Abstract
AIM: To investigate the influence of chitosan nanofiber scaffold on the production and infectivity of porcine endogenous retrovirus (PERV) expressed by porcine hepatocytes.
METHODS: Freshly isolated porcine hepatocytes were cultured with or without chitosan nanofiber scaffold (defined as Nano group and Hep group) for 7 d. The daily collection of culture medium was used to detect reverse transcriptase (RT) activity with RT activity assay kits and PERV RNA by reverse transcription-polymerase chain reaction (PCR) and real time PCR with the PERV specific primers. And Western blotting was performed with the lysates of daily retrieved cells to determine the PERV protein gag p30. Besides, the in-vitro infectivity of the supernatant was tested by incubating the human embryo kidney 293 (HEK293) cells.
RESULTS: The similar changing trends between two groups were observed in real time PCR, RT activity assay and Western blotting. Two peaks of PERV expression at 10H and Day 2 were found and followed by a regular decline. No significant difference was found between two groups except the significantly high level of PERV RNA at Day 6 and PERV protein at Day 5 in Nano group than that in Hep group. And in the in-vitro infection experiment, no HEK293 cell was infected by the supernatant.
CONCLUSION: Chitosan nanofiber scaffold might prolong the PERV secreting time in pig hepatocytes but would not obviously influence its productive amount and infectivity, so it could be applied in the bioartificial liver without the increased risk of the virus transmission.
Keywords: Chitosan nanofiber scaffold, Porcine hepatocyte, Porcine endogenous retrovirus, Bioartificial liver
INTRODUCTION
Although liver transplantation is currently recognized as the most effective treatment for acute liver failure and end-stage liver diseases, its application has been seriously limited because of the lack of donor organs[1,2]. Therefore, bioartificial liver (BAL) has been proposed as a temporary liver support for patients awaiting liver transplantation[3-5].
At present, porcine hepatocytes were still a major cell source because of their adequate resources, accessibility and characters similar to human hepatocytes[6-8]. However, the clinical application of BAL based on porcine hepatocytes was not very optimistic[4]. So how to increase the in-vitro function of hepatocytes with a suitable scaffold was always an attractive issue and some previous reports showed the enhanced function of procine hepatocytes with their scaffold, including the chitosan nanofiber scaffold developed in our institute[9-11].
Nevertheless, as xenogeneic cells, there were some problems with porcine hepatocytes. The security of the transmission of porcine endogenous retrovirus (PERV) has been one of the most essential concerns that can not be ignored, since PERV infection of human cells in vitro was widely reported[12]. Therefore, it raised new questions that whether the production and infectivity of PERV would be influenced and whether the risk of PERV infection in BAL would be increased when the function of porcine hepatocytes was enhanced with certain scaffolds.
Previously, we had proved the chitosan nanofiber scaffold could enhanced hepatocytes adhesion, viability and function in vitro[13]. This study was focused on the influence of chitosan nanofiber scaffold on PERV expression and infectivity in pig hepatocytes.
MATERIALS AND METHODS
Animals and reagents
Outbred white pigs with an average weight of 15-20 kg received humane care and all animal procedures were performed according to institutional and national guidelines and approved by the Animal Care Ethics Committee of Nanjing University and Nanjing Drum Tower Hospital. All cell culture-related reagents were purchased from GIBCO (USA). Lactobionic acid (LA) and chitosan (low molecular weight, brookfield viscosity 20 000 cps, 85% deacetylation) were purchased from Sigma-Aldrich (Saint Louis, USA). N-Hydroxysuccinimide was purchased from Thermo-Pierce (Rockford, USA). 1-Ethyl-3-(3-dimethyl aminopropyl) carbodiimide and N,N,N0N0-tetramethylethylenediamine were obtained from TCI (Tokyo, Japan). Poly(ethylenoxide) (PEO, MWz1_106) was supplied by Guoren Chemical Co. (Beijing, China). All other reagents were of analytical reagent grade.
Preparation of chitosan nanofiber scaffold via electrospinning
The chitosan nanofiber scaffold was prepared according to the previous reference[13]. In brief, Chitosan (Sigma) and PEO powders (9:1 w/w) were dissolved in formic acid/ethanol (7:3 v/v) to give 2.6% (w/v) at room temperature. The stock solution was then filled into a 5 mL glass syringe fitted with a 20 G needle and then expressed at 5 mL/h using a syringe pump. The nanofibers were collected on 24 mm diam. coverslips located at a fixed distance of 10-20 mm from the needle tip. A non-coated cover slip was also prepared and tested in the same manner as control.
Hepatocytes isolation, characterization and culture
Primary pig hepatocytes were harvested by a two-step in situ collagenase perfusion technique[14,15]. First, the pre-warmed (37°C) Ca2+ and Mg2+ free Hanks balanced salt solution was perfused into the livers of the anesthetized pigs in vivo via the portal vein at a flow rate of 80-
100 mL/min for 30-40 min, followed by 0.05% Type IV collagenase (37°C) at a rate of 10 mL/min for 40-50 min. The released cells were filtered through nylon mesh with 100-mm openings and washed via three centrifugations (50 g). The viability of the isolated primary hepatocytes determined by trypan blue exclusion was more than 95%. Nonparenchymal cells, as judged by their size (< 10 mm in diameter) and morphology (nonpolygonal or stellate), were less than 1%, which was also verified by immunocytochemical analysis of albumin and cytokeratin 18 (data not shown). Freshly isolated hepatocytes were seeded at a density of 106 cells/mL in the substratum of 2 mL RPMI-1640 without serum and incubated in 6-well microtiter plates with or without chitosan nanofiber scaffold (defined as Nano group and Hep group) at 37°C and 5% CO2. Culture medium was replenished daily without the growth medium containing low-glucose Dulbecco’s modified Eagle’s medium (DMEM-LG) supplemented with 10% fetal bovine serum (FBS) for 7 consecutive days. The daily collection of culture medium and cells were deposited at -80°C for later use.
Reverse transcription-polymerase chain reaction
Total RNA was extracted respectively from the centrifuged supernatant with Trizol (Invitrogen, US) and then treated with DNase I (Invitrogen, US) according to the manufacturer’s instructions. For each sample, 60 ng extracted RNA with the value of OD 260/280 among 1.60 and 2.00 was reversed transcribed to cDNA using the reverse transcriptase (RT) kits (Biouniquer, China) in accordance with the instructions. Then polymerase chain reaction (PCR) was completed with the protease-specific primers (forward, 5'-GCTACAACCATTAGGAAAACTAAAAG-3'; and reverse, 5'-AACCAGGACTGTATATCTTGATCAG-3'), polymerase-specific primers (forward, 5'-CTACAACCA TTAGGAAAACTAAAAG-3'; and reverse, 5'-AACCAGGACTGTATATCTTGATCAG-3'), and porcine glyceraldehyde 3-phosphate dehydrogenase-specific primers (forward, 5'-CATCACCATCTTCCAGGAG-3'; and reverse, 5'-TGCCCACAGCCTTGGCAGC-3') and its conditions were as follows: 50°C for 30 min and then 95°C for 5 min followed by 35 cycles of 94°C for 30 s, 55°C for 45 s, 72°C for 30 s, and a final extension step of 72°C for 5 min[16,17]. Amplified production was detected by 2% agarose gel electrophoresis and ethidium bromide (EB) staining.
PERV-specific real time PCR
Quantitative real time PCR was performed with the obtained cDNA, the Roche SuperScript III platinium system, the MX3000P thermocycler (Stratagene), and the primers specific for PERV protease gene (GenBank accession number U77599) (forward, 5'-AGTGCTGCTACAACCATTAGGAAA-3'; and reverse, 5'-AGGGATGACCAGAAACGAGTG-3') and for porcine β-actin gene (GenBank accession number DQ845171) (forward, 5'-GGACTTCGAGCAGGAGATGG-3', and reverse, 5'-AGGAAGGAGGGCTGGAAGAG-3').The conditions were as follows: 95°C for 10min followed by 40 cycles of 95°C for 15 s, 60°C for 1 min and a final cycle of 95°C for 1 min, 65°C for 30 s, 95°C for 30 s. PERV expression was normalized to the amount of the expression of the house-keeping genes β-actin and denoted with the relative value of protease/β-actin.
Western blotting analysis
The lysates of the cultured cells harvested every day were analysed by sodium dodecyl sulfate-10% polyacrylamide gel electrophoresis (SDS-10% PAGE) and Western blotting with polyvinylidene difluoride membranes (Millipore, Eschborn, Germany). The membranes were then incubated with a 1:1500 dilution of mouse anti-FeLV p27 IgG antibody (ABcam, San Francisco, US) which had been proven cross-reactivity with protein gag p30 of PERV[18] overnight at 4°C, followed by a 1:10 000 dilution of goat protein anti-mouse IgG conjugated horseradish peroxidase (KeyGEN, Nanjing, China) for 1 h at 37°C. Immunoreactive proteins on membranes were detected with the enhanced chemiluminescence system and exposured for 15 to 20 s on hyperfilm ECL (Kodak, US) Meanwhile, The protein β-actin was detected as controls. Software ImageJ was used to measure the lightness of each band.
In vitro infection experiments
The in vitro infection experiments were performed according to the reference with some modification[19,20]. Human embryo kidney 293 (HEK293) cells (as gifts from Professor Hua, Nanjing University) were passed overnight in 24-well plates and then incubated in the supernatant of the cell culture (0.5 mL/well) for 6 h in the presence of 8 mg/mL of polybrene after the supernatant was centrifuged at a speed of 5000 r/min for 5 min to remove the cells and cell debris. Meanwhile, the supernatant of porcine kidney 15 (PK15) cells and 0.8 g/mL polybrene was inoculated into the culture of HEK293 cells as positive control. After 4 h of exposure at 37°C, the incubating medium was removed and the cell monolayer was washed with phosphate buffered solution for two times. Then the cells were cultured with the DMEM-HG supplemented with 10% FBS and passed upon confluence for 1 mo before collection.
DNA extraction and PCR
Total DNA was extracted from the treated HEK293 cells with the DNA extracting kits (Axygen, California, US) according to their instructions. PCR was completed with the human β-actin primers (forward, 5'-GCTCGTCGTCGACAACGGCTC-3'; and reverse, 5'-CAAACATGATCTGGGTCATCTTCTC-3'), Sus scrofa cytochrome B (SsCytB) primers (forward, 5'-CATTGGAGTAGTCCTACTATTTACCG-3'; and reverse, 5'-GTAGGATTAGTATTATAAATAAGGCTCCT-3'), above mentioned protease-specific primers and polymerase-specific primers and its conditions consisted with the proceeding of PCR in reverse transcription-PCR (RT-PCR). Amplified production was detected by 2% agarose gel electrophoresis and EB staining.
RT activity assay
The RT activity of the supernatant from the cultured hepatocytes in both group and the treated HEK293, was detected by the C-type RT activity kits (Cavidi-Tech, Uppsala, Sweden) according to the quantitative and qualitative protocols of the manufacturer’s instructions, respectively. The RT activity was examined twice for each collected supernatant sample.
Statistics
Both culture conditions of this study were completed in quintuplicate. All values were expressed as mean ± SD. The two-tailed unpaired Student’s t-test was used to evaluate the statistical significance of differences which was set with a P-value less than 0.05.
RESULTS
PERV RNA in the supernatants
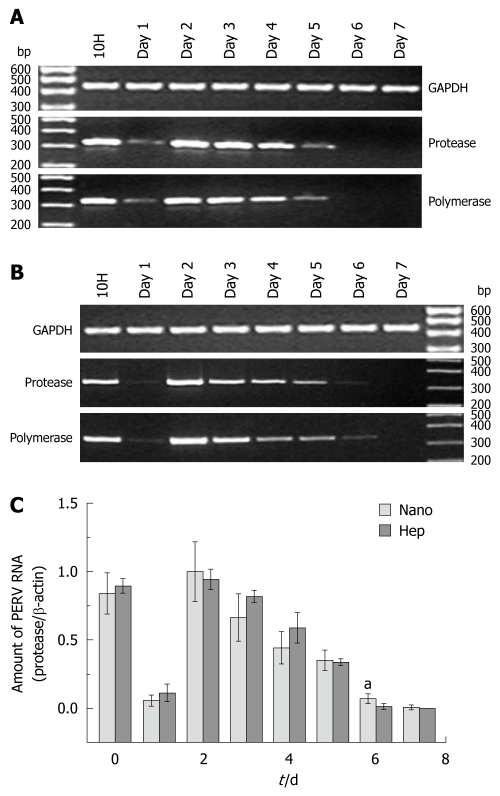
From the results of electrophoresis (shown in Figure 1A and B, representatively), it could be found that the changing trend of the protease sequence and the polymerase sequence was accordant. In real time PCR, the relative value of protease/β-actin was defined as the normalized amount of PERV RNA. Two PERV secreting peaks at 10H and Day 2 was observed, and then the amount of PERV RNA declined gradually after Day 2. No PERV expression was found after Day 6 in Hep group and Day 7 in Nano group, respectively (Figure 1C). There were significant differences at Day 6 between two groups (0.071 ± 0.0348 vs 0.014 ± 0.0193, P < 0.05).
Figure 1.
Porcine endogenous retrovirus RNA in the supernatants. A and B: The representative results of reverse transcription-polymerase chain reaction (PCR) electrophoresis with the RNA extracted from the supernatants in Hep group and Nano group, respectively; C: The results of real time PCR. aP < 0.05. PERV: Porcine endogenous retrovirus; GAPDH: Glyceraldehyde 3-phosphate dehydrogenase.
Western blotting
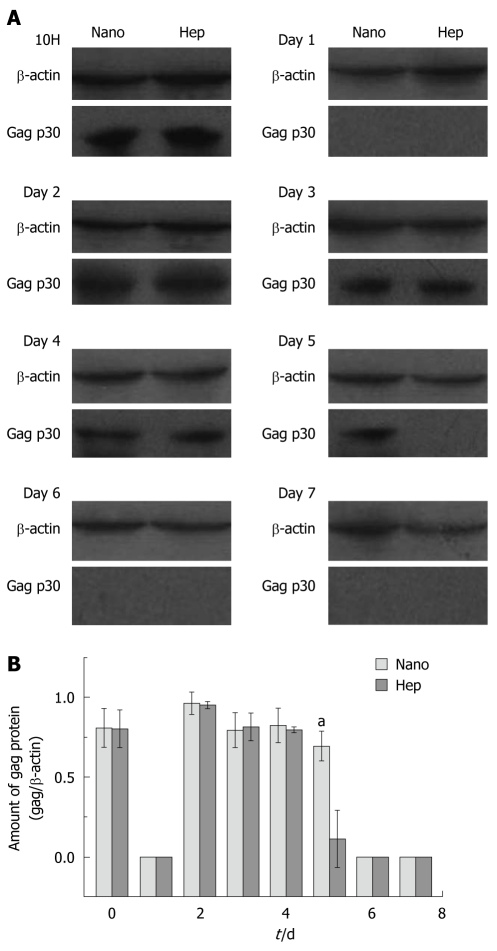
Figure 2A demonstrated a representative result in Western blotting. In Hep group, positive gag p30 proteins were found at 10H and from Day 2 to Day 4, while in Nano group positive bands were observed at 10H and from Day 2 to Day 5. The lightness of each band was measured by the software ImageJ and the amount of the expressed gag p30 protein was normalized with the relative value of gag/β-actin, which was depicted in Figure 2B. Except for the significant differences found at Day 5 (0.70 ± 0.0929 vs 0.11 ± 0.180, P < 0.01), there were no remarkable differences between two groups in other days.
Figure 2.
Western blotting of the porcine endogenous retrovirus gag protein in the cell lysates. A: Representative results of Western blotting with the cell lysates in both groups; B: The normalized protein amount in different days. aP < 0.05.
Infection of HEK293 cells in vitro
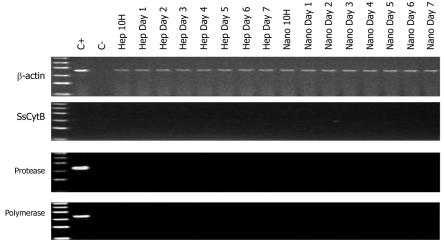
No PERV DNA sequence including protease and polymerase genes and SsCytB sequence were found in the HEK293 DNA (Figure 3). The DNA extracted from PK15 infected HEK293 cells and the pure water was used as positive control and negative control, respectively. And the RT activity in the culture supernatant of the treated HEK293 cells was all negative.
Figure 3.
Representative results of polymerase chain reaction electrophoresis with the DNA extracted from human embryo kidney 293 cells of in vitro infection experiments. C+: Porcine endogenous retrovirus-infected human embryo kidney 293 cells; C-: Pure water. The ladder ranged from 100 to 600 bp. “Hep” and “Nano” meant the incubating supernatant from the simple culture of hepatocytes and the hepatocyte culture on chitosan nanofiber scaffolds. SsCytB: Sus scrofa cytochrome B.
RT activity assay
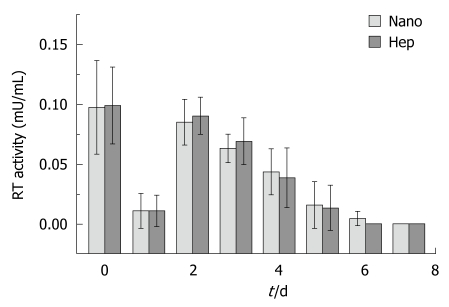
The RT activity in the supernatants from the cultured hepatocytes was demonstrated in Figure 4. No significant difference was found between two groups in each day. And there was no positive RT activity in the supernatant of the HEK293 cells incubated with the supernatant from Hep group or from Nano group.
Figure 4.
Reverse transcriptase activity of the cultured porcine hepatocytes on different substrata. RT: Reverse transcriptase.
DISCUSSION
Porcine hepatocytes were still a major cell source for BAL at present[6-8]. How to enhance their function was always an attractive issue and some progress has been made[9-11]. The chitosan nanofiber scaffold fabricated in our previous study had been proved its superior abilities on cell adhesion and biocompatibility, thereby greatly improved the functions of hepatocytes[13]. However, there had been a new and un-ignorable issue that whether the porcine hepatocytes with improved function would secrete more PERV with enhanced infectivity, which might limit its application in BAL.
PERV was first discovered in 1971 in porcine kidney (PK15) cells[21]. In 1997, Patience et al[22] found for the first time that PERV released from PK15 could infect HEK293 cells in vitro. And then PERV is known to exist regularly in the porcine genome and various porcine cells can excrete PERV particulates[12]. Meanwhile, it was found that PERV successfully infected a variety of human cells in vitro, such as endothelial cells, fibroblasts and bone marrow stromal cells and so on, and virus replication was observed in some of these cells as well[23-28]. Nyberg et al[19] reported that freshly isolated porcine hepatocytes secreted the PERV which didn’t infect HEK293 cells in vitro, at the same time, the production of PERV by cultured pig hepatocytes was unaffected by exposure to growth factors and cytokines present in human FHF sera. Fortunately, there was no evidence of PERV transmission into the patients treated with BAL so far[20,29-32]. However, a recent article[33] claimed PERV released from a BAL infected primary human cells by short-term contact of primary porcine liver cell supernatants with primary human cells, which increased the anxiety about the problem of PERV infection in the application of BAL.
In this study, we sought to find the influence of chitosan nanofiber scaffold on the production and infectivity of PERV expressed by porcine hepatocytes. PERV RNA, RT activity and PERV gag protein was detected for analysis of PERV production by porcine hepatocytes. At present, RT-PCR was the most specific and sensitive method for PERV detection[12]. And it had been identified that the RT activity was related with the retrovirus particles[34], so the result of low RT activity implied a small number of secreted PERV particles. In addition, in previous reports[18,35], sucrose gradient-purified PERV from the culture supernatant was used to qualitatively detect the protein gag p30 by Western blotting, but the condensed PERV was not suitable for hemi-quantitative analysis of the protein. Then we considered the supernatant as the samples directly and attempted to do all the test again. Much to our regret, no positive bands were observed. The possible reason may lay on the microamount of the virus. Therefore, the cell lysate was chosen as samples in Western blotting and the expressing level of the protein in cell lysate was detected to reflect the level of virus replication indirectly.
The results of real time PCR, RT activity assay and Western blotting presented a similar changing trend with some minor differences. Positive PERV RNA but no gag protein at Day 6 might result from less sensitivity of Western blotting. From these results, it could be seen that two peaks of PERV expression at 10H and Day 2 were followed by a regular decline in both groups, and no obviously increased expression level with chitosan nanofiber scaffold was found in first 5 d. The first secreting peak might be regarded as the expression of fresh isolated hepatocytes with some contents of the ruptured non-adherent cells. In order to reduce the influence of the ruptured non-adherent cells, 10H was chosen as the first time point for detecting. But the second peak should be the real secreting peak and the low PERV level in Day 1 might be attributed to the short supernatant collecting interval between 10H and Day 1. On the other hand, the level of PERV RNA at Day 6 and the amount of PERV protein at Day 5 in Nano group was significantly more than that in Hep group, implying a prolonged expression time, which might be due to the superior activity maintaining of hepatocytes cultured on chitosan nanofiber scaffold[13].
The most important concern was no doubt the effect of chitosan nanofiber scaffold on the infectivity of PERV secreted by hepatocytes. Nyberg et al[19] had reported PERV released from hepatocytes didn’t infect the HEK293 cells in vitro and its infectivity would not influenced by human fulminant hepatic failure sera. Likewise, our in vitro infection experiments demonstrated no PERV gene sequence and even no porcine specific SsCytB gene existed in the DNA of HEK293 cells, and no positive RT activity could be detected in the supernatant of treated HEK293 cells either, which implied no PERV transmission into the HEK293 cells and no microchimerism. So it could be concluded that there was no obvious infectivity of the PERV secreted by porcine hepatocytes.
In conclusion, porcine hepatocytes could express PERV for 5 d with normal culture condition, but the secreting time might be prolonged to 6 d when cells were cultured with chitosan nanofiber scaffold. Nevertheless, the productive amount and infectivity of the PERV expressed by porcine hepatocytes would not be obviously influenced by chitosan nanofiber scaffold. Therefore, it could be applied in BAL without enhanced risk of the virus infection.
COMMENTS
Background
Bioartificial liver (BAL) carrying porcine hepatocytes which were still a major cell source at present has been proposed as a temporary liver support for patients waiting for liver transplantation or even as a treatment for liver failure. However, the clinical application of BAL was not very optimistic. As a result of that, much progress had been made on cellular function improvement.
Research frontiers
Some scaffold materials were reported to be capable enhancing the in vitro function of hepatocytes. And chitosan nanofiber scaffold fabricated in our previous study had demonstrated superior abilities on cell adhesion and biocompatibility, thereby greatly enhanced the functions of hepatocytes. So it became a potential material which could be applied in BAL and a new bioreactor based on chitosan nanofiber scaffold was developed in our institute.
Innovations and breakthroughs
Although much progress had been made cellular function improvement, there were few researches on the influence of the materials on the expression and infectivity of porcine endogenous retrovirus (PERV) in porcine hepatocytes. So this study was focus on the safety of chitosan nanofiber scaffold, finding probable prolonging of PERV expression within chitosan nanofiber scaffold but no increased productive amount and infectivity.
Applications
The study identified no obvious influence of chitosan nanofiber scaffold on PERV expression and infectivity in porcine hepatocytes, and concluded that it could be applied in BAL without enhanced risk of the virus infection.
Terminology
PERV was a porcine specific C-type retrovirus which was capable infecting human cells in vitro. Chitosan nanofiber scaffold was a scaffold material made with chitosan by nanotechnology, and it was proved to enhance the hepatocyte function in vitro.
Peer review
Generally speaking, the paper presents a laboratory-based study in which a number of highly sophisticated procedures took place, especially in terms of the methods utilized to demonstrate the presence and infectivity of retroviral agent. In this sense, efforts made by the authors deserve particular attention, making this manuscript very interesting.
Footnotes
Supported by The Natural Science Foundation of Jiangsu Province, No. BK2006008; the foundation of Medical Center of Jiangsu Province, No.ZX200605
Peer reviewer: Mehmet Fatih Can, Assistant Professor, Gulhane School of Medicine, Department of Surgery, Etlik 06018, Ankara, Turkey
S- Editor Tian L L- Editor Ma JY E- Editor Zheng XM
References
- 1.Lee WM, Squires RH, Nyberg SL, Doo E, Hoofnagle JH. Acute liver failure: Summary of a workshop. Hepatology. 2008;47:1401–1415. doi: 10.1002/hep.22177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Riordan SM, Williams R. Perspectives on liver failure: past and future. Semin Liver Dis. 2008;28:137–141. doi: 10.1055/s-2008-1073113. [DOI] [PubMed] [Google Scholar]
- 3.Fiegel HC, Kaufmann PM, Bruns H, Kluth D, Horch RE, Vacanti JP, Kneser U. Hepatic tissue engineering: from transplantation to customized cell-based liver directed therapies from the laboratory. J Cell Mol Med. 2008;12:56–66. doi: 10.1111/j.1582-4934.2007.00162.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.McKenzie TJ, Lillegard JB, Nyberg SL. Artificial and bioartificial liver support. Semin Liver Dis. 2008;28:210–217. doi: 10.1055/s-2008-1073120. [DOI] [PubMed] [Google Scholar]
- 5.Gerlach JC, Zeilinger K, Patzer Ii JF. Bioartificial liver systems: why, what, whither? Regen Med. 2008;3:575–595. doi: 10.2217/17460751.3.4.575. [DOI] [PubMed] [Google Scholar]
- 6.Chamuleau RA, Deurholt T, Hoekstra R. Which are the right cells to be used in a bioartificial liver? Metab Brain Dis. 2005;20:327–335. doi: 10.1007/s11011-005-7914-4. [DOI] [PubMed] [Google Scholar]
- 7.Tsiaoussis J, Newsome PN, Nelson LJ, Hayes PC, Plevris JN. Which hepatocyte will it be? Hepatocyte choice for bioartificial liver support systems. Liver Transpl. 2001;7:2–10. doi: 10.1053/jlts.2001.20845. [DOI] [PubMed] [Google Scholar]
- 8.Kobayashi N, Okitsu T, Tanaka N. Cell choice for bioartificial livers. Keio J Med. 2003;52:151–157. doi: 10.2302/kjm.52.151. [DOI] [PubMed] [Google Scholar]
- 9.Hochleitner B, Hengster P, Bucher H, Ladurner R, Schneeberger S, Krismer A, Kleinsasser A, Barnas U, Klima G, Margreiter R. Significant survival prolongation in pigs with fulminant hepatic failure treated with a novel microgravity-based bioartificial liver. Artif Organs. 2006;30:906–914. doi: 10.1111/j.1525-1594.2006.00323.x. [DOI] [PubMed] [Google Scholar]
- 10.Hochleitner B, Hengster P, Duo L, Bucher H, Klima G, Margreiter R. A novel bioartificial liver with culture of porcine hepatocyte aggregates under simulated microgravity. Artif Organs. 2005;29:58–66. doi: 10.1111/j.1525-1594.2004.29014.x. [DOI] [PubMed] [Google Scholar]
- 11.Chu XH, Shi XL, Feng ZQ, Gu JY, Xu HY, Zhang Y, Gu ZZ, Ding YT. In vitro evaluation of a multi-layer radial-flow bioreactor based on galactosylated chitosan nanofiber scaffolds. Biomaterials. 2009;30:4533–4538. doi: 10.1016/j.biomaterials.2009.05.020. [DOI] [PubMed] [Google Scholar]
- 12.Wilson CA. Porcine endogenous retroviruses and xenotransplantation. Cell Mol Life Sci. 2008;65:3399–3412. doi: 10.1007/s00018-008-8498-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chu XH, Shi XL, Feng ZQ, Gu ZZ, Ding YT. Chitosan nanofiber scaffold enhances hepatocyte adhesion and function. Biotechnol Lett. 2009;31:347–352. doi: 10.1007/s10529-008-9892-1. [DOI] [PubMed] [Google Scholar]
- 14.Gu J, Shi X, Zhang Y, Ding Y. Heterotypic interactions in the preservation of morphology and functionality of porcine hepatocytes by bone marrow mesenchymal stem cells in vitro. J Cell Physiol. 2009;219:100–108. doi: 10.1002/jcp.21651. [DOI] [PubMed] [Google Scholar]
- 15.Gu J, Shi X, Zhang Y, Chu X, Hang H, Ding Y. Establishment of a three-dimensional co-culture system by porcine hepatocytes and bone marrow mesenchymal stem cells in vitro. Hepatol Res. 2009;39:398–407. doi: 10.1111/j.1872-034X.2008.00472.x. [DOI] [PubMed] [Google Scholar]
- 16.Moscoso I, Hermida-Prieto M, Mañez R, Lopez-Pelaez E, Centeno A, Diaz TM, Domenech N. Lack of cross-species transmission of porcine endogenous retrovirus in pig-to-baboon xenotransplantation with sustained depletion of anti-alphagal antibodies. Transplantation. 2005;79:777–782. doi: 10.1097/01.tp.0000152662.55720.83. [DOI] [PubMed] [Google Scholar]
- 17.van de Kerkhove MP, Germans MR, Deurholt T, Hoekstra R, Joziasse DH, van Wijk AC, van Gulik TM, Chamuleau RA, Roos A. Evidence for Galalpha(1-3)Gal expression on primary porcine hepatocytes: implications for bioartificial liver systems. J Hepatol. 2005;42:541–547. doi: 10.1016/j.jhep.2004.11.041. [DOI] [PubMed] [Google Scholar]
- 18.Czauderna F, Fischer N, Boller K, Kurth R, Tönjes RR. Establishment and characterization of molecular clones of porcine endogenous retroviruses replicating on human cells. J Virol. 2000;74:4028–4038. doi: 10.1128/jvi.74.9.4028-4038.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nyberg SL, Hibbs JR, Hardin JA, Germer JJ, Platt JL, Paya CV, Wiesner RH. Influence of human fulminant hepatic failure sera on endogenous retroviral expression in pig hepatocytes. Liver Transpl. 2000;6:76–84. doi: 10.1002/lt.500060105. [DOI] [PubMed] [Google Scholar]
- 20.Kuddus R, Patzer JF, Lopez R, Mazariegos GV, Meighen B, Kramer DJ, Rao AS. Clinical and laboratory evaluation of the safety of a bioartificial liver assist device for potential transmission of porcine endogenous retrovirus. Transplantation. 2002;73:420–429. doi: 10.1097/00007890-200202150-00017. [DOI] [PubMed] [Google Scholar]
- 21.Armstrong JA, Porterfield JS, De Madrid AT. C-type virus particles in pig kidney cell lines. J Gen Virol. 1971;10:195–198. doi: 10.1099/0022-1317-10-2-195. [DOI] [PubMed] [Google Scholar]
- 22.Patience C, Takeuchi Y, Weiss RA. Infection of human cells by an endogenous retrovirus of pigs. Nat Med. 1997;3:282–286. doi: 10.1038/nm0397-282. [DOI] [PubMed] [Google Scholar]
- 23.Takeuchi Y, Patience C, Magre S, Weiss RA, Banerjee PT, Le Tissier P, Stoye JP. Host range and interference studies of three classes of pig endogenous retrovirus. J Virol. 1998;72:9986–9991. doi: 10.1128/jvi.72.12.9986-9991.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wilson CA, Wong S, VanBrocklin M, Federspiel MJ. Extended analysis of the in vitro tropism of porcine endogenous retrovirus. J Virol. 2000;74:49–56. doi: 10.1128/jvi.74.1.49-56.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Specke V, Rubant S, Denner J. Productive infection of human primary cells and cell lines with porcine endogenous retroviruses. Virology. 2001;285:177–180. doi: 10.1006/viro.2001.0934. [DOI] [PubMed] [Google Scholar]
- 26.Wilson CA, Wong S, Muller J, Davidson CE, Rose TM, Burd P. Type C retrovirus released from porcine primary peripheral blood mononuclear cells infects human cells. J Virol. 1998;72:3082–3087. doi: 10.1128/jvi.72.4.3082-3087.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Martin U, Winkler ME, Id M, Radeke H, Arseniev L, Takeuchi Y, Simon AR, Patience C, Haverich A, Steinhoff G. Productive infection of primary human endothelial cells by pig endogenous retrovirus (PERV) Xenotransplantation. 2000;7:138–142. doi: 10.1034/j.1399-3089.2000.00052.x. [DOI] [PubMed] [Google Scholar]
- 28.Martin U, Kiessig V, Blusch JH, Haverich A, von der Helm K, Herden T, Steinhoff G. Expression of pig endogenous retrovirus by primary porcine endothelial cells and infection of human cells. Lancet. 1998;352:692–694. doi: 10.1016/S0140-6736(98)07144-X. [DOI] [PubMed] [Google Scholar]
- 29.Liu Q, Liu Z, Dalakas E. Prevalence of porcine endogenous retrovirus in Chinese pig breeds and in patients treated with a porcine liver cell-based bioreactor. World J Gastroenterol. 2005;11:4727–4730. doi: 10.3748/wjg.v11.i30.4727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang HH, Wang YJ, Liu HL, Liu J, Huang YP, Guo HT, Wang YM. Detection of PERV by polymerase chain reaction and its safety in bioartificial liver support system. World J Gastroenterol. 2006;12:1287–1291. doi: 10.3748/wjg.v12.i8.1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pitkin Z, Mullon C. Evidence of absence of porcine endogenous retrovirus (PERV) infection in patients treated with a bioartificial liver support system. Artif Organs. 1999;23:829–833. doi: 10.1046/j.1525-1594.1999.06444.x. [DOI] [PubMed] [Google Scholar]
- 32.Di Nicuolo G, van de Kerkhove MP, Hoekstra R, Beld MG, Amoroso P, Battisti S, Starace M, di Florio E, Scuderi V, Scala S, et al. No evidence of in vitro and in vivo porcine endogenous retrovirus infection after plasmapheresis through the AMC-bioartificial liver. Xenotransplantation. 2005;12:286–292. doi: 10.1111/j.1399-3089.2005.00226.x. [DOI] [PubMed] [Google Scholar]
- 33.Frühauf JH, Mertsching H, Giri S, Frühauf NR, Bader A. Porcine endogenous retrovirus released by a bioartificial liver infects primary human cells. Liver Int. 2009;29:1553–1561. doi: 10.1111/j.1478-3231.2009.02087.x. [DOI] [PubMed] [Google Scholar]
- 34.Pyra H, Böni J, Schüpbach J. Ultrasensitive retrovirus detection by a reverse transcriptase assay based on product enhancement. Proc Natl Acad Sci USA. 1994;91:1544–1548. doi: 10.1073/pnas.91.4.1544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Galbraith DN, Kelly HT, Dyke A, Reid G, Haworth C, Beekman J, Shepherd A, Smith KT. Design and validation of immunological tests for the detection of Porcine endogenous retrovirus in biological materials. J Virol Methods. 2000;90:115–124. doi: 10.1016/s0166-0934(00)00200-7. [DOI] [PubMed] [Google Scholar]