubulin, whereas no co-precipitation of the C-terminal region was observed. This is similar to the well characterized interaction between p115 and GM130, which was also only observed under conditions of p115 overexpression (24, 45). In support of these observations, fractionation of 293T cell lysates by sucrose gradient sedimentation revealed that p115 and γ-tubulin co-sediment in vivo with the microtubule polymerizing small and large complexes, γTuSC and γTuRC, respectively (supplemental Fig. S1D) (46). Together, our data provide evidence that p115 and γ-tubulin exist in the same complex and that the N-terminal armadillo-fold of p115 is critical for this association. Further studies will be necessary to characterize the nature of the association between p115 and γ-tubulin within this complex.
FIGURE 2.
Sequence requirements for p115-γ-tubulin interaction. A, schematic of the p115 homodimer: the C terminus (amino acids 636–962) contains 4 coiled-coil domains (CC1–CC4) and an acidic region (black line). Previously described interactions are noted in the figure and text. The N-terminal armadillo repeat (black square, amino acids 60–97) and the armadillo-fold domain (light gray, amino acids 1–321) were identified by a bioinformatics approach (www.ebi.ac.uk/interpro/). B, Western blot analysis of p115-γ-tubulin binding following incubation of equimolar amounts of GST-fused p115 truncation fragments with 500 μg of NRK interphase postnuclear cytosol. Binding efficiency of each construct is shown in the graph, and was quantified by densitometric measurement of γ-tubulin immunoreactive band intensity (ImageJ) normalized to the GST input (bottom panel). Low molecular weight bands are degraded GST peptides. For GST purification, we obtained a dimerized GST band of ∼50 kDa. Asterisks indicate the correct molecular weights of the purified proteins. C, Western blot (WB) of overexpressed 3XFLAG-p115 deletion constructs immunoprecipitated (IP) with γ-tubulin and α-tubulin antibodies from HeLa cells.
The p115 N Terminus Is Required for Golgi Structure in Interphase
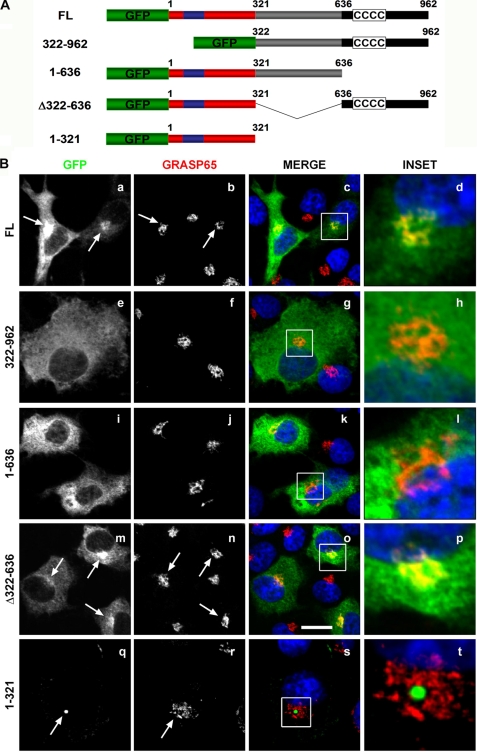
To characterize the targeting information in p115, we expressed a series of p115-GFP fusion proteins in COS7 cells (Fig. 3A). As previously observed, overexpressed full-length p115 was targeted to the juxtanuclear region of the cell (Fig. 3B, a) and co-localized with the Golgi marker protein, GRASP65 (Fig. 3B, a–d). Surprisingly, the C-terminal region (residues 322–962), which includes the giantin and GM130-acidic binding domain (23, 24), did not localize to the Golgi apparatus (Fig. 3B, e–h) but instead manifested a diffuse distribution throughout the cytoplasm (Fig. 3B, e), suggesting that the C terminus was not sufficient to effect Golgi localization. Unexpectedly, the N-terminal 636 amino acids, which contain the armadillo-fold, did not localize to the Golgi (Fig. 3B, i–l) or centrosomal (supplemental Fig. S2, i–l) regions and remained dispersed in the cytoplasm (Fig. 3B, i). Significantly, the minimal requirement for Golgi localization was a construct that included both the N-terminal 321 residues and the coiled-coil containing C terminus (construct Δ322–636) (Fig. 3B, m–p) suggesting that both the N- and C-terminal regions of p115 were required for Golgi targeting (Fig. 3B, o and p). In contrast, the minimal requirement for centrosomal targeting was the construct containing the armadillo-fold (amino acids 1–321). This construct localized to only a subset of the Golgi apparatus (Fig. 3B, q–t) and almost exclusively to the centrosome (supplemental Fig. S2, a–d). Full-length p115 (supplemental Fig. S2, e–h) and the Δ322–636 construct (supplemental Fig. S2, i–l) exhibited partial localization with the centrosomal marker as predicted. These findings suggest that the armadillo-fold (amino acids 1–321) is sufficient for centrosome targeting of p115, likely through its interaction with γ-tubulin, however, it requires the p115 C terminus for Golgi targeting.
FIGURE 3.
Localization of p115 to the Golgi apparatus requires both the N- and C-terminal domains. A, schematic of p115 constructs used in this experiment. B, COS7 cells were transfected with the indicated GFP-tagged p115 constructs for 24 h and processed for indirect immunofluorescence with an anti-GRASP-65 antibody. Arrows indicate localization of the transfected constructs (FL, 1–321, Δ322–636) to GRASP65 positive structures. The armadillo-fold (1–321) localizes to only a subset of the Golgi apparatus. Images are projected confocal Z-sections. Blue, Hoechst 33342. Scale bar, 10 μm.
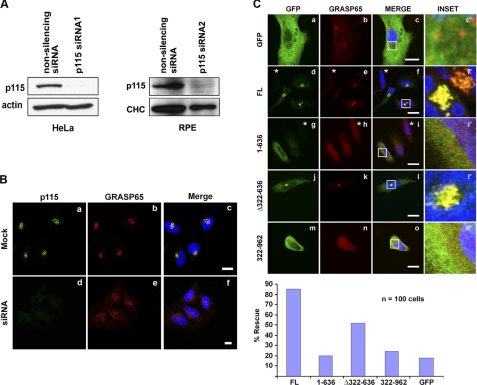
Previous studies showed that knockdown of p115 leads to Golgi fragmentation, and that the structure and function of the fragmented Golgi could be rescued by constructs lacking most of the C-terminal region but including the first coiled-coil domain of p115 (17). By using two previously published and well characterized siRNA sequences (17, 36), we achieved quantitative depletion of p115 in HeLa and RPE cells (Fig. 4A) that as expected, led to complete fragmentation of the Golgi apparatus (Fig. 4B and supplemental Fig. S3, A and B). We then tested the ability of siRNA-resistant, GFP-tagged bovine p115 deletion constructs to rescue the normal Golgi morphology in the p115-depleted cells (Fig. 4C). As predicted, the N-terminal 1–636 residues were unable to restore Golgi structure (Fig. 4C, g–i), much like the negative control, the GFP vector alone (Fig. 4C, a–c). The construct that included all four coiled-coil regions but lacked the N-terminal armadillo-like helical-fold (322–962; Fig. 2A) also failed to correct the fragmented phenotype (Fig. 4C, m–o). In contrast, both the full-length and Δ322–636 p115 constructs effectively restored Golgi morphology (Fig. 4C, d–f and j–l). These data demonstrated that Golgi integrity required the N-terminal, γ-tubulin interacting region of p115 in addition to the previously proposed, golgin interacting C terminus.
FIGURE 4.
Depletion of p115 results in complete vesiculation of the interphase Golgi, which is rescued by a construct containing both the N and C termini. A, Western blot of p115 protein levels in HeLa and RPE cells, following 4 days of either non-silencing or p115 siRNA treatment. Comparable knockdown levels and phenotypes were obtained with siRNA1 (17) and siRNA2 (36) in HeLa and RPE cells. Loading controls: actin and clathrin heavy chain (CHC). B, efficient p115 depletion results in a fragmented Golgi phenotype. C, HeLa cells were treated with the p115 siRNA for 3 days after which they were transfected for 24 h with the indicated GFP-tagged, siRNA-resistant bovine p115 constructs. Cells were fixed and processed for indirect immunofluorescence with the Golgi marker GRASP65. The rescue efficiency was quantified by scoring the Golgi morphology in 100 transfected cells. Asterisks denote fragmented Golgi in p115-depleted, untransfected/unrescued cells. Images are projected confocal Z-series. Blue, Hoechst 33342. Scale bars, 10 μm.
p115 Is Required for Spindle Maintenance and Completion of Cytokinesis
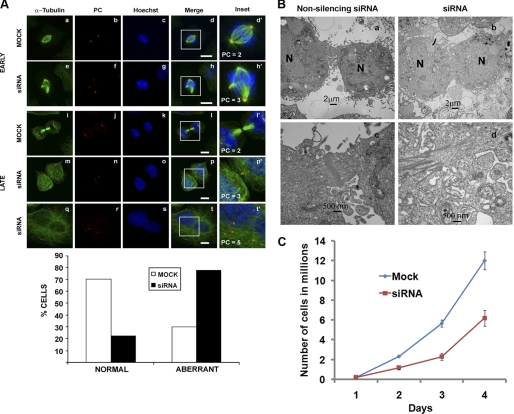
In the course of these experiments, we observed that the nuclei of p115-depleted cells were 40 to 50% larger than in control cells (supplemental Fig. S4A). We hypothesized therefore, that the increase in nuclear size was due to an inability of p115-depleted cells to either enter or complete mitosis. To investigate the possibility that p115-depleted cells display a cell cycle defect, cells were synchronized using an aphidicolin block (“Experimental Procedures”) and analyzed for spindle morphology, chromosome alignment, and centrosome integrity (Fig. 5A). Although in control transfected cells the chromosomes aligned at the metaphase plate and bipolar, focused spindles were readily evident (Fig. 5A, a–d), ∼80% of p115-depleted cells exhibited abnormal, multipolar spindles with a range of aberrant morphologies and misaligned chromosomes (Fig. 5A, e–h, and supplemental Fig. S4B). The multipolar spindles were likely a consequence of mitotically disrupted centrosomal integrity, as shown by multiple, smaller pericentrin positive foci (Fig. 5A, h′, p′, and t′) compared with two large pericentrin foci in control treated cells (Fig. 5A, d′ and l′). Pericentrin is a component of the pericentrosomal matrix, which also contains γ-tubulin as a component of the γTuRC and γTuSc microtubule nucleating complexes. Importantly, the centrosome defects were mitosis specific, because p115 depletion per se did not affect centrosome morphology or number in interphase cells, as shown by co-staining with pericentrin (supplemental Fig. S3C) and γ-tubulin (supplemental Fig. S3D). The aberrant chromosomal arrangement was also evident when the synchronized cells were examined by electron microscopy where there was no evidence of equatorially aligned chromosomes (supplemental Fig. S4C).
FIGURE 5.
p115-depleted cells display spindle and cytokinesis defects. A, following a 3-day mock or siRNA treatment, HeLa cells were synchronized in S-phase with aphidicolin then processed for immunofluorescence in early (metaphase; a–h) or late (cytokinesis; i–t) mitosis with antibodies against α-tubulin (green) or pericentrin (PC; red). PC positive foci are indicated in the insets. Images are projected confocal Z-sections. The percentage of cells displaying the normal versus aberrant spindle phenotype was quantified in both mock and siRNA-treated cells by counting ∼200 cells. B, electron micrographs of the cytokinetic furrow in synchronized non-silencing (a and c) or p115 siRNA-treated (b and d) HeLa cells collected at 10 h after release from the aphidicolin block (N, nucleus). C, growth curve of mock and p115 siRNA-treated cells. Equal numbers of cells were siRNA (red) or mock (blue) treated at day 0 and cells were counted every day for 4 days following treatment. Data are from three independent experiments. Error bars represent S.D. ± mean. Scale bars, 10 μm.
In addition, a subset of the p115 knockdown cells displayed a dramatic cytokinesis defect in which a normal cytokinetic bridge (Fig. 5A, i–l, and B, a and c) failed to form, and resulted in cell fusion (Fig. 5A, m–t, and B, b and d). This failed cytokinesis phenotype led to a significant decrease in growth rate over a period of 4 days following siRNA treatment (Fig. 5C). Despite the growth reduction, we did not observe mitotic arrest or any mitosis-related apoptosis4 and because the difference in growth was ∼2-fold at each time point, we conclude that the growth defect was a consequence of failed cytokinesis (Fig. 5, A and B, and see below) and daughter cell fusion.
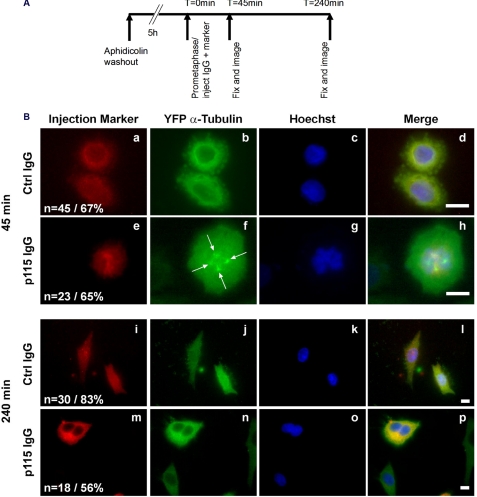
Because 2 to 4 days were required to achieve quantitative siRNA-mediated p115 knockdown, it was possible that the effects on spindle morphology and cytokinesis could be due to secondary effects accumulating during multiple cell cycles. To address this, prometaphase cells were injected with either preimmune serum or a purified monoclonal anti-p115 IgG (Fig. 6). The p115 antibody showed no detectable immunoreactivity in p115-depleted cells (Fig. 4B) and microinjection of this antibody in interphase cells led to Golgi fragmentation.4 Following injection in prometaphase, cells were allowed to progress through mitosis for either 45 or 240 min (Fig. 6A). At 45 min postinjection, cells injected with control IgG underwent mitosis with normal kinetics and were now in cytokinesis (Fig. 6B, a–d), the p115 IgG-injected cells exhibited mitotic delay and aberrant, multipolar spindles with misaligned chromosomes (Fig. 6B, e–h, arrows) similar to those observed in p115 siRNA-treated cells (Fig. 5B). Consistently, at 240 min following microinjection, the p115 IgG-injected cells appeared to have exited mitosis despite the apparent delay (Fig. 6B, f), however, the cells displayed the bi-nucleated phenotype indicative of failed cytokinesis (Fig. 6B, m–p). Given the immediate response to antibody injection, these results confirmed the requirement for p115 in spindle integrity and suggested that the p115 siRNA effects were not secondary to interphase effects on the Golgi or centrosome structure.
FIGURE 6.
Microinjection of a p115 inhibitory antibody results in multipolar spindles and binucleated cells. A, schematic showing the experimental procedure. HeLa cells stably expressing YFP-tagged α-tubulin were synchronized in S-phase with aphidicolin (see “Experimental Procedures”). After 5 h of release from the aphidocolin block, cells in prometaphase were injected with either p115 (7D1) or control IgG along with an injection marker. B, cells were then fixed at either 45 (a–h) or 240 min (i–p) postinjection and stained with Hoechst 33342 (blue). Number of injected, surviving cells is indicated in the figure (n) as well as the percentage of cells displaying the shown phenotype. Arrows, multipolar spindle triggered by microinjection of the p115 inhibitory antibody. Images are single sections acquired on an inverted microscope. Scale bars, 10 μm.
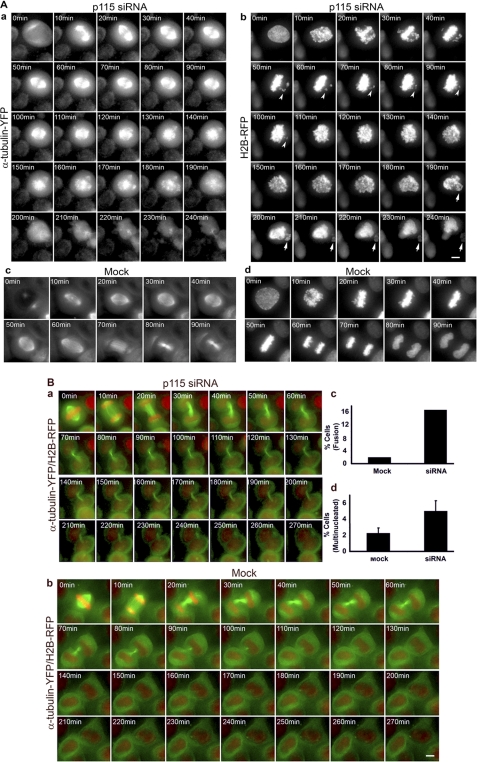
The presence of mitotic spindles, albeit disorganized, demonstrated that p115-depleted cells entered mitosis but were unable to maintain the spindle. To test this, we employed live cell imaging of mock or p115 siRNA-treated cells stably co-expressing YFP-tagged α-tubulin and RFP-tagged histone H2B (Fig. 7 and supplemental Movies S1–S6). This cell line allowed us to monitor both the mitotic spindle and chromosome segregation. Interestingly, in absence of p115, two mutually exclusive phenotypes were observed: 1) normal spindle assembly followed by spindle collapse and chromosome missegregation (Fig. 7A, compare a and c) and 2) a cytokinesis phenotype in which the spindle was formed and maintained normally, but the cytokinetic bridge failed, resulting in cell fusion and multinucleated cells (Fig. 7B, compare a and b).
FIGURE 7.
p115-depleted cells show spindle collapse and failed cytokinesis. A, time lapse of p115 siRNA-treated (a and b) or mock treated (c and d) HeLa cells stably expressing α-tubulin-YFP and H2B-RFP. a, spindle (α-tubulin-YFP) and b, chromosome (H2B-RFP) dynamics during mitosis progression show failure of chromosomes to align at the metaphase plate (arrowheads) followed by spindle collapse beginning at 120 min. Arrows indicate chromosome missegregation at late stages of mitosis (200 min) and during cytokinesis (240 min). Mock treated cells proceed through mitosis with normal kinetics as shown by spindle (c) and chromosome (d) behavior. 0 min = interphase. n > 120 cells. B, time lapse of cytokinesis in p115 siRNA (a) and mock treated HeLa (b) α-tubulin-YFP/H2B-RFP cells. a, siRNA-treated cells maintain a cytokinetic bridge for over 210 min and fuse at ∼220 min following mitotic exit. b, mock treated cells resolve the cytokinetic bridge at ∼90 min after mitotic exit. 0 min = late metaphase. Frames are deconvolved, projected Z-series captured using a DeltaVision RT deconvolution system (Applied Precision Inc.) at 5-min intervals; every other frame is shown. Scale bar, 5 μm. c, quantitation of the fusion phenotype, n > 180 cells. d, quantitation of the multinucleate phenotype, n > 750 cells. Error bars represent S.E. ± mean.
Mock-treated cells displayed normal mitosis progression whereby normal spindles were assembled and maintained and cells entered anaphase within ∼50 min of mitosis onset (Fig. 7A, c and d and supplemental Movies S3 and S4). In contrast, in a subset of p115-depleted cells, a spindle formed and was maintained for 80 min (Fig. 7A, a, 20–100 min, and supplemental Movie S1). Despite this seemingly normal spindle, chromosomes failed to efficiently align at the metaphase plate (Fig. 7A, compare b, arrowheads, and d, and supplemental Movie S2) and the spindle collapsed at ∼2 h following mitotic entry. Interestingly, spindle collapse did not result in mitotic arrest and cell death, suggesting it occurred after chromosome alignment and the metaphase-anaphase checkpoint (47). Because cells completed mitosis with an abnormal spindle (Fig. 7A, a, 120–240 min), this resulted in aberrant chromosome segregation where a small subset of chromosomes was partitioned into one daughter cell (Fig. 7A, b, arrows) and most of the duplicated genome was inherited by the second daughter cell.
In addition, our statistical analysis of siRNA (Fig. 7B, a, and supplemental Movie S5) and mock (Fig. 7B, b, and supplemental Movie S6) treated cells showed that p115 depletion resulted in ∼2.5 times more multinucleated cells (Fig. 7B, d) and almost 8 times more failed abscission (Fig. 7B, c). The higher frequency of failed abscission compared with the frequency of multinucleated cells could be explained by nuclear fusion and/or postmitotic cell death.
Taken together, our findings from three independent approaches suggest that p115 functions in the maintenance of the mitotic spindle and the abscission of the cytokinetic furrow. Both roles are likely to be dependent on p115-γ-tubulin association and involve maintenance of centrosome integrity in mitosis and Golgi structure/function during cytokinesis completion and then in interphase. These functions thus distinguish the mitotic role of p115 from that of other golgins such as GM130 or GRASP65 (30, 31).
DISCUSSION
Here we show that p115 is the first golgin to associate with the spindle poles throughout mitosis and implicate it in Golgi biogenesis and mitosis progression. Our studies were prompted by two observations: 1) only p115 and no other golgin remained at the spindle poles of mitotic cells and 2) the prediction that the p115 N terminus possesses an armadillo-like helical-fold, which could promote interactions with cytoskeletal proteins. Indeed, the N-terminal armadillo-fold mediated association with γ-tubulin, the main microtubule nucleating component and was essential for Golgi structure. Recently, the crystal structure of the p115 N terminus has been solved by two groups (42, 43) and these studies support our bioinformatics data on the existence of the armadillo domain.
The finding that the p115 N-terminal armadillo domain mediates association with a γ-tubulin-containing complex and is required for Golgi structure, together with the segregation of p115 with the spindle poles during mitosis suggest a mechanism whereby p115 maintains a centrosomal localization to provide a recruiting site for post-mitotic Golgi reassembly. Interestingly, the S. cerevisiae p115 homolog, Uso1p, lacks a classical armadillo domain and in contrast to mammalian cells, the yeast Golgi apparatus exists as dispersed ministacks (48). These differences support a model in which the association between the p115 armadillo-fold and a γ-tubulin-containing complex may be in part responsible for the pericentrosomal anchoring of the mammalian Golgi complex. Earlier studies suggested that Golgi protein GMAP210 recruits γ-tubulin to Golgi membranes and mediates the Golgi localization to the centrosome (49). Although the role of GMAP210 in centrosomal Golgi localization has been questioned (50, 51), it remains possible that other factors mediate interactions between Golgi components and the centrosome. Given the association of p115 with a γ-tubulin-containing complex (i.e. γTuRC), it would be of interest to determine whether components such as γ-complex proteins could act as adaptors for this association. In addition to the centrosomal pool, γ-tubulin has been proposed to exist in soluble form and on various membranes, at non-centrosomal microtubule nucleation sites (7). γ-Tubulin may be recruited to microtubule nucleation sites on Golgi membranes by a GM130/Golgi-tethered p115, however, this model remains to be tested further.
In p115-depleted cells, both the N-terminal armadillo-like helical-fold and the C-terminal region of p115 were required to restore normal Golgi morphology. Significantly, the armadillo-like helical domain (residues 1–321) was targeted almost exclusively to the centrosome, possibly due to its high affinity for γ-tubulin. Notably, no other p115-derived fragment showed centrosomal localization, suggesting a specific association of the 1–321 domain with centrosomal components. This implicates the armadillo-fold as the minimal sequence required for centrosome targeting. Although the details of these interactions are yet to be determined, failure of the extended N-terminal domain (residues 1–636) to target to the Golgi or the centrosome, together with its decreased γ-tubulin association compared with that of the full-length p115 suggest an important functional role for the proposed hairpin conformation of p115 (52).
Previous studies implicated Golgi proteins GM130 and GRASP65 in mitotic events (31, 53) and showed that their depletion leads to multiple centrosome-like structures, possibly due to centrosome overduplication in interphase. This was followed by aberrant spindle assembly, mitotic arrest, and apoptosis. In contrast, p115-depleted cells displayed normal interphase centrosomes and spindle assembly, which collapsed late in mitosis and interestingly, did not result in mitotic arrest or mitosis-related apoptosis. The spindle collapse coincided with fragmentation of the pericentrosomal matrix, as shown by the increased number of pericentrin positive foci and resulted in chromosome missegregation. The observation that spindles can form in the absence of p115 suggests that p115 is not required for interphase centrosome function or for its duplication cycle and that its role is mitosis specific. This is in contrast to the proposed role for GM130 in stabilizing interphase centrosomes (30) and suggests that p115 and GM130 play distinct roles outside of their overlapping function in Golgi structure.
Previous evidence suggested a role for the Golgi apparatus and Golgi-associated proteins in cytokinesis (54). We showed that p115 depletion led to a dramatic cell fusion phenotype, a novel observation for a golgin. Because p115 knockdown results in fragmentation of the interphase Golgi, the cytokinesis phenotype may be due to impaired post-Golgi trafficking of membranes and proteins to the cleavage furrow. Alternatively, Gromley et al. (34, 35) suggested that furrow abscission involves the exocyst complex and SNARE-mediated membrane fusion. Consistent with this model, the CC1 region of p115 interacts with the Golgi-associated SNARE, syntaxin5 (22), and interestingly, a hypomorphic allele of the Drosophila syntaxin 5 orthologue has been shown to produce a phenotype similar to that of p115 depletion (55). In this scenario, p115 could function to deliver SNAREs to the cytokinetic furrow (21, 22). Differentiating between a function for post-Golgi trafficking versus a direct p115 involvement in SNARE targeting to the furrow will provide an important advance in understanding the role of the trafficking machinery in cytokinesis.
Overall, our studies propose a model whereby the p115-γ-tubulin association may support microtubule nucleation in interphase and mitosis at the Golgi and the centrosome, respectively (supplemental Fig. S5). In this model, during interphase p115 interacts with Golgi-associated proteins (i.e. GM130) via its C terminus and γ-tubulin through its N-terminal armadillo-fold, thus providing a mechanism for γ-tubulin recruitment to Golgi membranes. This is consistent with our finding that both the C- and N-terminal domains of p115 are required for the Golgi structure and with previous findings that Golgi-nucleated microtubules maintain the structure of the Golgi ribbon (8). Therefore, p115 molecules could recruit γ-tubulin to the Golgi membrane for the establishment of non-centrosomal microtubule organizing centers, similarly to the GM130-mediated recruitment of the γ-TuRC-interacting protein, AKAP450, which has been proposed to be required for Golgi-dependent microtubule nucleation (9). It remains to be determined if p115 associates with centrosomal γ-tubulin or with the soluble γ-tubulin pool, however, both interactions could be relevant to Golgi positioning and structure in interphase and during post-mitotic reassembly. Whether a reformed, fully functional Golgi apparatus is required for sorting and targeting of membrane and proteins to the cytokinetic bridge awaits further investigation and will require high resolution spatial and temporal analysis of Golgi reassembly.
In addition, we show for the first time a role for p115 in the maintenance of the mitotic spindle under conditions of chromosome tension and propose that this function is likely to be mediated by the interaction between p115 and γ-tubulin. Furthermore, the mitotic spindle pole segregation of a golgin (i.e. p115) may be required to dictate efficient post-mitotic pericentrosomal recruitment and reassembly of the highly organized Golgi apparatus. Upon exit from mitosis, the centrosome-associated p115 could recruit Golgi-derived components via interaction of its C-terminal region with SNAREs and Golgi proteins such as GM130. This would establish a scaffold for the sequential assembly of the membranous cisternae. If this scenario is correct and the main γ-tubulin-dependent role of p115 is in Golgi biogenesis, its mitotic function in stabilizing the centrosome may have evolved as a result of its maintenance at the spindle poles.
We suggest that the cytokinesis and spindle phenotypes are independent of each other, and that although the cytokinesis defect may be a direct consequence of disrupted Golgi structure and function in p115-depleted cells, the spindle phenotype is likely due to centrosome instability in the absence of the p115-γ-tubulin association.
Supplementary Material
Acknowledgments
We thank Drs. Adam Linstedt, Don Cleveland, Gerry Waters, and Yukio Ikehara for their generous gift of reagents. We thank Leonid Novikov for technical assistance. We thank Drs. David Spray and Sylvia Suadicani for expert advice on microinjection and use of their facility. Special thanks to Drs. Margaret Kielian, Anne Muesch, and Samantha Zeitlin for helpful suggestions with the manuscript. A.R. would like to thank Soumit Roy and Drs. Don Cleveland, Andrew Holland, and Cristina Caescu for help completing this work.
Footnotes
This work was supported, in whole or in part, by National Institutes of Health Grant DK21860 (to D. S.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental “Experimental Procedures,” Figs. S1–S5, and Movies S1–S6.
A. E. Radulescu, S. Mukherjee, and D. Shields, unpublished data.
REFERENCES
- 1. Colanzi A., Corda D. (2007) Curr. Opin. Cell Biol. 19, 386–393 [DOI] [PubMed] [Google Scholar]
- 2. Colanzi A., Sutterlin C., Malhotra V. (2003) J. Cell Biol. 161, 27–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Altan-Bonnet N., Sougrat R., Lippincott-Schwartz J. (2004) Curr. Opin. Cell Biol. 16, 364–372 [DOI] [PubMed] [Google Scholar]
- 4. Barr F. A. (2004) J. Cell Biol. 164, 955–958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Lippincott-Schwartz J. (1998) Curr. Opin. Cell Biol. 10, 52–59 [DOI] [PubMed] [Google Scholar]
- 6. Lüders J., Stearns T. (2007) Nat. Rev. Mol. Cell Biol. 8, 161–167 [DOI] [PubMed] [Google Scholar]
- 7. Chabin-Brion K., Marceiller J., Perez F., Settegrana C., Drechou A., Durand G., Poüs C. (2001) Mol. Biol. Cell 12, 2047–2060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Efimov A., Kharitonov A., Efimova N., Loncarek J., Miller P. M., Andreyeva N., Gleeson P., Galjart N., Maia A. R., McLeod I. X., Yates J. R., 3rd, Maiato H., Khodjakov A., Akhmanova A., Kaverina I. (2007) Dev. Cell 12, 917–930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Rivero S., Cardenas J., Bornens M., Rios R. M. (2009) EMBO J. 28, 1016–1028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Miller P. M., Folkmann A. W., Maia A. R., Efimova N., Efimov A., Kaverina I. (2009) Nat. Cell Biol. 11, 1069–1080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Barr F. A., Puype M., Vandekerckhove J., Warren G. (1997) Cell 91, 253–262 [DOI] [PubMed] [Google Scholar]
- 12. Linstedt A. D., Hauri H. P. (1993) Mol. Biol. Cell 4, 679–693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Barroso M., Nelson D. S., Sztul E. (1995) Proc. Natl. Acad. Sci. U.S.A. 92, 527–531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Nakamura N., Rabouille C., Watson R., Nilsson T., Hui N., Slusarewicz P., Kreis T. E., Warren G. (1995) J. Cell Biol. 131, 1715–1726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Shorter J., Watson R., Giannakou M. E., Clarke M., Warren G., Barr F. A. (1999) EMBO J. 18, 4949–4960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Puthenveedu M. A., Linstedt A. D. (2001) J. Cell Biol. 155, 227–238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Puthenveedu M. A., Linstedt A. D. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 1253–1256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Waters M. G., Clary D. O., Rothman J. E. (1992) J. Cell Biol. 118, 1015–1026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Levine T. P., Rabouille C., Kieckbusch R. H., Warren G. (1996) J. Biol. Chem. 271, 17304–17311 [DOI] [PubMed] [Google Scholar]
- 20. Sapperstein S. K., Walter D. M., Grosvenor A. R., Heuser J. E., Waters M. G. (1995) Proc. Natl. Acad. Sci. U.S.A. 92, 522–526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Allan B. B., Moyer B. D., Balch W. E. (2000) Science 289, 444–448 [DOI] [PubMed] [Google Scholar]
- 22. Shorter J., Beard M. B., Seemann J., Dirac-Svejstrup A. B., Warren G. (2002) J. Cell Biol. 157, 45–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Nakamura N., Lowe M., Levine T. P., Rabouille C., Warren G. (1997) Cell 89, 445–455 [DOI] [PubMed] [Google Scholar]
- 24. Sönnichsen B., Lowe M., Levine T., Jämsä E., Dirac-Svejstrup B., Warren G. (1998) J. Cell Biol. 140, 1013–1021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Chiu R., Novikov L., Mukherjee S., Shields D. (2002) J. Cell Biol. 159, 637–648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Mukherjee S., Shields D. (2009) J. Biol. Chem. 284, 1709–1717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Mukherjee S., Chiu R., Leung S. M., Shields D. (2007) Traffic 8, 369–378 [DOI] [PubMed] [Google Scholar]
- 28. Guo Y., Punj V., Sengupta D., Linstedt A. D. (2008) Mol. Biol. Cell 19, 2830–2843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Sohda M., Misumi Y., Yoshimura S., Nakamura N., Fusano T., Ogata S., Sakisaka S., Ikehara Y. (2007) Traffic 8, 270–284 [DOI] [PubMed] [Google Scholar]
- 30. Kodani A., Sütterlin C. (2008) Mol. Biol. Cell 19, 745–753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Sütterlin C., Polishchuk R., Pecot M., Malhotra V. (2005) Mol. Biol. Cell 16, 3211–3222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Litvak V., Tian D., Carmon S., Lev S. (2002) Mol. Cell. Biol. 22, 5064–5075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Sisson J. C., Field C., Ventura R., Royou A., Sullivan W. (2000) J. Cell Biol. 151, 905–918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Gromley A., Yeaman C., Rosa J., Redick S., Chen C. T., Mirabelle S., Guha M., Sillibourne J., Doxsey S. J. (2005) Cell 123, 75–87 [DOI] [PubMed] [Google Scholar]
- 35. Goss J. W., Toomre D. K. (2008) J. Cell Biol. 181, 1047–1054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Sohda M., Misumi Y., Yoshimura S., Nakamura N., Fusano T., Sakisaka S., Ogata S., Fujimoto J., Kiyokawa N., Ikehara Y. (2005) Biochem. Biophys. Res. Commun. 338, 1268–1274 [DOI] [PubMed] [Google Scholar]
- 37. Radulescu A. E., Siddhanta A., Shields D. (2007) Mol. Biol. Cell 18, 94–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Shima D. T., Haldar K., Pepperkok R., Watson R., Warren G. (1997) J. Cell Biol. 137, 1211–1228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Wei J. H., Seemann J. (2009) J. Cell Biol. 184, 391–397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Seemann J., Pypaert M., Taguchi T., Malsam J., Warren G. (2002) Science 295, 848–851 [DOI] [PubMed] [Google Scholar]
- 41. Puri S., Telfer H., Velliste M., Murphy R. F., Linstedt A. D. (2004) J. Cell Sci. 117, 451–456 [DOI] [PubMed] [Google Scholar]
- 42. Striegl H., Roske Y., Kümmel D., Heinemann U. (2009) PLoS ONE 4, e4656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. An Y., Chen C. Y., Moyer B., Rotkiewicz P., Elsliger M. A., Godzik A., Wilson I. A., Balch W. E. (2009) J. Mol. Biol. 391, 26–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Coates J. C. (2003) Trends Cell Biol. 13, 463–471 [DOI] [PubMed] [Google Scholar]
- 45. Linstedt A. D., Jesch S. A., Mehta A., Lee T. H., Garcia-Mata R., Nelson D. S., Sztul E. (2000) J. Biol. Chem. 275, 10196–10201 [DOI] [PubMed] [Google Scholar]
- 46. Oegema K., Wiese C., Martin O. C., Milligan R. A., Iwamatsu A., Mitchison T. J., Zheng Y. (1999) J. Cell Biol. 144, 721–733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Musacchio A., Salmon E. D. (2007) Nat. Rev. Mol. Cell Biol. 8, 379–393 [DOI] [PubMed] [Google Scholar]
- 48. Malhotra V., Mayor S. (2006) Nature 441, 939–940 [DOI] [PubMed] [Google Scholar]
- 49. Ríos R. M., Sanchís A., Tassin A. M., Fedriani C., Bornens M. (2004) Cell 118, 323–335 [DOI] [PubMed] [Google Scholar]
- 50. Gillingham A. K., Tong A. H., Boone C., Munro S. (2004) J. Cell Biol. 167, 281–292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Barr F. A., Egerer J. (2005) J. Cell Biol. 168, 993–998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Beard M., Satoh A., Shorter J., Warren G. (2005) J. Biol. Chem. 280, 25840–25848 [DOI] [PubMed] [Google Scholar]
- 53. Kodani A., Kristensen I., Huang L., Sütterlin C. (2009) Mol. Biol. Cell 20, 1192–1200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Skop A. R., Bergmann D., Mohler W. A., White J. G. (2001) Curr. Biol. 11, 735–746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Xu H., Brill J. A., Hsien J., McBride R., Boulianne G. L., Trimble W. S. (2002) Dev. Biol. 251, 294–306 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.