Abstract
Mitochondrial complex I is the largest multimeric enzyme of the respiratory chain. The lack of a model system with facile genetics has limited the molecular dissection of complex I assembly. Using Chlamydomonas reinhardtii as an experimental system to screen for complex I defects, we isolated, via forward genetics, amc1–7 nuclear mutants (for assembly of mitochondrial complex I) displaying reduced or no complex I activity. Blue native (BN)-PAGE and immunoblot analyses revealed that amc3 and amc4 accumulate reduced levels of the complex I holoenzyme (950 kDa) while all other amc mutants fail to accumulate a mature complex. In amc1, -2, -5–7, the detection of a 700 kDa subcomplex retaining NADH dehydrogenase activity indicates an arrest in the assembly process. Genetic analyses established that amc5 and amc7 are alleles of the same locus while amc1–4 and amc6 define distinct complementation groups. The locus defined by the amc5 and amc7 alleles corresponds to the NUOB10 gene, encoding PDSW, a subunit of the membrane arm of complex I. This is the first report of a forward genetic screen yielding the isolation of complex I mutants. This work illustrates the potential of using Chlamydomonas as a genetically tractable organism to decipher complex I manufacture.
MULTIMERIC respiratory complexes I, III, and IV in the mitochondrial inner membrane generate the proton motive force that is critical for ATP production. Complex I, the largest respiratory complex, is a NADH-ubiquinone oxidoreductase with a hydrophilic peripheral arm protruding into the mitochondrial matrix and a membrane arm (Sazanov and Hinchliffe 2006; Efremov et al. 2010; Hunte et al. 2010). It is known that the assembly of multimeric enzymes is assisted by factors absent from the mature enzyme but nevertheless essential in promoting its assembly into an active form. In humans, complex I dysfunction is the cause of severe diseases (Distelmaier et al. 2009). Since only 40% of the nuclear mutations in complex I-linked diseases occur in structural subunits of complex I, it is generally accepted that the other 60% represent defects in factors recruited to assemble or regulate the complex (Loeffen et al. 2000; Thorburn 2004).
In recent years, the use of model organisms has contributed to the identification of factors involved in the assembly of respiratory complexes. Most of these studies have been carried out in the model organism Saccharomyces cerevisiae, which has a broad spectrum of genetic and molecular tools facilitating research (Barrientos 2003). However, given that S. cerevisiae and its related species have lost complex I subunits (Gray et al. 2001), they cannot be used as a model for the study of complex I assembly. Instead, the lack of complex I in S. cerevisiae and other eukaryotes has been instrumental in revealing candidate complex I assembly factors through subtractive phylogenetic analyses. In conjunction with mitochondrial proteomic data from complex I-bearing organisms, these analyses were recently used to identify a number of candidate assembly factors (Pagliarini et al. 2008). Three of these factors, C8ORF38, C20ORF7, and FOXRED1, were found to be mutated in complex I-deficient patients (Pagliarini et al. 2008; Sugiana et al. 2008; Fassone et al. 2010). A limitation of subtractive phylogenetics is that assembly factors are assumed to have been systematically lost from organisms lacking complex I, hence excluding the possibility of finding conserved genes that have acquired dual or novel function. Therefore, there is still a need for a genetic approach to discover loci controlling complex I assembly on the basis of loss-of-function phenotypes.
Fungi, animals, and vascular plants have been used extensively as experimental systems for the study of complex I defects (Remacle et al. 2008). Although such models were invaluable in documenting the impacts of specific mutations in complex I function, they were not developed for the isolation of complex I mutants through forward genetics.
Chlamydomonas reinhardtii, a unicellular green alga, is a very promising experimental system to address the question of complex I assembly (Remacle et al. 2008). It is an ideal organism to study mitochondrial biogenesis because complex I-less mutants are viable due to the operation of alternative NADH dehydrogenases (Remacle et al. 2001a). Moreover, complex I-deficient mutants can be maintained in phototrophic (light) or mixotrophic (light + carbon source) conditions because they are not defective in photosynthesis (Cardol et al. 2003). This provides an advantage to screen for complex I mutants, as they display robust growth under mixotrophic/phototrophic conditions but slow growth under heterotrophic conditions (dark + carbon source) (Remacle et al. 2001a; Cardol et al. 2002). In contrast, complex III or complex IV mutants display arrested growth in the dark (dark dier or dk). That a complex I mutant retains two respiratory complexes (III and IV) out of the three is believed to account for the “slow” growth phenotype. Indeed, in a complex I mutant, complexes III and IV still contribute to the formation of the proton electrochemical potential, and thus to ATP formation. However, in a complex III or complex IV mutant, the contribution of complex I to the proton gradient is assumed to be insufficient to sustain growth in the dark (Wikström 1984; Galkin et al. 2006). Several mutations in complex I subunits encoded by the mitochondrial genome and one nuclear mutation (amc2, assembly of mitochondrial complex I, not yet characterized molecularly) have been described in Chlamydomonas (Remacle et al. 2001a). Here, we used a mutagenesis approach to search for additional amc mutants of nuclear origin. Six amc mutants, defining five loci, were isolated. The amc mutants are affected to different extents in complex I activity and most of them result in the accumulation of subcomplexes, an indication that the assembly process of the membrane arm is compromised. In amc5 and amc7, the identification of molecular lesions in the NUOB10 gene encoding PDSW, a subunit of the membrane arm, validates our mutant screen strategy. This illustrates the potential of Chlamydomonas as an experimental model system to dissect the molecular basis of complex I assembly.
MATERIALS AND METHODS
Strains and culture conditions:
Strains were grown at 22–25° in Tris-acetate-phosphate (TAP), arginine-supplemented TAP liquid, or solid medium (Harris 1989) under continuous light (50 μE·m2·sec−1) as described in Howe and Merchant (1992). Wild-type (wt) strains 3A+ and 4C− were used for transformation (J. D. Rochaix, University of Geneva, Switzerland) and strains 137c+ and 137c− were used for backcrosses. Strain dn26 169 (mt−) and a mt+ derivative, renamed here amc2, were used for crosses. Genetic analyses were performed as in Harris (1989).
Insertional mutagenesis and identification of the amc mutants:
3A+ (mt+ arg7-8) or 4C− (mt− arg7-8) cells were electroporated with 100 ng of either a hygromycin B (iHyg) or paromomycin (iPm) resistance cassette as in Shimogawara et al. (1998). The iHyg and iPm cassettes were amplified by PCR from the pHyg3 (Berthold et al. 2002) or pSL18 (Depège et al. 2003) plasmids, respectively. Oligonucleotides Aph7-F (5′-TCGATATCAAGCTTCTTTCTTGC-3′) and Aph7-R (5′-AAGCTTCCATGGGATGACG-3′) were used for amplification of the iHyg cassette. Aph8-F2 (5′-TCAGGCAGACGGGCAGGTG-3′) and Aph8-R (5′-TCAGGCAGACGGGCAGGTG-3′) were used to amplify the iPm cassette. Transformants were selected on TAP + arginine solid media with 25 μg/ml of hygromycin B or paromomycin in the light. After a 7- to 10-day incubation under light, transformants were transferred individually to 96-well plates and grown in selective liquid media. Individual colonies were replica plated on solid TAP + arginine medium and incubated in the dark and in the light for 7 days before scoring.
Identification of flanking sequences by thermal asymmetric interlaced-PCR:
Amplification of insertion-linked sequence by thermal asymmetric interlaced (TAIL)-PCR was as described in Dent et al. (2005). Four different degenerate primers, AD1 and AD2 (Liu et al. 1995), RMD227 and RMD228 (Dent et al. 2005), were tested. iPm-specific primers were PmR1 (5′-GACCCCGCAGTGCACGCAAC-3′), PmR2 (5′-GCTTGAGACAGCGACAGAGGAGCC-3′), and PmR3 (5′-GCAAGTCAAATCTGCAAGCACG-3′) for the primary, secondary, and tertiary reactions, respectively. The following NUOB10-specific primers were used: PDSW-1F (5′-CACCTGGTGCACATTGCTGTA-3′), PDSW-2R (5′-TTCATGCTTGCCCGAGAAG-3′), PDSW-5F (5′-GACCAAGGGCTTTCTGACTG-3′), PDSW-7F (5′-AGCCTCAGGCATATTGCGC-3′), PDSW-6R (5′-ATCGCACATGACGGCAG-3′), PDSW-8R (5′-GTAGATGAAGCCCACGTCG-3′), PDSW-9F (5′-GTCATGAAGCGCCTGCAGG-3′), PDSW-10R (5′-GGAACTGGGTAAGGTTCTAAGC-3′), PDSW-11F (5′-CACTGCCTGAAAGCCTGCC-3′), and PDSW-12R (5′-GAGGAGAGCAGTGGCATCGTG-3′).
Protein extraction, SDS–PAGE, and immunoblot analyses:
Total protein was isolated from 2-day-old light-grown plates using the freeze/thaw method (Howe and Merchant 1992). SDS–PAGE was performed using standard protocols (Sambrook et al. 1989).
Measurement of respiratory enzyme activities:
Respiratory activities were measured as described previously (Remacle et al. 2001a, 2004; Cardol et al. 2002) using partially purified membranes extracted from 2- to 3-day-old cultures.
Antibody production:
Protein A purified polyclonal antibodies directed against complex I subunit-specific peptides were custom synthesized in rabbits by GeneScript (Piscataway, NJ) using the following peptide antigens: NUO7 (49 kDa), CGIDWDLRKTQPYDA; NUO6 (51 kDa), SLEGKQGKPRLKPPC; NUO8 (TYKY), YASDWENDPTFKRTC. To assess antibody specificity, protein extracts were competed with the peptides used to generate the polyclonal antibodies (Howe and Merchant 1992). For NUO7 (49 kDa), only the upper band detected with the antiserum is specific.
BN-PAGE and in-gel activity:
Separation of protein complexes was done by BN-PAGE (Schägger and Von Jagow 1991). Detection of in-gel NADH dehydrogenase activity was performed as in Rasmusson and Møller (1991).
RESULTS
Identification of complex I-deficient (amc) mutants via insertional mutagenesis:
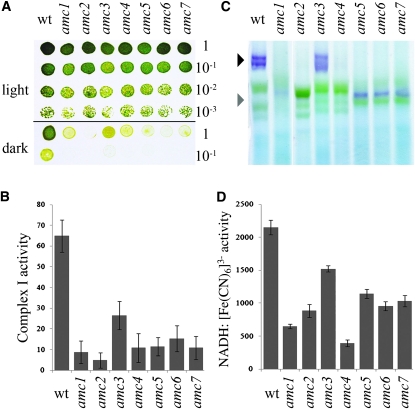
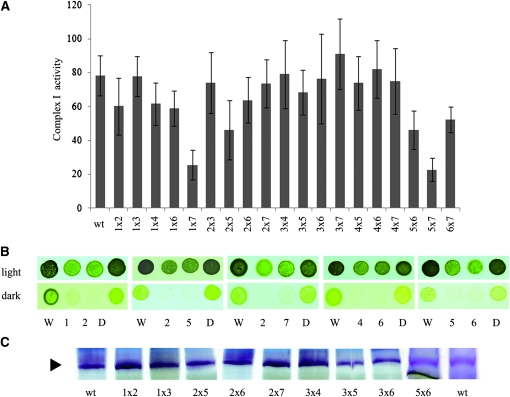
Only one nuclear mutation (amc2) resulting in a complex I defect has been previously described in Chlamydomonas (Remacle et al. 2001a; Cardol et al. 2002, 2008). To uncover additional AMC loci, we undertook a screen for nuclear mutants, resulting in complex I dysfunction. Out of ∼50,000 insertional transformants, 22 showed slow growth in the dark (sid for slow growth in the dark; Figure 1A), a feature of complex I mutants. A defect in complex I activity was verified for six sid mutants and we named them amc1 and amc3–7 (Figure 1B). The other sid mutants were either not significantly or only marginally reduced in complex I activity (P-value > 0.05) (not shown). In-gel staining for NADH dehydrogenase activity was performed for all amc mutants (Figure 1C). Note that wild-type complex I is often resolved in more than one purple band of high molecular weight, depending on solubilization conditions. The purple bands, observable at ∼950 kDa, correspond to the holoenzyme. These were only present in amc3 and amc4. In the case of amc3, while a wild-type–size complex I was detectable, the staining was reduced (Figure 1C and supporting information, Figure S1B). We also noted that very low levels of complex I accumulated in amc4 (Figure S1B and Figure S2A). This accumulation appeared to be dependent upon extraction, an indication that complex I is not stable in the presence of the amc4 mutation. All other amc mutants accumulated a 700-kDa subcomplex that stained for NADH dehydrogenase activity (Figure 1C and Figure S1, A and B). A similar 700-kDa subcomplex is also detected in Chlamydomonas nd4 or nd5 mitochondrial mutants (Remacle et al. 2006; Cardol et al. 2008) (Figure S1B). The subcomplex in amc2 is labile, as it was not always detected in independent extractions (Figure S1A).
Figure 1.—
Six novel sid mutants display a defect in complex I activity. (A) Tenfold dilution series of amc1–7 and wild-type (wt) cells were plated on acetate-containing medium and incubated 3 days in the light or 5 days in the dark. (B) Complex I-specific activity (nmol NADH reduced/min/mg of protein) is indicated as averages of 10 independent measurements. (C) BN-PAGE and in-gel NBT staining of NADH dehydrogenase activity. The black and gray arrowheads indicate the position of the mature 950-kDa complex I and the 700-kDa subcomplex, respectively. The purple bands indicate staining for NADH oxidase activity in complex I and subcomplexes. Note that complex I is often resolved into two or more purple bands of high molecular weight. The identity of such bands is unclear but they do not correspond to the supercomplex I + III2 (Cardol et al. 2008). The green bands correspond to photosystems I and II and their respective light harvesting complexes of the chloroplast (Rexroth et al. 2003; Cardol et al. 2006). (D) NADH:ferricyanide oxidoreductase activity (nmol K3[Fe (CN)6] reduced/min/mg of protein). The results represent the average of three independent determinations. (B and D) Error bars indicate standard error (SE).
To further evaluate the complex I NADH oxidoreductase function in the mutants, total NADH:ferricyanide oxidoreductase activity was determined. Total NADH:ferricyanide oxidoreductase activity reflects not only the activity of complex I but also that of other cellular NADH dehydrogenases (Quiles et al. 1996). However, because most of the NADH:ferricyanide oxidoreductase activity in Chlamydomonas is due to the NADH dehydrogenase activity of the peripheral arm of complex I, the level of this activity is reduced in complex I mutants. A strong reduction was observed in all mutants except amc3 (Figure 1D). Overall, the levels of reduction correlated with reduced in-gel staining for NADH dehydrogenase activity in the amc mutants (Figure 1, C and D). All amc mutants showed decreased whole-cell respiration when compared to wild type (Table 1). In accord with the complex I defect, the respiration in the amc mutants was rotenone insensitive.
TABLE 1.
Complex I-dependent oxygen consumption of amc and wild-type strains
| Respiration (μmol O2 h−1 mg−1 of chlorophyll) |
||
| Strain | −rotenone | +rotenone |
| wt | 61.0 ± 2.0 | 42.0 ± 4.0 |
| amc1 | 44.2 ± 3.2 | 39.8 ± 5.7 |
| amc2 | 42.7 ± 0.8 | 41.0 ± 5.4 |
| amc3 | 44.6 ± 5.0 | 40.1 ± 6.8 |
| amc4 | 43.9 ± 5.3 | 43.7 ± 7.8 |
| amc5 | 37.0 ± 4.2 | 33.4 ± 5.9 |
| amc6 | 36.2 ± 4.6 | 34.4 ± 3.1 |
Wild-type and amc strains representative of the six different AMC loci were grown mixotrophically (TAP + arginine, light). Oxygen consumption was measured in the dark, with or without the addition of 100 μM rotenone to specifically inhibit complex I. Values represent the average of three independent measurements and ± indicates standard error.
The amc mutants display complex I assembly defects:
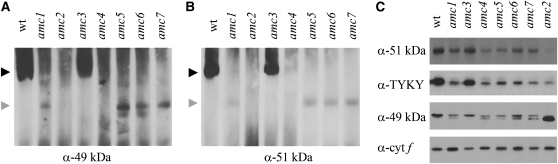
To verify that the 700-kDa subcomplex observed in amc1, -2, and -5–7 was a bona fide subcomplex of complex I, we carried out immunoblot analyses using antibodies against two subunits of the hydrophilic arm (49 and 51 kDa). As expected for an assembly intermediate of complex I, the 700-kDa subcomplex was detected by the two complex I-specific antibodies (Figure 2, A and B). In amc2, the putative subcomplex was very rarely detected (not shown). On the basis of the fact that the 700-kDa band was also rarely detected by in-gel staining, we reasoned that this subcomplex is highly unstable in amc2.
Figure 2.—
The amc mutants are deficient in complex I assembly. (A and B) Immunoblot analyses of membrane fractions, separated by BN-PAGE, using anti-49 kDa (A) and anti-51 kDa (B). A total of 150 μg of proteins was loaded per lane. The figures show the most representative blots of three independent experiments. The solid and shaded arrowheads indicate the position of mature complex I and the 700-kDa subcomplex, respectively. Equal loading was tested by Coomassie blue staining (not shown) and the presence of a nonspecific band revealed by the anti–49-kDa antiserum (Figure S2B for an example). (C) Membrane fractions were separated by SDS–PAGE and immunoblot analyses were performed using polyclonal antibodies against subunit-specific peptides. The subunits are 51 kDa, TYKY, and 49 kDa. A total of 10 μg of total protein per lane was loaded. Antibody specificity was evaluated by peptide competition assays (not shown). Only the top band is specific for the 49-kDa subunit. Plastid cytochrome f was used as a loading control. The results are representative from at least three independent membrane extractions.
Using an antiserum against TYKY, another subunit of the hydrophilic arm, we also consistently detected the 700-kDa subcomplex in amc1 and amc5–7 and in some instances in amc2 (not shown). Intermediate assembly subcomplexes of similar size were not observed in the wild type or in the amc3 and amc4 mutants. In addition, low levels of complex I was evidenced in amc4 using the anti–49-kDa antibody in instances where NADH dehydrogenase activity was also detected by in-gel staining (Figure S2, A and B).
Low accumulation of structural subunits has been observed in several cases of complex I deficiency (Saada et al. 2009). We decided to evaluate the steady-state levels of the three complex I subunits (49 kDa, 51 kDa, and TYKY) in membrane extracts of the amc mutants, separated under denaturing conditions. As shown in Figure 2C, the accumulation of TYKY and other subunits was severely reduced in all mutants, with the exception of amc3.
Impact of the amc mutations on respiratory complexes II, III, and IV:
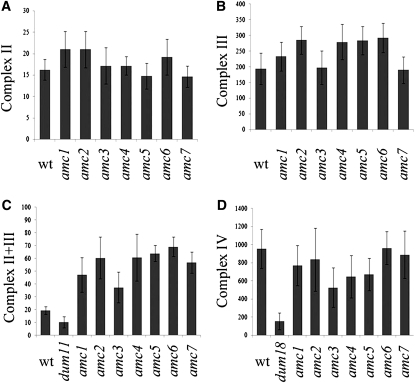
To determine whether the amc mutations resulted in a pleiotropic defect, activities of other respiratory complexes were measured. All amc mutants showed increased levels of complex II + III combined activity, an indication that they compensate for a defect in complex I (Figure 3C). Such a compensatory effect has already been observed in complex I mutants of mitochondrial origin (Remacle et al. 2001a; Cardol et al. 2002). Individually, complex II and complex III activities were not significantly affected (Figure 3, A and B). On average, the augmented complex II + III combined activity correlated with the level of reduction of complex I in all mutants. All amc mutants with the exception of amc3 displayed wild-type levels of complex IV activity (Figure 3D). However, we noted that the complex IV defect seen in amc3, unlike the complex I phenotype, was not inherited in the progeny (not shown). We concluded that all amc mutants resulted in isolated complex I deficiency.
Figure 3.—
Effects of the amc mutations on other respiratory complexes. (A) Complex II activity (succinate:2,6-dichlorophenolindophenol (DCIP) oxidoreductase). Three independent determinations were averaged and activities are expressed as nmol DCIP reduced/min/mg of protein. (B) Complex III activity (decylubiquinol:cytochrome c oxidoreductase). Averages from three independent measurements are shown in nmol of cytochrome c reduced/min/mg of protein. (C) Complexes II + III combined activities (succinate:cytochrome c oxidoreductase) of 10 independent determinations (nmol of cytochrome c reduced/min/mg of protein). dum11 is a complex III mutant (Dorthu et al. 1992). (D) Complex IV activity (cytochrome c oxidase). dum18 is a complex IV mutant (Remacle et al. 2001b). Average activities of 10 independent determinations are expressed in nmol cytochrome c oxidized/min/mg of protein. Error bars indicate SE.
Genetic analyses reveal six AMC loci:
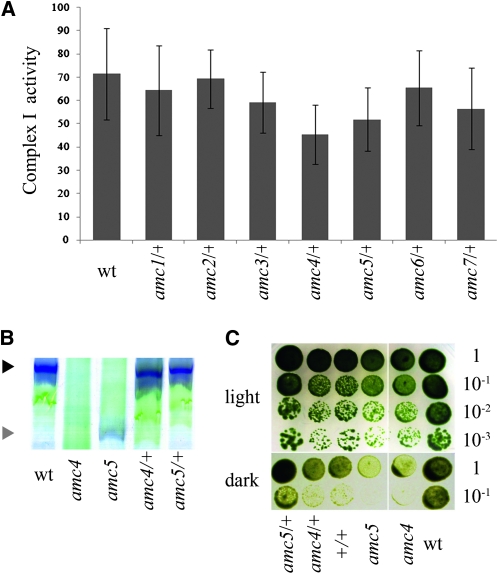
All amc/+ diploid strains displayed restored levels of complex I activity (Figure 4A). In addition, complex I assembly, assessed via BN-PAGE, was also wild type (not shown and Figure 4, B and C). Therefore, we concluded that all amc mutations were recessive. While all the amc mutations were found to be monogenic, the antibiotic resistance did not cosegregate with the sid phenotype for any except amc5 (not shown). We concluded that the AMC5 locus was tagged with the insertional marker. To establish the number of complementation groups defined by the amc mutations, we performed allelism tests. We were unable to obtain diploids from amc1 × amc5 and amc2 × amc4 crosses. The other 19 diploids could be constructed and were further analyzed. Although complex I activity levels did not always reach those of the wild-type strain, we observed restoration of a fully active complex I in most diploids (Figure 5A). This restoration correlated with the recovery of wild-type–like growth in the dark (Figure 5B) and NADH dehydrogenase activity (Figure 5C). Only two combinations of diploids (amc5 × amc7 and amc1 × amc7) showed low levels of complex I activity (Figure 5A).
Figure 4.—
The amc mutations are recessive. Heterozygous (amc/+) and homozygous (+/+) diploid strains were generated by crosses. (A) Complex I activities (nmol NADH oxidized/min/mg of protein). Columns represent the average of 10 independent determinations, with error bars representing SE. The +/+ diploid activity is not shown here and was 92 ± 8 nmol NADH oxidized/min/mg of protein. (B) BN-PAGE and in-gel NBT staining of NADH dehydrogenase activity. The black and gray arrowheads indicate the position of mature complex I and 700-kDa subcomplex, respectively. (C). Light/dark growth comparison of wild type (wt), amc4 and amc5 mutants, and their respective diploids (+/+, amc4/+, and amc5/+). Dilution series were plated on acetate containing medium and incubated 6 days in the light or 12 days in the dark. For B and C, only representative amc/+ diploids are shown. All amc/+ diploids exhibit restoration of the growth in the dark and complex I assembly phenotypes (not shown).
Figure 5.—
Complementation experiments define alleles of AMC loci. (A) Complex I activity of the wild type (wt) and the 19 diploids strains (amc × amc). The average of 10 independent replicas is indicated. The error bars represent standard error. The activity is expressed in nmol NADH oxidized/min/mg of protein. (B) Light/dark growth comparison of wild type (W), amc mutants (1, 2, and 4–7) and respective heterozygous diploids (D). Cells were plated on acetate-containing medium and incubated 3 days in the light or 5 days in the dark. (C) BN-PAGE and in-gel NBT staining of NADH dehydrogenase activity. The black arrowhead indicates the position of mature complex I. For B and C, only representative amc × amc diploids are shown. All diploids exhibiting restoration of the growth in the dark phenotype also showed restoration of complex I assembly as assessed by NBT staining (not shown). Only the top part of the BN gel is shown in C.
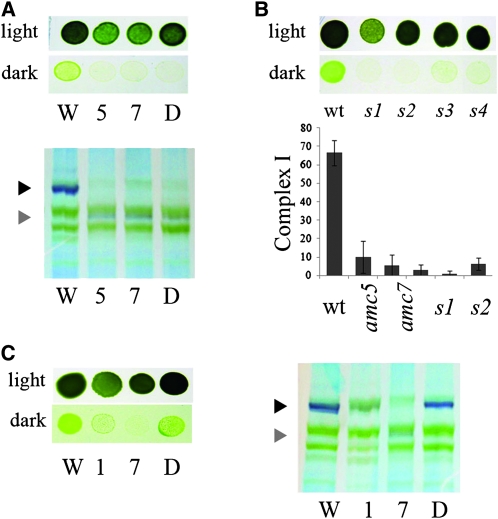
The amc5 × amc7 diploid accumulated the same subcomplex as each single mutant and displayed a sid phenotype (Figure 6A). Moreover, all the progeny (100 meiotic products) from the amc5 to amc7 cross had a sid phenotype and was deficient in complex I activity (Figure 6B). Thus, we concluded that amc5 and amc7 are alleles of the same locus.
Figure 6.—
amc5 and amc7 define alleles of the same AMC locus. (A, top) Light/dark growth comparison of wild type (W), amc5 and amc7, and the amc5/amc7 diploid (D). (A, bottom) Detection of complex I and the 700-kDa subcomplex via NADH dehydrogenase activity on membrane fractions separated by BN-PAGE. (B, top) Light vs. dark growth of wild type (wt) and representative spores (s1–s4) from the amc5 × amc7 cross. Note that the strains used in our study are not isogenic and it is possible that other genetic factors segregate and modulate the slow growth phenotype (compare s1 and s2 to s3 and s4). (B, bottom) Complex I activities (nmol NADH oxidized/min/mg of protein) of wild type (wt), amc5, amc7, and two sid spores (s1 and s2) originating from the amc5 × amc7 cross. Columns represent the average of 10 independent determinations, with SE as the error bars. (C, left) Light vs. dark growth of wild type (W), amc1 and amc7 mutants, and the amc1/amc7 diploid (D). (C, right) Detection of complex I and the 700-kDa subcomplex via NADH dehydrogenase activity on membrane fractions separated by BN-PAGE. (A, B, and C) Cells were plated on acetate-containing medium and incubated 3 days in the light or 5 days in the dark. (A and C) The black and gray arrowheads indicate the position of mature complex I and the 700-kDa subcomplex, respectively.
The amc1 × amc7 diploid displayed a low level of complex I activity (Figure 5A) but was restored for the growth in the dark and complex I assembly when assessed by BN-PAGE (Figure 6C). We concluded that amc1 and amc7 define distinct loci.
We were not able to generate meiotic products of the amc1 × amc5 cross. However, if amc5 and amc7 define the same locus and amc1 and amc7 are alleles of distinct loci, then amc1 and amc5 are nonallelic. Similarly, because we were unable to generate amc2 × amc4 diploids, we analyzed the segregation of the meiotic spores originating from this cross. Several spores from the amc2 × amc4 cross displayed faster growth in the dark (Figure S3, top). Wild-type levels of complex I activity were also confirmed in spores exhibiting a wild-type–like growth phenotype in the dark (Figure S3, bottom). Therefore, we concluded that amc2 and amc4 belong to different complementation groups. In summary, the seven amc mutants define six nuclear loci.
The AMC5/7 locus encodes the NUOB10/PDSW encoding gene:
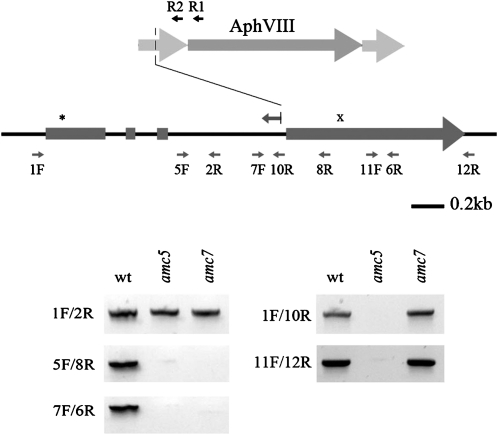
Since the amc5 mutant was tagged with the insertional marker (iPm), we sought to identify the interrupted locus by TAIL-PCR. The iPm was mapped to intron 3 of the NUOB10 gene encoding PDSW (according to the nomenclature of bovine complex I), a noncore subunit of the membrane arm of complex I (Hirst et al. 2003) (Figure 7 and Figure S4). Mitochondrial complex I is composed of core subunits required for the catalytic activity and noncore subunits believed to play a role in the assembly and/or regulation of the activity of the enzyme (Remacle et al. 2008). While core subunits are common to both bacterial and mitochondrial complex I, noncore subunits are only found in the eukaryotic enzymes. On the basis of the failure to amplify the region with specific primers, the insertion of the marker occurred at the 3′ end of intron 3 and was accompanied by a deletion of the entire coding sequence and the 3′-UTR downstream of the integration site (Figure 7). In amc7, PCR analyses revealed that a molecular lesion had also occurred in the NUOB10 gene. (Figure 7). Unfortunately, despite several attempts, we were unable to transform the amc5/amc7 mutants with a NUOB10-containing cosmid (not shown). Almost all the amc mutants were found to be very recalcitrant to transformation, regardless of the transformation method employed.
Figure 7.—
The amc5 and amc7 mutants carry molecular lesions in the NUOB10 gene encoding the noncore subunit PDSW. (Top) Schematic representation of the NUOB10 gene structure showing the four exons (rectangles) and three introns (thin lines). The asterisk denotes the position of the ATG codon while the “x” symbol indicates the position of the stop codon. The insertional cassette (AphVIII) is represented as an arrow (dark shading) flanked by the promoter and terminator regions (light shading). The positions of primers used to diagnose the molecular lesion are indicated by small arrows. The leftward arrow with a vertical line above intron 3 indicates the extent of the NUOB10 sequence obtained by TAIL-PCR (Figure S3). R1 and R2 indicate the positions of the AphVIII-specific primers used in the TAIL-PCR to recover the sequence flanking the cassette. A deletion of the promoter occurred upon integration of the cassette in the NUOB10 locus. (Bottom) Molecular lesions in NUOB10 were analyzed by PCR using NUOB10-specific primers. PCR products were stained by ethidium bromide and imaged using an imaging system (Kodak Image Station 2000R).
The AMC1 locus does not map to a gene encoding a subunit of the membrane arm:
As amc1 also accumulated a 700-kDa subcomplex, we reasoned that the mutation could affect 1 of the 12 nuclear-encoded subunits of complex I present in the membrane arm (Lazarou et al. 2009), the NUOP1 and NUOP4 genes encoding small hydrophobic subunits (Cardol et al. 2004) or the NUOAF1 gene encoding the CIA30 assembly factor whose defect results in the accumulation of a subcomplex (Kuffner et al. 1998; Vogel et al. 2005; Dunning et al. 2007). The sequencing of these loci did not reveal any mutation that could account for the phenotype of amc1 (Table 2). The finding of a wild-type NUOB10 sequence in amc1 further confirms that this mutation is not allelic to amc5 and amc7 mutations. We concluded that the amc1 mutation did not map to a gene encoding a complex I subunit that is part of the membrane arm.
TABLE 2.
Sequencing of complex I structural genes and NUOAF1 in the amc1 mutant
| Sequence obtained |
|||||||
| Chlamydomonas subunit | Accession no. | Subcomplex | H. sapiens | B. taurus | bp from ATG | bp from STOP | Sequencing result |
| ACP1 | AAQ73138 | α, βS | NDUFAB1 | SDAP | −57 | +32 | wt |
| NDUFA11 | AAS58499 | α, λ | NDUFA11 | B14.7 | −99 | +372 | wt |
| NUO17 | AAS48192 | β | NDUFB11 | ESSS | −109 | +130 | wt |
| NUOA8 | AAQ55460 | α, λ | NDUFA8 | PGIV | −8 | +43 | wt |
| NUOB10 | AAQ55459 | βL, βS | NDUFB10 | PDSW | −57 | +611 | wt |
| NUOB12 | AAS48194 | β | NDUFB3 | B12 | −140 | +338 | wt |
| NUOB18 | AAQ73135 | βS | NDUFB7 | B18 | −13 | 0 | wt |
| NUOB22 | AAQ73134 | βS | NDUFB9 | B22 | −3 | +20 | wt |
| NUOP2 | AAS48193 | α, βS | NDUFB4 | B15 | −182 | +69 | wt |
| NUOP1 | AAS58501 | ND | NI | NI | −133 | +570 | wt |
| NUOP4 | AAS58498 | ND | NI | NI | −18 | +215 | wt |
| NUOA1 | AAS48198 | α | NDUFA1 | MWFE | −63 | +488 | wt |
| NUOA9 | AAQ55458 | α | NDUFA9 | 39 | −233 | +177 | wt |
| NUOB16 | AAQ64637 | α, λ | NDUFA13 | B16.6 | −151 | +656 | wt |
| NUOB8 | AAQ63699 | α, λ | NDUFA2 | B8 | −75 | +527 | wt |
| Assembly factor | |||||||
| NUOAF1 | ACN88152 | NDUFAF1 | CIA30 | −13 | +288 | wt | |
The assignment of the subunits in the various subcomplexes (α, β, βS, βL, and λ) is based on the predicted organization shown in Lazarou et al. (2009). NI, not identified; ND, not determined. The location of NUOP4, only identified in Chlamydomonas complex I, has not yet been determined experimentally (Cardol et al. 2004). NUOP1 was found in the membrane arm of Arabidopsis complex I (Klodmann et al. 2010).
DISCUSSION
The assembly of mitochondrial complex I is an intricate process involving coordination of multiple factors to yield a holoenzyme with at least 40 subunits, one FMN molecule, and eight FeS clusters. We aimed to further dissect this process and pursued a forward genetics approach in the green alga C. reinhardtii to uncover mutations (amc) resulting in loss of complex I function. We took advantage of the fact that in this alga, mutations inactivating complex I are not lethal and result in a slow growth in the dark phenotype, which facilitates visual screening of candidate mutants (Remacle et al. 2008). We isolated six new complex I-deficient mutants, four of them resulting in the formation of intermediate assembly subcomplexes. On the basis of our genetic analyses, all amc mutations are recessive and fall into six complementation groups (Table 3). The finding that amc5 and amc7 are the only recovered allelic mutations indicates that our screen is not saturated. Even though all amc mutations result in a defect in complex I activity, they appear different when examined at the level of complex I assembly (Table 3). While amc1, -2, -5/7, and -6 accumulated a complex I assembly intermediate in the form of a subcomplex, amc3 and amc4 did not accumulate any subcomplexes. The 700-kDa subcomplex was shown to contain the 51 kDa, 49 kDa, and TYKY subunits that are components of the hydrophilic arm of complex I. Defects in ND4 and ND5 subunits of the membrane arm also yield a 700-kDa subcomplex (Remacle et al. 2006; Cardol et al. 2008). Overall, the accumulation of the subcomplex indicates that amc1, -2, -4, -5/7, and -6 are disrupted for the assembly of the membrane arm of complex I. The amc4 mutant appears to accumulate lower amounts of a wild-type–size complex I and is probably not deficient in the assembly process per se but rather for the stability of the holoenzyme. The fact that amc3 displays a reduced complex I activity but no change in the abundance of the TYKY, 49-, and 51-kDa subunits suggests that the enzyme is fully assembled but less active.
TABLE 3.
Phenotype and molecular genetics of amc mutants
| Strain | CI activity | Fully assembled (950 kDa) | Subcomplex (700 kDa) | Allelism | Monogenic | Tagged |
| wt | 100 | ++++ | — | — | — | — |
| amc1 | 12 | — | + | No | Yes | No |
| amc2 | 4 | — | + | No | Yes | No |
| amc3 | 41 | +++ | — | No | Yes | No |
| amc4 | 14 | + | — | No | Yes | No |
| amc5 | 14 | — | ++ | Yes (amc7) | Yes | Yes |
| amc6 | 22 | — | ++ | No | Yes | No |
| amc7 | 14 | — | ++ | Yes (amc5) | Yes | No |
Complex I activity (CI) is indicated as % of wild type (100). +, ++, +++, ++++, and — indicate the relative levels of fully assembled complex I or 700-kDa subcomplexes.
The amc5/7 locus was cloned and shown to correspond to the NUOB10/PDSW encoding gene (Figure 7). The NUOB10/PDSW family is a poorly characterized protein family characterized by the presence of a C(X)11C motif (Cardol et al. 2004). The bovine PDSW has been found in the membrane arm (Hirst et al. 2003) and the Neurospora ortholog is an integral membrane protein, on the basis of its lack of extractability in alkali-treated samples (Videira et al. 1993). In Arabidopsis, NUOB10/PDSW is found in the membrane arm of complex I, associated with the hydrophobic subunit ND5 (Klodmann et al. 2010). This result is in accord with the fact that single mutants affected in ND5 (dum23, Figure S1) or NUOB10/PDSW (amc5, Figure S1) generate the same subcomplex, lacking the distal part of the membrane domain (this study and Hirst et al. 2003; Cardol et al. 2008). The finding that the amc5 and amc7 mutants carry mutations in the NUOB10 gene validates our screening strategy for complex I defects.
The identities of the other AMC gene products revealed through our mutant screen are unknown and can only be speculated upon at the present time. One possibility is that the AMC loci correspond to genes encoding bona fide subunits. Alternatively, AMC loci could encode factors required for expression or assembly of complex I. Orthologs for identified assembly factors, NDUFAF3 (Saada et al. 2009), CIA30 (Dunning et al. 2007), C20ORF7 (Sugiana et al. 2008; Gerards et al. 2009), IND1 (Bych et al. 2008; Sheftel et al. 2009), C8ORF38 (Pagliarini et al. 2008), FOXRED1 (Fassone et al. 2010), GLDH (Pineau et al. 2008), and candidate assembly factors (C7ORF10, MCCC2, AMACR, LYRM5, DCI, and IVD) (Pagliarini et al. 2008) are present in the predicted proteome of Chlamydomonas. Whole-genome sequencing of the amc mutants should enable the rapid identification of the molecular lesions causing complex I defects. Such a technology is currently being tested in Chlamydomonas (O. Vallon, personal communication).
To our knowledge, this is the first description of a mutant screen in any organism yielding mutants with an isolated complex I deficiency. The variety of assembly defects in the amc mutants suggests the corresponding gene products (either structural subunits or assembly factors) act at different steps of the assembly process. This further stresses the value of Chlamydomonas as an experimental model to address the question of complex I assembly. This question has now received considerable attention in plants, where complex I displays unique features compared to its mammalian/fungal counterpart (Braun and Zabaleta 2007; Pineau et al. 2008; Klodmann et al. 2010) and also in humans, as most of complex I-linked diseases still have no molecular explanation (Lazarou et al. 2009).
Acknowledgments
We thank M. Cerda-Richards and A. Martzolff for technical help and R. Lamb and S. Cline for extensive editing of the manuscript. This research was supported by a grant from the United Mitochondrial Disease Foundation (to P.H and C.R.), F.R.S.-FNRS 1.5.255.08, 2.4638.05, and 2.4601.08, Action de la Recherche Concertée ARC07/12-04 and FP7-funded Sunbiopath project (GA 245070) (to C.R.). V.L. is supported by a Fonds de la Recherche Scientifique - Fonds National de la Recherche Scientifique, Formation à la Recherche dans l’Industrie et l’Agriculture fellowship. This work is also sponsored by the Action Centre National de la Recherche Scientifique (C.N.R.S)–US 2008-2010 grant.
LITERATURE CITED
- Barrientos A., 2003. Yeast models of human mitochondrial diseases. IUBMB Life 55: 83–95 [DOI] [PubMed] [Google Scholar]
- Berthold P., Schmitt R., Mages W., 2002. An engineered Streptomyces hygroscopicus aph 7 gene mediates dominant resistance against hygromycin B in Chlamydomonas reinhardtii. Protist 153: 401–412 [DOI] [PubMed] [Google Scholar]
- Braun H.-P., Zabaleta E., 2007. Carbonic anhydrase subunits of the mitochondrial NADH dehydrogenase complex (complex I) in plants. Physiol. Plant. 129: 114–122 [Google Scholar]
- Bych K., Kerscher S., Netz D. J., Pierik A. J., Zwicker K., et al. , 2008. The iron-sulphur protein Ind1 is required for effective complex I assembly. EMBO J. 27: 1736–1746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardol P., Matagne R. F., Remacle C., 2002. Impact of mutations affecting ND mitochondria-encoded subunits on the activity and assembly of complex I in Chlamydomonas. Implication for the structural organization of the enzyme. J. Mol. Biol. 319: 1211–1221 [DOI] [PubMed] [Google Scholar]
- Cardol P., Gloire G., Havaux M., Remacle C., Matagne R., et al. , 2003. Photosynthesis and state transitions in mitochondrial mutants of Chlamydomonas reinhardtii affected in respiration. Plant Physiol. 133: 2010–2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardol P., Vanrobaeys F., Devreese B., Van Beeumen J., Matagne R. F., et al. , 2004. Higher plant-like subunit composition of mitochondrial complex I from Chlamydomonas reinhardtii: 31 conserved components among eukaryotes. Biochim. Biophys. Acta 1658: 212–224 [DOI] [PubMed] [Google Scholar]
- Cardol P., Lapaille M., Minet P., Franck F., Matagne R. F., et al. , 2006. ND3 and ND4L subunits of mitochondrial complex I, both nucleus encoded in Chlamydomonas reinhardtii, are required for activity and assembly of the enzyme. Eukaryot. Cell 5: 1460–1467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardol P., Boutaffala L., Memmi S., Devreese B., Matagne R. F., et al. , 2008. In Chlamydomonas, the loss of ND5 subunit prevents the assembly of whole mitochondrial complex I and leads to the formation of a low abundant 700 kDa subcomplex. Biochim. Biophys. Acta 1777: 388–396 [DOI] [PubMed] [Google Scholar]
- Dent R. M., Haglund C. M., Chin B. L., Kobayashi M. C., Niyogi K. K., 2005. Functional genomics of eukaryotic photosynthesis using insertional mutagenesis of Chlamydomonas reinhardtii. Plant Physiol. 137: 545–556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Depège N., Bellafiore S., Rochaix J. D., 2003. Role of chloroplast protein kinase Stt7 in LHCII phosphorylation and state transition in Chlamydomonas. Science 299: 1572–1575 [DOI] [PubMed] [Google Scholar]
- Distelmaier F., Koopman W. J. H., van den Heuvel L. P., Rodenburg R. J., Mayatepek E., et al. , 2009. Mitochondrial complex I deficiency: from organelle dysfunction to clinical disease. Brain 132: 833–842 [DOI] [PubMed] [Google Scholar]
- Dorthu M. P., Remy S., Michel-Wolwertz M. R., Colleaux L., Breyer D., et al. , 1992. Biochemical, genetic and molecular characterization of new respiratory-deficient mutants in Chlamydomonas reinhardtii. Plant Mol. Biol. 18: 759–772 [DOI] [PubMed] [Google Scholar]
- Dunning C. J., McKenzie M., Sugiana C., Lazarou M., Silke J., et al. , 2007. Human CIA30 is involved in the early assembly of mitochondrial complex I and mutations in its gene cause disease. EMBO J. 26: 3227–3237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Efremov R. G., Baradaran R., Sazanov L. A., 2010. The architecture of respiratory complex I. Nature 465: 441–445 [DOI] [PubMed] [Google Scholar]
- Fassone E., Duncan A. J., Taanman J. W., Pagnamenta A. T., Sadowski M. I., et al. , 2010. FOXRED1, encoding an FAD-dependent oxidoreductase complex-I-specific molecular chaperone, is mutated in infantile-onset mitochondrial encephalopathy. Hum. Mol. Genet. 19: 4837–4847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galkin A., Dröse S., Brandt U., 2006. The proton pumping stoichiometry of purified mitochondrial complex I reconstituted into proteoliposomes. Biochim. Biophys. Acta 1757: 1575–1581 [DOI] [PubMed] [Google Scholar]
- Gerards M., Sluiter W., van den Bosch B. J., de Wit E., Calis C. M., et al. , 2009. Defective complex I assembly due to C20orf7 mutations as a new cause of Leigh syndrome. J. Med. Genet. 47: 507–512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray M. W., Burger G., Lang B. F., 2001. The origin and early evolution of mitochondria. Genome Biol. 2(6):REVIEWS1018.1–1018.5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris E. H., 1989. The Chlamydomonas Sourcebook: A Comprehensive Guide to Biology and Laboratory Use. Academic Press, San Diego: [DOI] [PubMed] [Google Scholar]
- Hirst J., Carroll J., Fearnley I. M., Shannon R. J., Walker J. E., 2003. The nuclear encoded subunits of complex I from bovine heart mitochondria. Biochim. Biophys. Acta 1604: 135–150 [DOI] [PubMed] [Google Scholar]
- Howe G., Merchant S., 1992. The biosynthesis of membrane and soluble plastidic c-type cytochromes of Chlamydomonas reinhardtii is dependent on multiple common gene products. EMBO J. 11: 2789–2801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunte C., Zickermann V., Brandt U., 2010. Functional modules and structural basis of conformational coupling in mitochondrial complex I. Science 329: 448–451 [DOI] [PubMed] [Google Scholar]
- Klodmann J., Sunderhaus S., Nimtz M., Jansch L., Braun H. P., 2010. Internal architecture of mitochondrial complex I from Arabidopsis thaliana. Plant Cell 22: 797–810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuffner R., Rohr A., Schmiede A., Krull C., Schulte U., 1998. Involvement of two novel chaperones in the assembly of mitochondrial NADH:Ubiquinone oxidoreductase (complex I). J. Mol. Biol. 283: 409–417 [DOI] [PubMed] [Google Scholar]
- Lazarou M., Thorburn D. R., Ryan M. T., McKenzie M., 2009. Assembly of mitochondrial complex I and defects in disease. Biochim. Biophys. Acta 1793: 78–88 [DOI] [PubMed] [Google Scholar]
- Liu Y. G., Mitsukawa N., Oosumi T., Whittier R. F., 1995. Efficient isolation and mapping of Arabidopsis thaliana T-DNA insert junctions by thermal asymmetric interlaced PCR. Plant J. 8: 457–463 [DOI] [PubMed] [Google Scholar]
- Loeffen J. L., Smeitink J. A., Trijbels J. M., Janssen A. J., Triepels R. H., et al. , 2000. Isolated complex I deficiency in children: clinical, biochemical and genetic aspects. Hum. Mutat. 15: 123–134 [DOI] [PubMed] [Google Scholar]
- Pagliarini D. J., Calvo S. E., Chang B., Sheth S. A., Vafai S. B., et al. , 2008. A mitochondrial protein compendium elucidates complex I disease biology. Cell 134: 112–123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pineau B., Layoune O., Danon A., De Paepe R., 2008. L-galactono-1,4-lactone dehydrogenase is required for the accumulation of plant respiratory complex I. J. Biol. Chem. 283: 32500–32505 [DOI] [PubMed] [Google Scholar]
- Quiles M. J., Albacete M. E., Sabater B., Cuello J., 1996. Isolation and partial characterization of the NADH dehydrogenase complex from barley chloroplast thylakoids. Plant Cell Physiol. 37: 1134–1142 [Google Scholar]
- Rasmusson A. G., Møller I. M., 1991. NAD(P)H dehydrogenases on the inner surface of the inner mitochondrial membrane studied using inside-out submitochondrial particles. Physiol. Plant. 83: 357–365 [Google Scholar]
- Remacle C., Baurain D., Cardol P., Matagne R. F., 2001a. Mutants of Chlamydomonas reinhardtii deficient in mitochondrial complex I: characterization of two mutations affecting the nd1 coding sequence. Genetics 158: 1051–1060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remacle C., Duby F., Cardol P., Matagne R. F., 2001b. Mutations inactivating mitochondrial genes in Chlamydomonas reinhardtii. Biochem. Soc. Trans. 29: 442–446 [DOI] [PubMed] [Google Scholar]
- Remacle C., Gloire G., Cardol P., Matagne R. F., 2004. Impact of a mutation in the mitochondrial LSU rRNA gene from Chlamydomonas reinhardtii on the activity and the assembly of respiratory-chain complexes. Curr. Genet. 45: 323–330 [DOI] [PubMed] [Google Scholar]
- Remacle C., Cardol P., Coosemans N., Gaisne M., Bonnefoy N., 2006. High-efficiency biolistic transformation of Chlamydomonas mitochondria can be used to insert mutations in complex I genes. Proc. Natl. Acad. Sci. USA 103: 4771–4776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remacle C., Barbieri M. R., Cardol P., Hamel P. P., 2008. Eukaryotic complex I: functional diversity and experimental systems to unravel the assembly process. Mol. Genet. Genomics 280: 93–110 [DOI] [PubMed] [Google Scholar]
- Rexroth S., Meyer zu Tittingdorf J. M., Krause F., Dencher N. A., Seelert H., 2003. Thylakoid membrane at altered metabolic state: challenging the forgotten realms of the proteome. Electrophoresis 24: 2814–2823 [DOI] [PubMed] [Google Scholar]
- Saada A., Vogel R. O., Hoefs S. J., van den Brand M. A., Wessels H. J., et al. , 2009. Mutations in NDUFAF3 (C3ORF60), encoding an NDUFAF4 (C6ORF66)-interacting complex I assembly protein, cause fatal neonatal mitochondrial disease. Am. J. Hum. Genet. 84: 718–727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J., Fritsch E. F., Maniatis T., 1989. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- Sazanov L. A., Hinchliffe P., 2006. Structure of the hydrophilic domain of respiratory complex I from Thermus thermophilus. Science 311: 1430–1436 [DOI] [PubMed] [Google Scholar]
- Schägger H., von Jagow G., 1991. Blue native electrophoresis for isolation of membrane protein complexes in enzymatically active form. Anal. Biochem. 199: 223–231 [DOI] [PubMed] [Google Scholar]
- Sheftel A. D., Stehling O., Pierik A. J., Netz D. J., Kerscher S., et al. , 2009. Human ind1, an iron-sulfur cluster assembly factor for respiratory complex I. Mol. Cell. Biol. 29: 6059–6073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimogawara K., Fujiwara S., Grossman A., Usuda H., 1998. High-efficiency transformation of Chlamydomonas reinhardtii by electroporation. Genetics 148: 1821–1828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugiana C., Pagliarini D. J., McKenzie M., Kirby D. M., Salemi R., et al. , 2008. Mutation of C20orf7 Disrupts Complex I Assembly and Causes Lethal Neonatal Mitochondrial Disease. Am. J. Hum. Genet. 83: 468–478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorburn D. R., 2004. Mitochondrial disorders: prevalence, myths and advances. J. Inherit. Metab. Dis. 27: 349–362 [DOI] [PubMed] [Google Scholar]
- Videira A., Azevedo J. E., Werner S., Cabral P., 1993. The 12.3 kDa subunit of complex I (respiratory-chain NADH dehydrogenase) from Neurospora crassa: cDNA cloning and chromosomal mapping of the gene. Biochem. J. 291(Pt 3): 729–732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel R. O., Janssen R. J., Ugalde C., Grovenstein M., Huijbens R. J., et al. , 2005. Human mitochondrial complex I assembly is mediated by NDUFAF1. FEBS J. 272: 5317–5326 [DOI] [PubMed] [Google Scholar]
- Wikström M., 1984. Two protons are pumped from the mitochondrial matrix per electron transferred between NADH and ubiquinone. FEBS Lett. 169: 300–304 [DOI] [PubMed] [Google Scholar]