Abstract
Neuregulin-1 (NRG1) variations have been shown to modulate schizophrenia candidate endophenotypes related to brain structure and function. The aim of this study was to determine the effect of NRG1 on several oculomotor schizophrenia endophenotypes. The effects of 5 core single-nucleotide polymorphisms (SNPs) within the NRG1 gene to oculomotor parameters in a battery of oculomotor tasks (saccade, antisaccade, smooth eye pursuit, fixation) were investigated in a sample of 2243 young male military conscripts. Additive regression models, bootstrap and permutation techniques, were used as well as structural equation modeling and haplotype analysis. A deficit in global smooth eye pursuit performance measured using the root-mean-square error (RMSE) was related to the risk allele of SNP8NRG243177, and a deficit in global smooth eye pursuit performance measured using the saccade frequency was related with the risk allele of SNP8NRG433E1006. Structural equation modeling confirmed a global effect of NRG1 genotype on smooth eye pursuit performance using the RMSE, while the effect on saccade frequency was not confirmed. Haplotype analysis further confirmed the prediction from the structural equation modeling that a combination of alleles corresponding to the Icelandic high-risk haplotype was related to a deficit in global pursuit performance. NRG1 genotype variations were related to smooth eye pursuit variations both at the SNP level and at the haplotype level adding to the validation of this gene as a candidate gene for the disorder.
Keywords: schizophrenia candidate genes, oculomotor, saccade, antisaccade, NRG1, endophenotypes
Introduction
Several candidate genes have been investigated for association with schizophrenia. Neuregulin-1 (NRG1), a gene located in chromosome 8p12–21, is one of the most promising among them and subject of considerable research since the original report of Stefansson et al1 linking NRG1 to schizophrenia. Neuregulin is a protein that has a crucial role in functions including neuronal growth, myelination, migration, and synaptic plasticity.2,3 After the initial report, a number of studies have been published on the relationship between the NRG1 gene and schizophrenia, and their results are not consistent. Some studies using both linkage and association confirmed Stefansson's original finding,4–8while others failed to replicate the original finding.9–11
The reason for this inconsistency is thought to be the polygenic and multifactorial etiology as well as the clinical heterogeneity of the disorder. In order to deal with these problems, contemporary research attention is directed toward the use of endophenotypes, described by some researchers as the “key to unlocking schizophrenia.”12,13 Schizophrenia endophenotypes as initially defined by Gottesman and Gould14 are quantitative, heritable, trait-related, laboratory-assessed deficits that are identified in patients as well as their unaffected relatives. The idea behind the use of endophenotypes is that specific deficits are associated with specific protein changes that are due to discrete genetic abnormalities.15
A wide range of schizophrenia endophenotypes have been evaluated, and their relationship with various candidate genes has been assessed, including structural and functional brain abnormalities as well as sensory processing and neurophysiological and neuropsychological measures. Although recent studies have shown that variations in NRG1 gene are linked to some of these endophenotypes,16–20 no previous reports have associated this gene to oculomotor dysfunction endophenotypes that are among the most extensively studied candidate endophenotypes for schizophrenia.21
In the present study, we examined the possible association of 5 NRG1 gene single-nucleotide polymorphisms (SNPs) that were identified in the core schizophrenia-related haplotype originally reported by Stefansson et al1 with oculomotor endophenotypes in a large sample of young Greek army conscripts (assessed in the Athens Study for Psychosis Proneness and Incidence of Schizophrenia, ASPIS). In previous studies, we have reported on the relation of catechol-O-methyltransferase gene variations to cognitive and psychological endophenotypes from this sample.22–25 In a recent study, we have also reported on the effects of other schizophrenia candidate genes including NRG1 on schizotypy and cognitive endophenotypes in the ASPIS sample.18 Our aim in this study was to investigate if NRG1 gene variations were related to variations in oculomotor parameters thus validating the role of NRG1 as a schizophrenia candidate gene.
Participants and Methods
Participants
This study used the oculomotor dataset from the ASPIS sample described in our previous studies.26–31 The ASPIS sample consists of 2243 young male conscript subjects aged 18–24 years who were recruited from the Greek Air Force in the years 1998–2000. These individuals performed a battery of eye movement tasks (smooth eye pursuit, saccade, antisaccade, visual fixation) and cognitive tasks,25 while they also completed questionnaires for a detailed psychometric analysis.32 DNA has been extracted from mouthwash provided by each individual.22 All data from the ASPIS sample have been codified using a unique number code for each subject, and the link to personal identification of these subjects has been destroyed.
Eye Movement Measurement Apparatus and Task Procedures
A detailed description of the apparatus for eye movement measurements was presented in previous reports.22,29 Eye movements were recorded from the right eye only using the IRIS SCALAR infrared device. A 12-bit A/D converter was used for data acquisition (Advantech PC-Lab Card 818L). Eye movement data were sampled at 600 Hz and stored in the PC hard disk for off-line data processing.
The subjects performed the following tasks: smooth eye pursuit,31 saccade,26 antisaccade,26,29 and 3 fixation tasks (with no distracting stimuli, with distracting stimuli, and fixation with no visual target30). The detailed task procedures are described in the Supplementary methods.
Eye Movement Parameters
The details for measuring the eye movement parameters for each task are also described in the Supplementary methods. For the smooth eye pursuit task, the root-mean-square error (RMSE) between the eye position and the target position in the closed loop pursuit task at each target speed (10°/s, 20°/s, and 30°/s) was measured for each subject. The RMSE is a global measure of pursuit accuracy.33 The gain was also measured for each subject at each target speed that is the ratio of eye velocity to target velocity. The gain is considered a specific measure of the integrity of the pursuit system.33 Finally, the frequency of saccades during pursuit was also measured for each subject at each target speed. All types of saccades occurring during smooth eye pursuit were pooled together for this measure that is also considered a global measure of pursuit accuracy.33 In an effort to reduce the number of oculomotor variables that would be tested for their relation to the genotype variables in the original exploratory analysis using the allele load regression model, we extracted a single factor for each one of the 3 pursuit measures (RMSE, gain, saccade frequency). Factor analysis is the standard statistical analysis to extract the common variance between different variables, and the rational for its use was that if pursuit performance was indeed related to genetic variability between subjects this effect should be more pronounced by excluding other sources of variability such as the intrasubject variability related to the different measures at different speeds. The specific effects of target speed were then assessed if the original analysis using the factor score resulted in a significant effect. A principal component analysis (performed using the STATISTICA 7.0 software) was used to extract a single factor combining the 3 RMSE variables (one for each target speed). The factor explained 70.7% of the common variance for the 3 variables. The same analysis was performed for the 3 gain variables (one for each target speed), and the factor explained 72.1% of the common variance. Finally, a factor combining the 3 saccade frequency variables of smooth eye pursuit explained 46% of the common variance. The 3 factor scores were used in the analysis instead of the individual scores for each speed.
For the saccade and antisaccade tasks, the median latency of saccades and antisaccades and the corresponding coefficient of variation of this latency were also measured, for each subject, as described in the Supplementary methods. For the saccade task, the amplitude gain was also measured. The percentage of antisaccade errors, the median latency and its coefficient of variation for error prosaccades, and the latency and its coefficient of variation for correction antisaccades that followed an error were also measured in the antisaccade task (Supplementary methods, see also Smyrnis33 for a detailed description of these parameters).
For each of the 3 fixation tasks, the saccade frequency was also measured for each subject at each 1 of the 3 fixation tasks (see Supplementary methods). These 3 variables were combined in one factor explaining 70.7% of their common variance. This factor was used in the analysis of the relation of fixation to the SNPs instead of the saccade frequency separately for each fixation task.
Table 1 presents all the oculomotor variables that were measured in the ASPIS sample. We used the data from the meta-analysis of Calkins et al34 to categorize these variables. The variables defined as positive were those for which there was evidence for their heritability in schizophrenia in the above-mentioned meta-analysis. The variables defined as negative were those for which there was evidence that they are not heritable in schizophrenia in the same meta-analysis. All other variables for which there is little evidence for their candidacy and they were not included in the meta-analysis were defined as exploratory.
Table 1.
Oculomotor Variables and Their Categorization
| Oculomotor Variable | N | Mean (SD) | Category |
| Smooth eye pursuit | |||
| RMSE 10°/s | 1765 | 149 (94) | — |
| RMSE 20°/s | 1765 | 196 (109) | — |
| RMSE 30°/s | 1765 | 241 (125) | — |
| RMSE factor | 1765 | — | Positive |
| Gain 10°/s | 1994 | 0.91 (0.16) | — |
| Gain 20°/s | 1994 | 0.86 (0.19) | — |
| Gain 30°/s | 1994 | 0.72 (0.24) | — |
| Gain factor | 1994 | — | Positive |
| Saccade frequency 10°/s | 1765 | 1.72 (1.29) | — |
| Saccade frequency 20°/s | 1765 | 2.39 (1.43) | — |
| Saccade frequency 30°/s | 1765 | 3.27 (1.77) | — |
| Saccade frequency factor | 1765 | — | Positive |
| Saccade | |||
| Median latency | 1078 | 177 (21) | Negative |
| CV latency | 1078 | 0.23 (0.08) | Exploratory |
| Amplitude gain | 1078 | 0.97 (0.21) | Negative |
| Antisaccade | |||
| Error rate | 2006 | 23.9% (17.6%) | Positive |
| Median latency correct | 2004 | 262 (39) | Positive |
| CV latency correct | 2004 | 0.25 (0.09) | Exploratory |
| Median latency errors | 1994 | 201 (38) | Negative |
| CV latency errors | 1994 | 0.27 (0.17) | Exploratory |
| Median latency correction | 1988 | 131 (56) | Exploratory |
| CV latency correction | 1988 | 1.03 (4.03) | Exploratory |
| Fixation | |||
| Saccade frequency undistracted | 1728 | 0.30 (0.35) | — |
| Saccade frequency distracted | 1810 | 0.39 (0.35) | — |
| Saccade frequency no target | 1831 | 0.52 (0.38) | — |
| Saccade frequency factor | 1515 | — | Positive |
Note: RMSE, root-mean-square error; CV, coefficient of variation. RMSE measurements are in arbitrary units of AD converter measuring eye movement and target movement signals; all saccade frequency measurements are in saccade number per second (Hz); all median latencies and CV latencies are measured in milliseconds.
DNA Extraction and Genotyping
Mouthwash samples for DNA extraction were chosen as described previously22 to obtain a better procedure acceptance rate. The following 5 SNPs were genotyped for NRG1: SNP8NRG221132, SNP8NRG221533, SNP8NRG241930, SNP8NRG243177, and SNP8NRG433E1006 as described in our previous work.18
All genotyping was performed blind to phenotype measures.18 Genotyping for NRG1 was performed by K-Biosciences (Herts, UK; http://www.kbioscience.co.uk/) using a competitive allele-specific polymerase chain reaction system.
The frequencies in the ASPIS sample of each one of the 5 SNPs that were genotyped were not significantly deviant from Hardy-Weinberg equilibrium18 (and Supplementary table 1).
Data Analysis
The major problem analyzing this dataset was the issue of multiple comparisons in the interpretation of both positive and negative results. In order to address this issue, we proceeded in the analysis in a step fashion. In the first step, an exploratory analysis was used to identify significant relations between each of the oculomotor variables and each of the SNPs. The exploratory analysis used the allele load regression model. According to the classification of the oculomotor variables into positive, negative, and exploratory, a valid outcome of this exploratory analysis should be to identify significant relations only for positive and/or exploratory variables but not for negative variables. The next step was to confirm the positive results of the exploratory analysis using 2 nonparametric procedures, a bootstrap and a permutation test. The next step involved another approach using a global analysis of the data with structural equation modeling. This analysis bypassed the multiple comparison issue. Finally, a haplotype analysis was performed to further validate the result of the structural equation modeling.
Allele Load Model.
The effect of each one of the 5 NRG1 SNPs on each of the primary oculomotor variables was investigated using an allele load regression model. The reason for selecting this model was based on the sensitivity of this model at detecting significant effects of NRG1 SNPs and phenotypic variables as demonstrated in our previous study.18 In this model, a simple linear regression was performed for each pair of SNP and primary oculomotor variable (table 1). The SNP was the independent predictor variable, and the oculomotor variable was the dependent variable. According to the allele load model, the SNP variable could take 1 of 3 values: 0 if the rare allele was absent, 1 if 1 copy of the rare allele was present, and 2 if 2 copies of the rare allele were present for the particular individual. Thus, the regression model was the following:
| (1) |
where b was the coefficient of the allele load effect, and its significance was tested by a t test with significance level set at 0.05.
Bootstrap and Permutation.
All significant effects in the exploratory allele load regression analysis were confirmed using 2 nonparametric procedures, a bootstrap and a permutation test.
Let us suppose that the allele load regression analysis showed a significant t test for the b of one particular SNP and primary oculomotor variable. In the bootstrap procedure, a new sample with the same number of subjects was drawn at random with replacement from the group of individuals having 0 copies of the rare allele. The same procedure was used to draw a new sample of individuals having 1 copy of the rare allele and again for individuals having 2 copies. This new bootstrap sample was then used to run an allele load regression of the form shown in formula 1, and the regression b coefficient was computed. The bootstrap procedure was repeated for 1.000.000 times resulting in a distribution of bootstrap b values. The percentile where b = 0 was computed on this distribution. Let us suppose then that the allele load regression b for the original data as was computed in model (1) was positive, and this percentile was lower than the fifth percentile of the bootstrap distribution of b. This would mean that the process of randomly resampling the original populations of individuals with 0, 1, and 2 copies of the rare allele of the SNP would result by chance in a relation with a regression b that would be 0 or lower than 0 with a P value below .05. This is equivalent to a t test testing the null hypothesis that the original regression b value is significantly larger from 0 at the 0.05 level. In the case where the original allele load regression b was negative, the 95th percentile was used as the cutoff in the distribution of bootstrap b.
In the permutation procedure, a new sample of the same number of subjects was drawn by randomly shuffling the data and assigning a new value of the oculomotor variable to each subject. The result of this process was that a random value of the oculomotor variable was assigned to each original allele load value. Then, the same regression model (1) as was used for the original data was computed for the permutated data, and the process was repeated 1.000.000 times to result in a permutation distribution of b values. The percentile corresponding to the original regression b was computed on this distribution. Let us suppose then that the allele load regression b for the original data was positive, and this percentile was higher than the 95th percentile. This would mean that the process of randomly shuffling the original data would result by chance to a regression with b equal to or higher than the original regression b with a P value below .05. This is again equivalent to a t test testing the null hypothesis that the original regression b value is significantly larger than 0 at the 0.05 level. In the case where the original regression b would be negative, the fifth percentile of the permutation b distribution was used as the cutoff.
Structural Equation Modeling.
Structural equation modeling was used to model the effects of the NRG1 SNPs on the smooth eye pursuit phenotype variables. This analysis provides a tool for studying simultaneously the effect of multiple independent variables on multiple dependent variables using a set of assumptions for how the 2 sets of variables are connected. Thus, this analysis bypasses the problem of multiple testing and the consequent problem of adjusting significance levels. In order to define the structural equation model, one has to identify the “exogenous” variables that are the independent factors. The exogenous variables in turn affect the “endogenous” variables that are the dependent variables. We chose as exogenous variables the SNP allele loads for all 5 NRG1 SNPs. These affected the endogenous variables that were the oculomotor variables. The model also allows the specification of “latent” variables that reflect a conceptual reduction of measured variables, called “manifest” variables. Thus, for the exogenous manifest SNP variables, this reduction led to the identification of a single latent genotype variable reflecting the variations in all 5 NRG1 SNPs. The same reduction led to the identification of 3 endogenous latent oculomotor variables for smooth eye pursuit namely the RMSE, gain, and saccade frequency. Each endogenous latent variable reflected the variation of the corresponding three manifest variables, namely, the measurements at the 3 pursuit speeds. The details for model application and the comparison of different models are presented in the Supplementary methods and Smyrnis et al.25
Haplotype Analysis.
The phase 2.1 software35,36 was used to derive haplotype estimates for each subject in the ASPIS population. A set of the most common haplotypes was defined in the population as those haplotypes that exceeded the 5% frequency. Supplementary table 2 presents the frequencies for the most common haplotypes in the ASPIS sample. Then, for each subject, we used the best pair subset of haplotypes given as an output in phase 2.1 to derive an estimate of haplotype load (0, no copies of the particular haplotype; 1, 1 copy; and 2: 2 copies) for each one of the most common haplotypes. The effects of a particular haplotype load on an oculomotor variable were then tested using the following linear regression model:
| (2) |
where b was the coefficient of the specific haplotype load effect, and its significance was tested by a t test with significance level at 0.05.
Results
Allele Load Model and Bootstrap-Permutation Tests
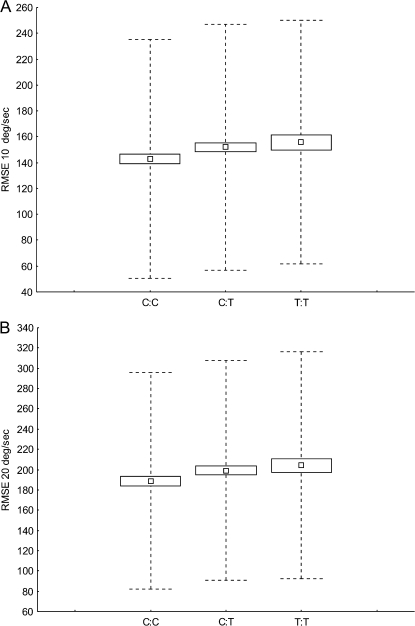
The allele load regression analysis of the 5 SNPs on positive, negative, and exploratory oculomotor variables resulted in 2 significant effects (Supplementary table 3 presents the resulting P values for all regressions performed). The first was of SNP8NRG243177on the smooth eye pursuit RMSE factor score (β = −.06, F1,1500 = 5.14, P = .023). It should be noted that the RMSE factor score was negatively correlated with the score for each individual pursuit speed; thus, a negative β in the regression indicates that the rare allele T of SNP8NRG243177 is related to an increase in RMSE. The effect was confirmed using both the bootstrap test (P = .01) and the permutation test (P = .012). Further analysis for each target speed separately showed that the effect was significant at the low target speed of 10°/s (β = .053, F1,1500 = 4.15, P = .041) and at the medium target speed of 20°/s (β = .054, F1,1500 = 4.35, P = .037) but not at the high target speed of 30°/s(β = .041, F1,1500 = 2.48, P = .11) (figure 1).
Fig. 1.
Plots Presenting the Modulation of the Mean Root-Mean-Square Error (RMSE) (Small Boxes) ± SEM (Large Boxes) and SDs (Dotted Error Bars) of Smooth Eye Pursuit With the Allele Variations of SNP8NRG243177. The upper plot presents the modulation of RMSE at 10°/s pursuit speed, and the lower plot presents the modulation of RMSE at 20°/s pursuit speed.
The second significant effect was of SNP8NRG433E1006 on the smooth eye pursuit saccade frequency (β = −.05, F1,1500 = 4.6, P = .032). It should be noted that the saccade frequency factor score was negatively correlated with the score for each individual speed; thus, a negative beta in the regression indicates that the rare allele A of SNP8NRG433E1006 was related to an increase in saccade frequency. The effect was significant using both the bootstrap test (P = .016) and the permutation test (P = .017). Further analysis for each target speed separately showed that the effect was significant at the low target speed of 10°/s (β = .05, F1,1500 = 3.94, P = .047) and at the medium target speed of 20°/s(β = .055, F1,1500 = 4.78, P = .028) but not at the high target speed of 30°/s(β = .001, F1,1500 = 0.002, P = .96).
Structural Equation Modeling
In this analysis, we formulated a hypothesis of how genetic variability was related to phenotype variability. Using the a priori information that only smooth eye pursuit phenotypes seemed to have a relation to NRG1 genotype variation, we tested 3 models. In all models, the exogenous latent variable was the NRG1 genotype structure based on the measurement of the 5 SNPs. In the first general model, all smooth eye pursuit performance variables (RMSE for each target speed, saccade frequency for each target speed and gain for each target speed, total of 9 variables) were grouped together in one latent pursuit performance variable. In the second analytic model, 3 latent variables were formed: RMSE, saccade frequency, and gain. Each latent variable loaded on the corresponding 3 variables for each target speed. Finally, in the third specific model, 2 of the 3 latent variables of the previous model were used based on the positive findings of the SNP analysis, namely, the RMSE and the saccade frequency.
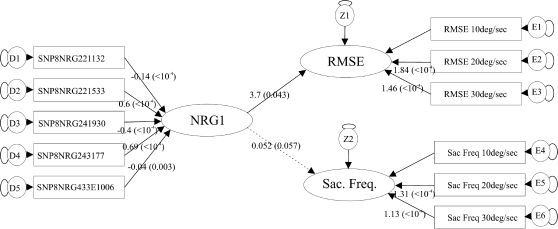
The structural equation model estimation process converged normally for all 3 models that were estimated. The comparison of the 3 models (see Supplementary methods and Supplementary table 4) showed that the specific model was the one that provided the best fit to the data in accordance with the prediction from the SNP analysis. Figure 2 presents a diagram of the resulting specific model parameters and their level of significance. It can be seen that all SNP exogenous variables loaded significantly on the NRG1 genotype latent variable. Also, the exogenous RMSE and saccade frequency variables loaded significantly on their corresponding latent variables. Finally, the model showed that the relation of the NRG1 latent variable to the RMSE latent variable was significant, while the relation of the NRG1 to the saccade frequency latent variable did not reach significance. An even more specific relation of each SNP with the RMSE in smooth eye pursuit can be defined by examining the sign of the corresponding parameter values. We defined the allele load for each SNP using the number of rare alleles (see “Participants and Methods”). Thus, a negative parameter value in the model for a particular SNP indicates that an increase in the RMSE is related to an increase in the number of copies of the common allele. The opposite is true for a positive parameter value, namely, that an increase in the RMSE is related to an increase in the number of copies of the rare allele. Looking then at figure 2, one can conclude that an increase in RMSE of smooth eye pursuit, indicating worse pursuit performance, is related to the common allele G of SNP8NRG221132, the rare allele C of SNP8NRG221533, the common allele G of SNP8NRG241930, the rare allele T of SNP8NRG243177, and finally the common allele G of SNP8NRG433E1006. A combination of these alleles corresponds to the haplotype GCGTG.
Fig. 2.
Path Diagram of the Structural Equation Model Describing the Relation of Neuregulin-1 (NRG1) and Smooth Eye Pursuit Global Performance Variables. The exogenous manifest variables (squares) were the rare allele loads of the additive model for each SNP. These variables loaded (arrows) on one exogenous latent variable (ellipse) named “NRG1.” The endogenous manifest variables (squares), namely, the smooth pursuit root-mean-square error (RMSE) and saccade frequency (sac freq) variables for the 3 pursuit speeds (10°/s, 20°/s, and 30°/s) also loaded (arrows) on 2 latent endogenous variables (ellipses) named “RMSE” and “sac freq.” Finally, the exogenous latent variable “NRG1” loaded (arrows) on the 2 endogenous latent variables “RMSE” and “sac freq.” The model also included residual variables for all exogenous manifest variables (depicted as δ 1–5), residual variables for all endogenous manifest variables (depicted as ϵ 1–7,) and disturbances for all endogenous latent variables (depicted as ζ 1 and 2) that all loaded on their respective variable (small elliptic lines). Solid arrows indicate significant factor estimates, while dotted arrows indicate nonsignificant factor estimates at the level of P = .05. The factor loading estimates and their respective P values (in parentheses) are depicted with a number on the line.
Haplotype Analysis
Haplotype analysis was performed using all 5 SNPs (see “Participants and Methods”). The GCGTC was the most frequent haplotype in the ASPIS sample (Supplementary table 2; Stefanis et al18). The effect of this haplotype on the factor score of the RMSE of pursuit was significant (β = −.05, F1,1857 = 5.26, P = .022), while the effect on the factor of pursuit saccade frequency was not (β = −.002, F1,1655 = 0.08, P = .92). Because the factor score is negatively correlated to individual RMSE scores, this analysis confirmed the prediction from the structural equation modeling, namely, that increasing GCGTC haplotype load would be related to increasing RMSE corresponding to decreasing global pursuit performance while it would have no significant effect on saccade frequency in pursuit.
Discussion
The ASPIS sample provided a collection of oculomotor variables measured in different tasks in a population of apparently healthy young Greek army conscripts who were also genotyped. Although limited by age and gender, this sample could be thought of as representative of the Greek population as far as genetic variation is concerned.22 In this study, we used the ASPIS oculomotor dataset to examine their relation to SNP variations in NRG1.
A major methodological issue one faces in this analysis is that of multiple comparisons among many different SNPs and many different phenotypic variables leading to the identification of false-positive results. We addressed this issue both in terms of variable selection methods and in terms of multiple steps in the analysis.
The selection of phenotypes was based on the categorization of all oculomotor variables. We used a recent meta-analysis that measured the heritability of several eye movement parameters in different eye movement tasks34 to define a set of 6 positive parameters (3 for the smooth eye pursuit task, 2 for the antisaccade task, and 1 for the 3 fixation tasks). Similarly, we defined a set of 3 negative parameters (2 for the saccade task and 1 for the antisaccade task) that were found to have no heritability in schizophrenia. Finally, we defined a third set of exploratory parameters for which no data on heritability were available in the same meta-analysis. The rational for this categorization was that a meaningful relation of NRG1 genotype variations to schizophrenia-related eye movement endophenotypes should be observed for the positive variables and should be absent for the negative variables. A relation observed for the exploratory variables would also not be considered as a positive finding but only as tentative relation bearing further exploration.
The analysis proceeded in multiple steps. The first step was an exploratory analysis using the allele load model. Two SNPs were related to 2 positive variables measuring smooth eye pursuit, while all other relations were not significant as would be expected by our original categorization of the oculomotor phenotypic variables. The first positive relation was of the T load of SNP8NRG243177 to an increase in pursuit RMSE suggesting a deficient global pursuit performance. This relation is particularly interesting considering the relevant literature both on the particular SNP and on the particular phenotype.
SNP8NRG243177 was shown to be a functional polymorphism and the high risk T allele was related to higher levels of type IV messenger RNA expression.37 It was also shown that the rare T allele of this particular SNP was correlated to decreased activation of frontal and temporal regions increased incidence of psychotic symptoms and decreased premorbid IQ in a group of young individuals at high risk for developing schizophrenia.17 In our previous study,18 we showed that increased T load of SNP8NRG243177 was associated with a decrease in performance (measured using d′) in a spatial working memory task (n-back). In yet another study, the rare allele T of this same SNP was associated with decreased white matter density and connectivity in the anterior limb of the internal capsule in healthy subjects.38 Finally, Mata et al16 observed that the rare allele T of SNP8NRG243177 was related to an increase in the lateral ventricular volume in patients with schizophrenia experiencing their first psychotic episode.
Let us now consider the RMSE phenotype. Since the pioneer work of Diefendorf and Dodge39 reporting abnormalities of the gaze movement patterns of the mentally ill, a deficit in global pursuit performance is the most consistently replicated deficit in oculomotor function in schizophrenia.40,41 In the meta-analysis on heritability of different oculomotor variables, global pursuit performance was one of the most significant variables showing a heritability effect size of 0.5.34 Among pursuit indexes of performance, global indexes such as RMSE have the most powerful effect at dissociating patients with schizophrenia and their relatives from normal control subjects.41
The second effect that we observed was a positive relation of G load of the SNP8NRG433E1006 SNP to increased saccade frequency in the smooth eye pursuit task. In our previous study,18 we observed a significant effect of this SNP on verbal working memory performance (verbal n-back) as well as on sustained attention performance (Continuous Performance Test, Identical Pairs version). A problem with this SNP was the fact that G homozygotes are extremely rare (only 4 subjects in our ASPIS sample). Concerning the particular phenotype, the pursuit saccade frequency is considered a global index of pursuit performance that effectively dissociates patients with schizophrenia from normal control subjects.41 This index, though, showed a rather weak heritability in the meta-analysis of Calkins et al34 with an effect size of 0.1. Furthermore, this variable exhibits large intersubject variability that may be due to the inclusion of different types of saccadic eye movements in its definition and measurement.33
The next step in the analysis was to confirm the results of the exploratory allele load model regression analysis. Because the β’s of the original regressions were very small (explaining less than 1% of the common variance), the possibility is raised that these are false-positive effects that would disappear using a more stringent significance criterion. We used 2 complementary nonparametric procedures to test the hypothesis that these small effects are in fact random. The bootstrap test explores how probable it would be to observe a smaller β from that observed in the original analysis by randomly resampling the original samples. The permutation test explores the complementary hypothesis, namely, how probable it would be to observe the same or larger β by randomly shuffling the original sample. The results showed that the original β’s would be observed by resampling and would not be observed by randomly shuffling the original sample with a P approaching .99 confirming the significance of the relations.
In summary, then this analysis confirmed a positive association of 2 SNPs within the core NRG1 haplotype that were related to variations in smooth eye pursuit global performance. Although the a priori categorization of the phenotypes and the use of bootstrap and permutation to validate the significant effects were all efforts in the direction of reducing the risk of random effects, the possibility remains that these significant effects were indeed random due to the multiple comparisons performed. Also, this multiple comparison analysis might possibly miss a global relation between the set of genotype and phenotype variables.
The next step in the analysis was structural equation modeling.25 The 5 core SNPs of the NRG1were used to construct a latent genotype variable that in turn was tested against latent phenotype variables that were constructed from the oculomotor variables. Different constructs of the pursuit performance variables were tested, and the model that best described the data was the one that included 2 phenotype pursuit latent variables, one combining the 3 RMSE variables and one combining the 3 saccade frequency variables. The model confirmed a significant effect of the latent genotype variable on RMSE and a nonsignificant effect on saccade frequency. The specific loadings of the 5 SNPs on the latent genotype variable suggested that a combination of common and rare alleles that was identical to those described in the core Icelandic haplotype by Stefansson et al1 resulted in larger RMSE or equivalently to a deficit in global pursuit performance in the ASPIS sample. It should be noted here that the original haplotype also included 2 microsatelites that were not included in this study. In order to confirm this prediction, we performed a haplotype analysis that showed that the particular Icelandic haplotype load in the ASPIS sample was related to a significant increase in RMSE, while it had no effect on pursuit saccade frequency. We believe that these results are in favor of the use of structural equation modeling as a powerful tool in association studies of SNPs to endophenotypes that can confirm a robust relation among the 2 sets of variables without the drawbacks of multiple comparisons that can detect nonexisting relations or miss important relations on a global scale.
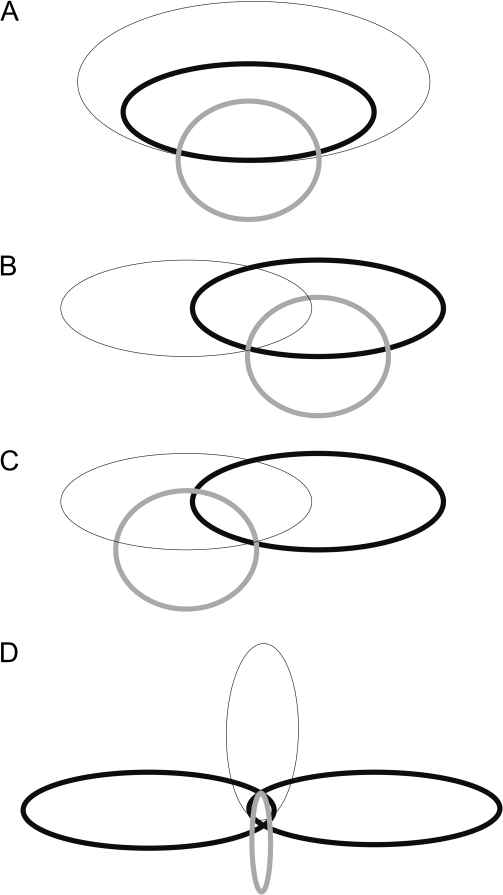
This study used a large sample of apparently healthy young men to test for a relation between a schizophrenia candidate gene and specific schizophrenia endophenotypes. The original definition of an endophenotype would predict that the relation of the endophenotype and the candidate gene should be stronger than that of the clinical syndrome and the gene. This prediction is illustrated graphically in figure 3A. The population having the clinical syndrome is depicted with the black hairline ellipse. Within this population is the population of patients who have the endophenotype deficit (thick black line ellipse). Finally, the population of individuals who have the specific allele variation is depicted with the thick gray line circle. The relation depicted in this figure suggests that the effect of the genotype is intensified in the subpopulation of patients with a deficit of the endophenotype variable. The results of this study as well as the results of our previous study on the relation of cognitive endophenotypes and NRG118 are not in accordance with this prediction. Although we observed significant relations, their magnitude is very small in the order of magnitude that is observed for the relation of the gene to the clinical syndrome or even smaller. One potential problem with figure 3A is the relation of the endophenotype to the clinical syndrome. Not only it is known that not all patients have a smooth eye pursuit deficit but it is also known that this deficit is not encountered only in schizophrenia. Thus, in a large population of apparently healthy individuals, there could be individuals with a deficit in smooth eye pursuit who are not related to the susceptibility to schizophrenia. The relation then could be modified as shown in figures 3B or 3C. In both figures, it is assumed that the endophenotype is partly related to the clinical syndrome. In figure 3B, the genotype is specifically related to the endophenotype, and through this relation the relation to the clinical syndrome is mediated. This relation though results to the same prediction as the relation in figure 3A, namely, a larger effect of the genotype to the endophenotype than to the clinical syndrome. On the other hand, the relation depicted in figure 3C predicts that the genotype has multiple effects that are related to the clinical syndrome one of which is also partly related to the particular endophenotype. Thus, the relation of the genotype to the particular endophenotype would actually be smaller than the relation to the clinical syndrome. Furthermore, it could be hypothesized that the particular genotype has multiple effects in more than one domain of cognitive function thus affecting multiple endophenotypes as depicted in figure 3D. The different endophenotypes would be related to the genotype and the clinical syndrome. We tested this prediction using the data from this and our previous study.18 The particular prediction here was that the working memory endophenotype deficit and the smooth eye pursuit endophenotype deficit that were related to NRG1 and also related to schizophrenia would be different endophenotypes that would not correlate strongly between them. Table 2 demonstrates that the correlation coefficients for the correlations between working memory and smooth eye pursuit endophenotypes are very small. Most important, the sign of these correlations is opposite to what would be expected showing that better performance in working memory tasks (larger d′ scores) was correlated to worse performance in smooth eye pursuit (lower gain, higher RMSE, and higher saccade frequency). Thus, the relation of deficit in smooth eye pursuit and the deficit in working memory function to the NRG1 variations is not due to a general covariation of these 2 measures in the population. Indeed, it can be said that these results could be viewed as predicting a very specific relation of these 2 phenotypes with the NRG1 phenotype and schizophrenia susceptibility as depicted in figure 3D. The prediction from figure 3D would be that a high-risk group combining deficits in independent endophenotypes might show a strong relation to the NRG1 genotype variations and schizophrenia susceptibility favoring the hypothesis that this gene affects many independent functional systems in the brain leading to schizophrenia susceptibility.
Fig. 3.
Schematic Representation of Different Types of the Relation Between Clinical Phenotype (Black Hairline) and Endophenotypes (Black Thick Lines) as Well as Genotype (Gray Thick Line) (See “Discussion”).
Table 2.
Pearson Correlation Coefficients for the Relation of Cognitive to Smooth Eye Pursuit Phenotypes
| CPT d′ | V-n-back d′ | S-n-back d′ | |
| RMSE factor | 0.06 | 0.13 | 0.05 |
| Gain factor | −0.03 | −0.06 | −0.02 |
| Saccade frequency factor | 0.02 | 0.07 | 0.04 |
Note: CPT, continuous performance test; RMSE, root-mean-square error.
In conclusion then, this study showed that NRG1 genotype variations were related to smooth eye pursuit variations both at the SNP level and at the haplotype level providing validation that this gene is a candidate gene for the disorder.
Funding
General Secretariat of Research and Technology of the Greek Ministry of Development (EKBAN 97 to N.C.S.)
Supplementary Material
Supplementary methods and tables 1–4 are available at http://schizophreniabulletin.oxfordjournals.org.
References
- 1.Stefansson H, Sigurdsson E, Steinthorsdottir V, et al. Neuregulin 1 and susceptibility to schizophrenia. Am J Hum Genet. 2002;71:877–892. doi: 10.1086/342734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Falls DL. Neuregulins: function, forms and signalling strategies. Exp Cell Res. 2003;284:14–30. doi: 10.1016/s0014-4827(02)00102-7. [DOI] [PubMed] [Google Scholar]
- 3.Birchmeier C. ErbB receptors and the development of the nervous system. Exp Cell Res. 2009;315:611–618. doi: 10.1016/j.yexcr.2008.10.035. [DOI] [PubMed] [Google Scholar]
- 4.Stefansson H, Thorgeirsson TE, Gulcher JR, Stefansson K. Neuregulin1 in schizophrenia: out of Iceland. Mol Psychiatry. 2003;8:639–640. doi: 10.1038/sj.mp.4001384. [DOI] [PubMed] [Google Scholar]
- 5.Petryshen TL, Middletton FA, Kirby A, et al. Support for involvement of neuregulin 1 in schizophrenia pathophysiology. Mol Psychiatry. 2005;10:328, 366–374. doi: 10.1038/sj.mp.4001608. [DOI] [PubMed] [Google Scholar]
- 6.Thompson PA, Christoforou A, Morris SW, et al. Association of neuregulin 1 with schizophrenia and bipolar disorder in a second cohort from the scottish population. Mol Psychiatry. 2007;12:94–104. doi: 10.1038/sj.mp.4001889. [DOI] [PubMed] [Google Scholar]
- 7.Munafo MR, Thiselton DL, Clark TG, Flint J. Association of the NRG1 gene and schizophrenia: a meta-analysis. Mol Psychiatry. 2006;11:539–546. doi: 10.1038/sj.mp.4001817. [DOI] [PubMed] [Google Scholar]
- 8.Munafo MR, Attwood AS, Flint J. Neuregulin 1 genotype and schizophrenia. Schizophr Bull. 2008;34:9–12. doi: 10.1093/schbul/sbm129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Thiselton DL, Webb BT, Neale BM, et al. No evidence for linkage or association of neuregulin 1 (NRG1) with disease in the Irish study of high density schizophrenia families (ISHDSF) Mol Psychiatry. 2004;9:777–783. doi: 10.1038/sj.mp.4001530. [DOI] [PubMed] [Google Scholar]
- 10.Iwata N, Suzuki T, Ikeda M, et al. No association with the neuregulin 1 haplotype to Japanese schizophrenia. Mol Psychiatry. 2004;9:126–127. doi: 10.1038/sj.mp.4001456. [DOI] [PubMed] [Google Scholar]
- 11.Sanders AR, Duan J, Levinson DF, et al. No significant association of 14 candidate genes with schizophrenia in a large European ancestry sample: implications of psychiatric genetics. Am J Psychiatry. 2008;165:497–506. doi: 10.1176/appi.ajp.2007.07101573. [DOI] [PubMed] [Google Scholar]
- 12.Holden C. Neuroscience: deconstructing schizophrenia. Science. 2003;299:333–335. doi: 10.1126/science.299.5605.333. [DOI] [PubMed] [Google Scholar]
- 13.Calkins ME, Dobie DJ, Cadenhead KS, et al. The consortium on the genetics of endophenotypes in schizophrenia (COGS): model recruitment, assessment and endophenotyping methods for a multi-site collaboration. Schizophr Bull. 2007;33:33–48. doi: 10.1093/schbul/sbl044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gottesman II, Gould TD. The endophenotype concept in psychiatry: etymology and strategic intentions. Am J Psychiatry. 2003;160:636–645. doi: 10.1176/appi.ajp.160.4.636. [DOI] [PubMed] [Google Scholar]
- 15.Braff DL, Freedman R, Schork NJ, Gottesman II. Deconstructing schizophrenia: an overview of the use of endophenotypes in order to understand a complex disorder. Schizophr Bull. 2007;33:21–32. doi: 10.1093/schbul/sbl049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mata I, Perez-Iglesias R, Roiz-Santiañez R, et al. A neuregulin 1 variant is associated with increased lateral ventricle volume in patients with first-episode schizophrenia. Biol Psychiatry. 2009;65:535–540. doi: 10.1016/j.biopsych.2008.10.020. [DOI] [PubMed] [Google Scholar]
- 17.Hall J, Whalley HC, Job DE, et al. A neuregulin 1 variant associated with abnormal cortical function and psychotic symptoms. Nat Neurosci. 2006;9:1477–1478. doi: 10.1038/nn1795. [DOI] [PubMed] [Google Scholar]
- 18.Stefanis NC, Trikalinos TA, Avramopoulos D, et al. Impact of schizophrenia candidate genes on schizotypy and cognitive endophenotypes at the population level. Biol Psychiatry. 2007;62:784–792. doi: 10.1016/j.biopsych.2006.11.015. [DOI] [PubMed] [Google Scholar]
- 19.Bramon E, Dempster E, Frangou S, et al. Neuregulin-1 and the P300 waveform—a preliminary association study using a psychosis endophenotype. Schizophr Res. 2008;103:178–185. doi: 10.1016/j.schres.2008.03.025. [DOI] [PubMed] [Google Scholar]
- 20.Hong LE, Wonodi I, Stine OC, Mitchell BD, Thaker GK. Evidence of missense mutations on the neuregulin 1 gene affecting function of prepulse inhibition. Biol Psychiatry. 2008;63:17–23. doi: 10.1016/j.biopsych.2007.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Calkins ME, Iacono WG. Eye movement dysfunction in schizophrenia: a heritable characteristic for enhancing phenotype definition. Am J Med Genet. 2000;97:72–76. doi: 10.1002/(sici)1096-8628(200021)97:1<72::aid-ajmg10>3.0.co;2-l. [DOI] [PubMed] [Google Scholar]
- 22.Avramopoulos D, Stefanis NC, Hantoumi I, Smyrnis N, Evdokimidis I, Stefanis CN. Higher scores of self reported schizotypy in healthy young males carrying the COMT high activity allele. Mol Psychiatry. 2002;7:706–711. doi: 10.1038/sj.mp.4001070. [DOI] [PubMed] [Google Scholar]
- 23.Stefanis NC, Van Os J, Avramopoulos D, et al. Variation in catechol-o-methyltransferase val158 met genotype associated with schizotypy but not cognition: a population study in 543 young men. Biol Psychiatry. 2004;56:510–515. doi: 10.1016/j.biopsych.2004.06.038. [DOI] [PubMed] [Google Scholar]
- 24.Stefanis NC, van Os J, Avramopoulos D, Smyrnis N, Evdokimidis I, Stefanis CN. Effect of COMT Val158Met polymorphism on the Continuous Performance Test, Identical Pairs Version: tuning rather than improving performance. Am J Psychiatry. 2005;162:1752–1754. doi: 10.1176/appi.ajp.162.9.1752. [DOI] [PubMed] [Google Scholar]
- 25.Smyrnis N, Avramopoulos D, Evdokimidis I, Stefanis CN, Tsekou H, Stefanis NC. Effect of schizotypy on cognitive performance and its tuning by COMT val158 met genotype variations in a large population of young men. Biol Psychiatry. 2007;61:845–853. doi: 10.1016/j.biopsych.2006.07.019. [DOI] [PubMed] [Google Scholar]
- 26.Evdokimidis I, Smyrnis N, Constantinidis TS, et al. The antisaccade task in a sample of 2,006 young men. I. Normal population characteristics. Exp Brain Res. 2002;147:45–52. doi: 10.1007/s00221-002-1208-4. [DOI] [PubMed] [Google Scholar]
- 27.Constantinidis TS, Smyrnis N, Evdokimidis I, et al. Effects of direction on saccadic performance in relation to lateral preferences. Exp Brain Res. 2003;150:443–448. doi: 10.1007/s00221-003-1454-0. [DOI] [PubMed] [Google Scholar]
- 28.Smyrnis N, Evdokimidis I, Stefanis NC, Constantinidis TS, Avramopoulos D, Theleritis C. The antisaccade task in a sample of 2,006 young males. II. Effects of task parameters. Exp Brain Res. 2002;147:53–63. doi: 10.1007/s00221-002-1207-5. [DOI] [PubMed] [Google Scholar]
- 29.Smyrnis N, Evdokimidis I, Stefanis NC, et al. Antisaccade performance of 1,273 men: effects of schizotypy, anxiety, and depression. J Abnorm Psychol. 2003;112:403–414. doi: 10.1037/0021-843x.112.3.403. [DOI] [PubMed] [Google Scholar]
- 30.Smyrnis N, Kattoulas E, Evdokimidis I, et al. Active eye fixation performance in 940 young men: effects of IQ, schizotypy, anxiety and depression. Exp Brain Res. 2004;156:1–10. doi: 10.1007/s00221-003-1759-z. [DOI] [PubMed] [Google Scholar]
- 31.Smyrnis N, Evdokimidis I, Mantas A, et al. Smooth pursuit eye movements in 1,087 men: effects of schizotypy, anxiety, and depression. Exp Brain Res. 2007;179:397–408. doi: 10.1007/s00221-006-0797-8. [DOI] [PubMed] [Google Scholar]
- 32.Stefanis NC, Smyrnis N, Avramopoulos D, Evdokimidis I, Ntzoufras I, Stefanis CN. Factorial composition of self-rated schizotypal traits among young males undergoing military training. Schizophr Bull. 2004;30:335–350. doi: 10.1093/oxfordjournals.schbul.a007083. [DOI] [PubMed] [Google Scholar]
- 33.Smyrnis N. Metric issues in the study of eye movements in psychiatry. Brain Cogn. 2008;68:341–358. doi: 10.1016/j.bandc.2008.08.022. [DOI] [PubMed] [Google Scholar]
- 34.Calkins ME, Iacono WG, Ones DS. Eye movement dysfunction in first-degree relatives of patients with schizophrenia: a meta-analytic evaluation of candidate endophenotypes. Brain Cogn. 2008;68:436–461. doi: 10.1016/j.bandc.2008.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stephens M, Smith NJ, Donnelly P. A new statistical method for haplotype reconstruction from population data. Am J Hum Genet. 2001;68:978–989. doi: 10.1086/319501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stephens M, Scheet P. Accounting for decay of linkage disequilibrium in haplotype inference and missing-data imputation. Am J Hum Genet. 2005;76:449–462. doi: 10.1086/428594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Law AJ, Lipska BK, Weickert CS, et al. Neuregulin 1 transcripts are differentially expressed in schizophrenia and regulated by 5’ SNPs associated with the disease. Proc Natl Acad Sci U S A. 2006;103:6747–6752. doi: 10.1073/pnas.0602002103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.McIntosh AM, Moorhead TW, Job D, et al. The effects of a neuregulin 1 variant on white matter density and integrity. Mol Psychiatry. 2008;13:1054–1059. doi: 10.1038/sj.mp.4002103. [DOI] [PubMed] [Google Scholar]
- 39.Diefendorf AR, Dodge R. An experimental study of the ocular reactions of the insane from photographic records. Brain. 1908;31:451–489. [Google Scholar]
- 40.Levy DL, Holzman PS, Matthysse S, Mendell NR. Eye tracking dysfunction and schizophrenia: a critical perspective. Schizophr Bull. 1993;19:461–536. doi: 10.1093/schbul/19.3.461. [DOI] [PubMed] [Google Scholar]
- 41.O'Driscoll GA, Brandy LC. Smooth pursuit in schizophrenia: a meta-analytic review of research since 1993. Brain Cogn. 2008;68:359–370. doi: 10.1016/j.bandc.2008.08.023. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.