Abstract
Heterozygosity is a major challenge to efficient, high-quality genomic assembly and to the full genomic survey of polymorphism and divergence. In Drosophila melanogaster lines derived from equatorial populations are particularly resistant to inbreeding, thus imposing a major barrier to the determination and analyses of genomic variation in natural populations of this model organism. Here we present a simple genome sequencing protocol based on the whole-genome amplification of the gynogenetically derived haploid genome of a progeny of females mated to males homozygous for the recessive male sterile mutation, ms(3)K81. A single “lane” of paired-end sequences (2 × 76 bp) provides a good syntenic assembly with >95% high-quality coverage (more than five reads). The amplification of the genomic DNA moderately inflates the variation in coverage across the euchromatic portion of the genome. It also increases the frequency of chimeric clones. But the low frequency and random genomic distribution of the chimeric clones limits their impact on the final assemblies. This method provides a solid path forward for population genomic sequencing and offers applications to many other systems in which small amounts of genomic DNA have unique experimental relevance.
THE power of genetic and genomic studies of outbreeding diploid multicellular organisms often depends practically, if not conceptually, on the facility with which inbred or pure breeding lines can be established. Many experimentally successful systems offer efficient, practical schemes of close inbreeding. In Drosophila, full-sib mating is such a scheme, but it can require many generations to achieve theoretical genome-wide homozygosity. In practice, often large regions (>500 kbp) exhibit residual heterozygosity in such inbred lines (Falconer 1989, p. 101). This is usually attributed to natural selection favoring the maintenance of complementing linked deleterious mutations. In the case of Drosophila melanogaster, this is especially challenging for samples from equatorial populations (data not shown). In D. melanogaster, balancer chromosomes are often used to create stocks (pure breeding for specific chromosomes) in a few generations of crosses. While this approach can and has been extended to the simultaneous “extraction” of several chromosomes, the practical limits to such schemes are substantial in whole-genome studies. Furthermore, genomes from natural populations harbor recessive lethal and sterile variants that further lower the prospects of establishing large numbers of independent stocks that are effectively homozygous for most of the genome.
While many genome sequences of inbred lines of Drosophila have been determined with the new sequencing technologies (http://www.dpgp.org, http://www.hgsc.bcm.tmc.edu/project-species-i-Drosophila_genRefPanel.hgsc; Blumenstiel et al. 2009), the assembly of sequences of a highly heterozygous diploid genome remains a challenge. The assembly of a consensus or composite genome sequence requires considerably more raw sequence data and auxiliary analyses than does the comparable task for a homozygous or haploid genome (El-Sayed et al. 2005; Vinson et al. 2005). Furthermore, the utility of such composite genome sequences for population genomic analyses is very limited. Indeed, Drosophila population geneticists routinely leveraged the system’s genetic tools to focus on a single haplotype (inbred or haploid genome) from each independently sampled but genetically complex isofemale lines. With the increasing interest in the population genomics of D. melanogaster generated by the new sequencing technologies, any new method to genetically isolate completely homozygous or haploid genomes will have great utility. Here we present a remarkably simple approach that meets that need.
Fuyama (1984) described a recessive male sterile mutant, ms(3)K81, which he isolated from a natural population. He sought a male sterile that would help in his search for genetic variation in D. melanogaster for parthenogenetic development. Fuyama’s description of his initial mutant and that by Yasuda et al. (1995) of several additional induced mutations concur that ms(3)K81 is a unique mutation: fertilization occurs normally, but subsequent events that normally lead to a developing diploid zygote are blocked. Loppin et al. (2005) identified CG14251 as the ms(3)K81 and noted that ms(3)K81 is a diverged paralog of the loosely linked CG6874. Recently, Gao et al. (2010) demonstrated that HipHop (CG6874) is an essential component of the telomere capping complex that also contains HP1 and HOAP, while Dubruille et al. (2010) identified the protein encoded by ms(3)K81 (K81) as part of the telomere capping complex for the sperm genome. Loss-of-function phenotypes of HipHop and K81 are typical of telomere capping defects, i.e., telomere fusion and chromatin bridges at anaphase. In the first nuclear divisions of eggs fertilized by ms(3)K811/ms(3)K811 males, the paternally derived chromosomes lack HOAP as well as K81 and form such bridges (Dubruille et al. 2010). The great majority of eggs from females mated to ms(3)K811/ms(3)K811 males fail to develop; the few that do develop never become well-formed first instar larvae. Cytological studies of the “escaping zygotes” revealed that the nuclei of the blastoderm are haploid, apparently derived from the maternal pronucleus. Yasuda et al. (1995) confirmed that the observed frequencies of haploid development were similar for the available alleles of ms(3)K81 and not sensitive to maternal genotypes. This ms(3)K81-dependent phenotype is similar to that caused by the female sterile mutation maternal haploid (Loppin et al. 2001). Both of these mutations cause catastrophic mitotic failure of the paternal chromosomes during the early syncytial rounds of nuclear replication and division, leading predominantly to early arrest of development (Sullivan et al. 1993; Fogarty et al. 1997) with a low percentage of eggs proceeding through cellularization to develop as gynogenetic haploids. The reciprocal phenotype is seen in eggs from I-R dysgenesis in which the maternal chromosomes suffer catastrophic mitosis and the surviving embryos are androgenic haploids (Orsi et al. 2010).
We reasoned that the otherwise normal-appearing nuclei of such a single haploid embryo derived from a cross of a female from any stock of interest to a ms(3)K81 male might yield sufficient quantity and quality of genomic DNA for a subsequent whole-genome amplification (WGA) and construction of a representative genomic library suitable for sequencing. Here we present evidence supporting this conjecture. Whole-genome shotgun sequencing ideally covers the genome with reads with a Poisson variance (Lander and Waterman 1988). However, inherent biases associated with library construction and sequencing on the Solexa/Illumina platform increase this variance (Bentley et al. 2008). Whole-genome amplification also has inherent quantitative biases in coverage, in addition to the increased numbers of chimeric clones (Dean et al. 2002; Lasken and Stockwell 2007). While we do observe the expected increase in the variance in read depth relative to that observed for unamplified genomic DNAs, and chimeric clones increase in frequency as expected, we find that high-quality genome assemblies can be obtained from a single lane of an Illumina GA IIx flow cell with the standard 2 × 76 bp paired-end sequencing protocol. The paternal genome of the apparently haploid progeny of crosses to ms(3)K81/ms(3)K81 males was nearly universally excluded: we observed a single interesting and readily detectable exception among >150 sequenced haploids.
MATERIALS AND METHODS
Drosophila stocks and matings:
The stocks that were used in this study are referred to as y; cn bw sp, ms(3)K81, and GA187. The first is the inbred stock from which the D. melanogaster reference genome sequence is derived (Adams et al. 2000) and has the genotype y1; Gr22b1 Gr22d1 cn1 CG33964R4.2 bw1 sp1; LysC1 MstProx1 GstD51 Rh61. The second stock has the genotype ms(3)K811/TM3, Sb1 Ser1. Both are available at the Bloomington Drosophila Stock Center. The third stock, GA187, is an isofemale line established from a single inseminated female collected by B. Ballard and S. Charlat in Franceville, Gabon, in March 2002. Diploid adult genomic DNA was prepared from 1 g of flash-frozen female and male y; cn bw sp adult flies using the CsCl protocol described in Bingham et al. (1981). Virgin females of y; cn bw sp and GA187 (isofemale line from Gabon) were collected and crossed to males homozygous for the ms(3)K811 allele (Fuyama 1984). After mating overnight, the flies were transferred to vials containing an oviposition substrate and left there for 24 hr. Mated flies were then transferred to fresh vials and maintained there for 10 days to monitor for the presence of viable offspring. The presence of viable larvae was taken as evidence of non-virginity or misclassification of the male genotype.
Embryo collection and genome amplification:
Embryos were allowed to develop on the oviposition substrate for 12–24 hr after mated flies were removed. Embryos were then harvested and dechorionated as described in Rothwell and Sullivan (2007a,b). Dechorionated embryos were examined under a stereomicroscope at ×40 power, and well-developed embryos (as evidenced by visible abdominal segmentation) were collected and stored individually in 3 μl of 1× PBS at −80° until use.
A single embryo was thawed on ice and then subjected to multiple displacement amplification (Dean et al. 2002) using the QIAGEN REPLI-g Midi kit following the manufacturer’s instructions. Briefly, buffer D2 was added and the embryo was crushed and ground thoroughly with a pipette tip. After a 10-min incubation on ice, Stop Solution was added, followed by the amplification master mix. Amplification proceeded for 16 hr at 30° after which the DNA polymerase was deactivated by heating at 65° for 3 min. Analysis of the WGA products produced under these standard conditions in >100 independent experiments shows that total yield ranges between 25 and 50 μg, typically closer to 50 μg. The majority of the fragments consistently appear to be >12 kb on a 1% agarose gel, as reported in the product literature.
A time-course series performed indicated that whole-genome amplification reactions under the conditions that we used were largely complete after 6 hr (data not shown). Reasoning that unbalanced depletion of primer and nucleotide species late in the incubation period might introduce undesirable amplification biases, we compared the genome sequencing results of a WGA incubated for 16 hr with an aliquot of the same WGA allowed to incubate for only 4 hr, a point at which our time-course experiment indicated that amplification should still be proceeding uninhibited. We found that libraries prepared from the two WGA time points were virtually indistinguishable in terms of mean coverage, variance in coverage, mean GC content, and number of putative chimeras (data not shown).
Illumina sequencing:
Concentration of the multiple displacement amplified DNA was estimated using the Quant-iT PicoGreen dsDNA Assay Kit (Invitrogen) and verified by agarose gel electrophoresis. Approximately 5 μg of DNA was used as starting material for paired-end sequencing library construction following the Illumina protocol. Fragmentation was done by sonication using the Diagenode Bioruptor at high power for 15 cycles of 30 sec on/30 sec off (the same for both unamplified and amplified genomic DNA). The adapter-ligation product was gel-purified to select molecules ∼400 bp in length and then quantified using an Agilent Bioanalyzer. A total of 10 ng of size-selected ligation product was used as template for 10 cycles of library enrichment PCR. The enriched library was purified using Ampure XP beads (Beckman Coulter) and sequenced on a single lane of a flow cell with an Illumina GAIIx running the Illumina software (Table S1).
One library-accession, ycnbwsp_7-HE, was prepared from the same WGA reaction as ycnbwsp_8-HE except that the amplified DNA was “de-branched” following the method described by Zhang et al. (2006). A total of 25 μl of the WGA reaction was ethanol-precipitated and resuspended in 50 μl of 1× RepliPHI reaction buffer containing 1 mm dNTP (but no primers). Four hundred units of RepliPHI phi29 DNA polymerase (Epicentre) was added, and the reaction was incubated for 2 hr at 37° and then for 3 min at 65° to inactivate the enzyme. The product was purified by phenol/chloroform extraction, ethanol-precipitated, and resuspended in 200 μl of 1× reaction buffer (30 mm sodium acetate, pH 4.5, 50 mm NaCl, 1 mm ZnCl2). A total of 200 U of S1 nuclease (USB) was added, and the reaction was incubated for 30 min at 37°. DNA was then purified by phenol/chloroform extraction, ethanol-precipitated, and resuspended in 75 μl of TE. Library construction then proceeded as described above, beginning with concentration estimation and sonication.
Genome assemblies:
The assemblies analyzed in this article were created as follows. The reads were aligned to the five major chromosome arms of the Berkeley Drosophila Genome Project’s Release 5 D. melanogaster reference genome sequence (BDGPr5) using ELANDv2 from the Illumina CASAVA pipeline indicated in Table S1. Repetitive reads that align to multiple locations are removed by default. Paired ends with a distance of >1000 bp were identified as outliers and also removed. These filtered alignments were then passed to MAQ v 0.7.1 for consensus sequence determination (Li et al. 2008). For “diploid” assemblies where the rate of heterozygous loci is explicitly evaluated, the prior probability of a heterozygous site was set to the default value of 10−3. Otherwise, for haploid or inbred genomes the prior probability of a heterozygous site is set to 0 to remove the heterozygous genotypes from model selection. Such assemblies are referred to as “diploid” and “haploid,” respectively. All remaining parameters were kept at the program’s default settings. The empirical quality score of a set of sites in the (consensus) assembly, Q, is calculated as −10log10(e), where e is the measured proportion of sites that are different from the BDGP Build 5 reference genome. The variance and correlation coefficient in per-base-pair Illumina read depth was calculated over the assemblies at sites spaced 1000 bp apart to ensure independence.
Five genomic libraries were created, sequenced, and MAQ-assembled using both haploid and diploid parameters as described above (also see Table S1). Four were created from y; cn bw sp, the inbred stock from which the reference genome sequence was derived (Adams et al. 2000). The first, ycnbwsp-da_2, was derived from a library of genomic DNA isolated from adults from the stock. The ycnbwsp_3-HE, ycnbwsp_7-HE, and ycnbwsp_8-HE libraries were created from whole-genome amplifications of the genomic DNA of single haploid embryos of the cross y; cn bw sp virgin females × ms(3)K811/ms(3)K811 males. The GA187-he library is derived from the whole-genome-amplified DNA of an apparently haploid progeny of the GA187 virgin females to the ms(3)K811/ms(3)K811 males.
RESULTS
To determine how effectively the genome sequence of a single haploid Drosophila embryo could be determined, a simple protocol was developed based on whole-genome amplification and routine Solexa/Illumina sequencing of individual rare embryos from crosses involving ms(3)K81 males. The only fundamental differences among the five haploid genomic sequences described and analyzed here are the maternal parents (Table S1). Two are derived from the same embryo and differ only by the application of a post-WGA de-branching protocol.
Comparing haploid amplified to unamplified diploid DNA:
To evaluate the impact of ms(3)K81-induced haploid development and whole-genome amplification on variation in read depth and assembly quality, we compared the four independent haploid genome assemblies (one from inbred diploid adults and three from haploid embryos) to the reference genome sequence. We evaluated the empirical consensus error rates. We also evaluated the variation in assembled read depth and its impact on the quality of the resulting consensus sequence. Finally, we evaluated the rates of chimeric clones, which have been reported in sequences derived from WGA (Lasken and Stockwell 2007).
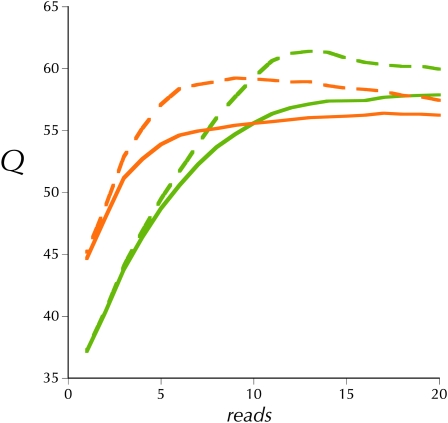
Figure 1 presents the upper and lower estimates of the mean consensus quality score, Q, as a function of read depth for the haploid MAQ asssemblies of both ycnbwsp-da_2 and ycnbwsp_3-HE. The upper estimate is simply the rate, at different read depths, of non-reference basecalls in the consensus. If we assume that all non-reference basecalls shared among the assemblies of both independent libraries are simply mutations fixed in our y; cn bw sp stock, then these can be removed from the error rate estimate to yield the lower estimate in Figure 1. The haploid extracted sample shows a moderately increased error rate at high coverage values, but our overall estimate of Q is still well above what is considered high quality for a consensus sequence.
Figure 1.—
Empirical consensus quality, Q, at different read depths for ycnbwsp_2 (olive) and ycnbwsp_3-HE (orange). Dashed lines are the upper estimates and solid lines are the lower estimates (see text).
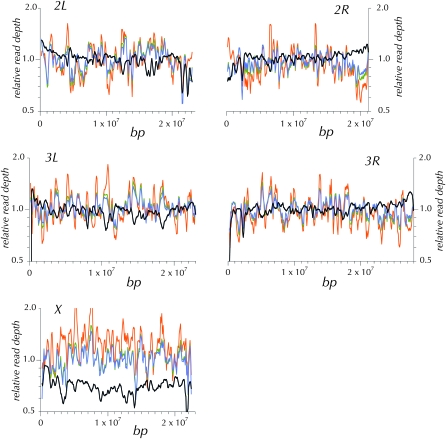
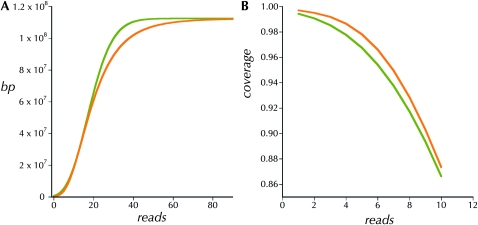
Under an ideal model, read depth is sampled from the Poisson distribution with a coefficient of variation of 1.0 (Lander and Waterman 1988). Inflation of the ratio of the standard deviation over the mean above this ideal indicates an increase in areas with more extreme read depths. By this criterion neither the unamplified adult nor the WGA haploid embryo protocol is ideal. But whole-genome amplification does add a moderate amount to the coefficient of variation (Table 1). This increased variation in read depth is illustrated in 300-kbp windows along the chromosomes in Figure 2. The patterns of variation in read depth are highly correlated between the two WGA haploid embryo assemblies (Table 1). But these two are weakly correlated with that from the ycnbwsp_2 assembly based on unamplified genomic DNA (ycnbws_2). Note also in Figure 2 that ycnbwsp_3-HE exhibits several obvious regions of distinctly different relative depth from ycnbwsp_7-HE and ycnbwsp_8-HE (e.g., chromosome 2R: 6.5 × 10+6 bp; chromosome 3L: 9 × 10+6 bp; and chromosome X: 3 × 10+6 bp). In terms of the practical goal, the primary concern is that this additional variance may cause a significant degradation in the yield of high-quality consensus sequence resulting primarily from an increase in areas with low read depth. Figure 3 presents the cumulative depth distribution for ycnbwsp-da_2 and ycnbwsp_3-HE. The additional variance does not significantly affect the low end of the distribution (Figure 3B). Assuming that five reads is a minimal depth to reliably determine the sequence of a haploid genome (Figure 1) (Keightley et al. 2009), we see in Figure 3B that >97.8% of ycnbwsp_3-HE and 96.7% of ycnbwsp_2 are covered with five or more reads.
TABLE 1.
Library assembly statistics
| SNPs |
Read depth |
|||||||
| Diploid |
Haploid | Correlation |
||||||
| +/− | +/+ | − | Mean | σ/μ | 3-HE | 7-HE | 8-HE | |
| ycnbwsp_2 | 6.7 × 10−6 | 2.4 × 10−6 | 2.6 × 10−6 | 18.57 | 0.455 | 0.0656 (0.0598–0.0714) | 0.2001 (0.1945–0.2057) | 0.2620 (0.2565–0.2674) |
| ycnbwsp_3-HE | 1.5 × 10−5 | 3.0 × 10−6 | 3.6 × 10−6 | 22.58 | 0.634 | − | 0.7472 (0.7446–0.7498) | 0.6904 (0.6874–0.6935) |
| ycnbwsp_7-HE | 1.7 × 10−5 | 2.9 × 106 | 3.3 × 10−6 | 21.04 | 0.426 | − | − | 0.7621 (0.7597–0.7646) |
| ycnbwsp_8-HE | 1.9 × 10−5 | 2.8 × 10−6 | 3.1 × 10−6 | 17.45 | 0.637 | − | − | − |
The proportion of sites in the assemblies with read depth of ≥10 that were called as different from the reference sequence (SNPs), either as heterozygotes (+/−) or as homozygotes (−/−) in a MAQ diploid assembly or as a divergent allele (−), in a MAQ haploid assembly are shown. Three properties of the distributions of read depth are presented: the mean, the coefficient of variation, and the correlation between pairs of assemblies at sites spaced 1000 bp apart (see text). The approximate 5% and 95% confidence intervals are in parentheses.
Figure 2.—
The pattern of read depth across the chromosome arms in overlapping 300-kbp windows. For each genome, the observed read depth is normalized to the autosomal average. The three WGA haploid genomes—ycnbwsp_3-HE (orange), ycnbwsp_7-HE (olive), and ycnbwsp_8-HE (blue)—show considerably more variation and are strongly correlated with one another and not with the unamplified ycnbwsp_2 (black) created from diploid adults without WGA. The reduced depth of X in the ycnbwsp_2 assembly reflects the fact that both female and male adult flies were the source of the DNA.
Figure 3.—
(A) The cumulative coverage of the unique portion of the reference genome as a function of read depth in the two libraries, ycnbwsp_2 (olive) and ycnbwsp_3-HE (orange). (B) The proportion of the unique reference genome covered at low minimum read depths in the two libraries, ycnbwsp_2 (olive) and ycnbwsp_3-HE (orange).
Finally, it has been reported in the literature that WGA can create a significant number of chimeric DNA fragments primarily via a short-range (<10 kbp) intramolecular self-priming mechanism (Lasken and Stockwell 2007; Rodrigue et al. 2009). Obviously, such chimeras are potential sources of assembly errors. The comparison of ycnbwsp_2, ycnbwsp_3-HE, ycnbwsp_7-HE, and ycnbwsp_8-HE in Table 2 indeed shows an increased but still small proportion of clearly chimeric clones, i.e., the “inverted” classes (F+ and F−) and the direct class (R−) that Lasken and Stockwell (2007) reported, as well as the “interchromosomal” class. The “too small” and “too large” classes are likely to be a mixture of chimeric clones and those outside the arbitrary boundaries of the size-selected population of molecules. The inverted classes, F+ and F−, were interpreted by Lasken and Stockwell (2007) as arising from intramolecular self-priming. Consistent with their model and their observations, we observed that these inverted chimeras predominantly involve reads that map within 10 kbp of one another (data not shown).
TABLE 2.
Distribution of mapped genomic positions and orientations in the reference sequence of the first (R1) and second (R2) reads
| Chimeric |
|||||||
| Library | R+: R1> <R2 | R−: <R2 R1> | F+: R1> R2> | F−: R2> R1> | Interchromosomal | Too large | Too small |
| ycnbwsp_2 | 0.98089 | 0.00218 | 0.00007 | 0.00007 | 0.00089 | 0.00151 | 0.00144 |
| ycnbwsp_3-HE | 0.95122 | 0.00116 | 0.00538 | 0.00584 | 0.00732 | 0.00270 | 0.02639 |
| ycnbwsp_7-HE | 0.97839 | 0.00055 | 0.00265 | 0.00253 | 0.00195 | 0.00355 | 0.01037 |
| ycnbwsp_8-HE | 0.97394 | 0.00068 | 0.00250 | 0.00276 | 0.00158 | 0.00553 | 0.01300 |
These reads are from the clones of libraries constructed from whole-genome-amplified and unamplified DNA (see text). The proportions are based on the numbers reported by the Illumina CASAVA software (Summary.xml and anomaly.txt files). The second column, R+, is the proportion of normal (nonchimeric) clones in which the two reads from opposite ends of the clone are on alternative strands and the expected distance apart. In the R− column are clones in which the two reads map in divergent orientation. The F+ and F− columns list the proportions of clones in which the two reads map on the same strand and typically map within 10 kbp. These inverted chimeric clones (F+ and F−) are thought to arise via self-priming and have been reported to be more abundant in libraries derived from WGA DNA (Lasken and Stockwell 2007).
Zhang et al. (2006) reported a technique that significantly reduced the frequency of chimeras that they observed in their shotgun sequencing of cloned DNA from whole-genome-amplified single prokaryotic cells (without de-branching, their chimera rate was reported as 19.3%; with de-branching, it was reduced to 6.25%). However, we did not observe any significant reduction in the number of chimeras in ycnbwsp_7-HE compared with ycnbwsp_8-HE, which utilized the same WGA without de-branching (Table 2).
It is important to note that these additional chimeric sequences are randomly distributed across the genome (data not shown), allowing any error contributions to be effectively eliminated by sufficient clone depth in the assembly consensus algorithms. We did not observe an improvement in the consensus error rates of the MAQ assemblies when these chimeric reads were filtered from the assemblies (data not shown).
Evidence of haploidy:
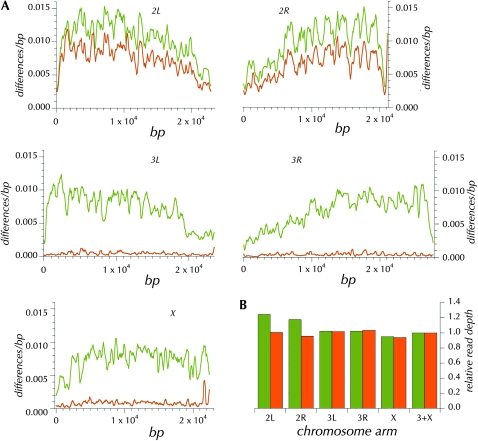
The failure of the ms(3)K811/ms(3)K811 paternally derived genome to be synchronously replicated with the maternally derived genome may be totally attributable to the absence of the telomere capping complex (Dubruille et al. 2010). In any case, developmental arrest occurs in the great majority of eggs fertilized by ms(3)K811/ms(3)K811 males. But a small percentage of eggs continue to develop and are reported to be haploid on the basis of clear cytogenetic imaging (Fuyama 1984; Yasuda et al. 1995). We created diploid MAQ assemblies and examined the proportions of homozygous and heterozygous non-reference genotypes as a means of detecting evidence of residual amounts of the genome from the ms(3)K811/ms(3)K811 parent. As Table 1 shows, there is little evidence for heterozygous SNP calls in diploid assemblies created from haploid WGA or from inbred diploid adults. In our laboratory, we have sequenced >150 such haploid genomes derived from independent isofemale lines from natural populations with similar patterns of uniformly low estimates of heterozygosity. Only one exception has been observed. Figure 4A shows 100-kbp windows of the diploid assembly derived from this putatively haploid embryo, a progeny of a cross of ms(3)K811/ms(3)K811 males to virgin females from the isofemale line GA187. While chromosomes X, 3, and 4 exhibit the standard result (many SNPs but few called as heterozygous), chromosome 2 shows mostly heterozygous SNPs, typical of what we have observed when sequencing outbred diploids (data not shown). The sensitivity of this assay to detect mixed genotypes is underscored by a large increase in heterozygotes accompanied by only a moderate increase in additional read depth on the second chromosome (see below).
Figure 4.—
(A) The pattern of differences from the reference sequence in 100-kbp windows across the GA187 genome (olive) and the pattern of heterozygous sites in the GA187_1-HE diploid MAQ assembly (orange). (B) Relative read depth in the GA187_1-HE (olive) and ycnbwsp_3-HE (orange) assemblies for the different chromosome arms (scaled by the autosomal average).
To identify the origin of the second chromosomes, the sequence of two short segments (736 and 962 bp) on chromosome 2 (chr2L: 16304275–16305233 and chr2R: 12637285–12638020) was determined for 32 individual females from the ms(3)K811/ms(3)K811 stock using standard PCR-based double-stranded Sanger sequencing. No polymorphism was detected among the sequences from this stock. The genotype in the diploid assembly of GA187-he_1 was consistent with a paternally contributed second chromosome; i.e., each site was either heterozygous or homozygous for the allele determined for the ms(3)K811/ms(3)K811 stock. The genotypes at these two second-chromosome loci are more similar to the reference sequence, since at only 2 of the 12 heterozygous sites the paternally derived allele was different from the reference sequence (Table S2). Thus we can conclude on the basis of variation in these two regions that the embryo contained both maternally and paternally derived second chromosomes.
Formally, this embryo could have been a complete second-chromosome diploid. Or only a portion of the cells in the embryo might have been diploid. Alternatively, the embryo could have been thoroughly haploid but chimeric with respect to the origin of the second chromosome. These hypotheses can be distinguished on the basis of the read depth of the second relative to the X and third chromosome. The right two columns of Table S2 show that the read depth is substantially higher for GA187 alleles than for ms(3)K81 alleles identified in the two second-chromosome segments. Finally, the overall read depth (shown in Figure 4B) on the second chromosome relative to that on the X plus the third chromosomes indicates an excess of reads mapping to the second chromosome, although clearly not twofold. Thus partial (mosaic) diploidy for the second chromosome is well supported.
DISCUSSION
Determining the full sequence of a single haploid Drosophila genome has many potential applications. Surveying population genomic variation among inbred lines sampled independently from tropical populations has been a struggle for us because of ineffective inbreeding. Neither sib mating nor use of balancer chromosomes yielded sufficient numbers of highly inbred stocks when the original isofemale lines were derived from equatorial populations. Therefore, we investigated a radically different approach based on a genetic cross that yields partially developed haploid embryos suitable for whole-genome amplification. Three critical observations indicate that this approach has potentially high utility. First, we have confirmed the observations of Fuyama (1984) and Yasuda et al. (1995) that all tested maternal strains yield these haploid embryos at a low but reliably reproducible rate. We have successfully conducted this procedure on >150 independent isofemale lines (data not shown). Thus it seems likely that mating with ms(3)K811/ms(3)K811 males is a robust method for obtaining these haploid embryos. Second, the genomic assemblies produced from Illumina sequencing of whole-genome-amplified haploid embryos do indeed have a substantially bigger variance in coverage across the genome. While this increased variance does reduce the sequencing efficiency, with the recent Illumina reagents and protocols a single lane of sequence data yields a serviceable genome sequence. Third, while the whole-genome amplification leads to an increase in the frequency of particular chimeric clones, the overall proportion of such anomalous clones remains small, and they are readily eliminated from the consensus sequence.
The mechanism(s) leading to haploid embryos from a cross with ms(3)K811/ms(3)K811 males is thought to involve mitotic failure of the paternally derived genome leading to developmental arrest because of a checkpoint. A small fraction of embryos escape this checkpoint and go on to develop as apparent gynogenetic haploids with many of the tissues and structures of first instar larvae. Our analysis of the genomic evidence of haploidy clearly shows that the paternally derived genome is not detectable in the MAQ assemblies. We observed only one exception, an embryo apparently mosaically diploid for the entire second chromosome. This single observation in >150 haploid embryos suggests that elimination of the paternal chromosomes depends on their presumed terminal fusion and mitotic failure. On a more practical level, its low frequency of occurrence and readily detected properties support the utility of the described method in surveying genomic variation among stocks.
We hope that the substantial loss of efficiency due to the increased variance in read depth stimulates improvements in the WGA reagents and protocols to make future applications more effective. WGA also introduces a well-defined class of chimeric clones into the sequencing libraries. Because of the small spatial scale of a major chimera-generating mechanism (predominantly intramolecular and <10 kbp), such chimeric molecules are a severe impediment to development and application of larger insert (3–10 kbp) libraries. Such large-insert libraries have great promise in both resequencing and de novo sequencing applications. Again, we hope that these observations concerning chimeric clones motivate improvements in WGA protocols.
The method presented here surmounts a critical barrier to the systematic survey of full genomic variation in D. melanogaster. The success of this approach may foster the pursuit of many other applications where the determination of the sequence from a unique, small amount of genomic DNA has potentially high experimental value. Finally, it is worth emphasizing that the methods for whole-genome amplification have been developed and optimized mainly to support genotyping assays. We expect that the effectiveness and value of the genomic sequencing methods such as the one described here can be greatly increased by advances in whole-genome amplification technology oriented specifically toward genomic sequencing.
Acknowledgments
We acknowledge the support of National Institutes of Health grant HG02942.
LITERATURE CITED
- Adams M. D., Celniker S. E., Holt C. A., Evans J. D., Gocayne J. D., 2000. The genome sequence of Drosophila melanogaster. Science 287: 2185–2195 [DOI] [PubMed] [Google Scholar]
- Bentley D. R., Balasubramanian S., Swerdlow H. P., Smith G. P., Milton J., et al. , 2008. Accurate whole human genome sequencing using reversible terminator chemistry. Nature 456: 53–59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bingham P. M., Levis R., Rubin G. M., 1981. Cloning of DNA sequences from the white locus of D. melanogaster by a novel and general method. Cell 25: 693–704 [DOI] [PubMed] [Google Scholar]
- Blumenstiel J. P., Noll A. C., Griffiths J. A., Perera A. G., Walton K. N., et al. , 2009. Identification of EMS-induced mutations in Drosophila melanogaster by whole-genome sequencing. Genetics 182: 25–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dean F. B., Hosono S., Fang L., Wu X., Faruqi A. F., et al. , 2002. Comprehensive human genome amplification using multiple displacement amplification. Proc. Natl. Acad. Sci. USA 99: 5261–5266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubruille R., Orsi G. A., Delabaere L., Cortier E., Couble P., et al. , 2010. Specialization of a Drosophila capping protein essential for the protection of sperm telomeres. Curr. Biol. 20: 2090–2099 [DOI] [PubMed] [Google Scholar]
- El-Sayed N. M., Myler P. J., Bartholomeu D. C., Nilsson D., Aggarwal G., et al. , 2005. The genome sequence of Trypanosoma cruzi, etiologic agent of Chagas disease. Science 309: 409–415 [DOI] [PubMed] [Google Scholar]
- Falconer D. S., 1989. Introduction to Quantitative Genetics, Ed. 3 Longmans Green/John Wiley & Sons, Harlow, Essex, UK/New York [Google Scholar]
- Fogarty P., Campbell S. D., Abu-Shumays R., Phalle B. D. S., Yu K. R., et al. , 1997. The Drosophila grapes gene is related to checkpoint gene chk1/rad27 and is required for late syncytial division fidelity. Curr. Biol. 7: 418–426 [DOI] [PubMed] [Google Scholar]
- Fuyama Y., 1984. Gynogenesis in Drosophila melanogaster. Jpn. J. Genet. 59: 91–96 [Google Scholar]
- Gao G., Walser J., Beaucher M. L., Morciano P., Wesolowska N., et al. , 2010. HipHop interacts with HOAP and HP1 to protect Drosophila telomeres in a sequence-independent manner. EMBO J. 29: 819–829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keightley P. D., Trivedi U., Thomson M., Oliver F., Kumar S., et al. , 2009. Analysis of the genome sequences of three Drosophila melanogaster spontaneous mutation accumulation lines. Genome Res. 19: 1195–1201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lander E. S., Waterman M. S., 1988. Genomic mapping by fingerprinting random clones: a mathematical analysis. Genomics 2: 231–239 [DOI] [PubMed] [Google Scholar]
- Lasken R. S., Stockwell T. B., 2007. Mechanism of chimera formation during the multiple displacement amplification reaction. BMC Biotechnol. 7: 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Ruan J., Durbin R., 2008. Mapping short DNA sequencing reads and calling variants using mapping quality scores. Genome Res. 18: 1851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loppin B., Berger F., Couble P., 2001. Paternal chromosome incorporation into the zygote nucleus is controlled by maternal haploid in Drosophila. Dev. Biol. 231: 383–396 [DOI] [PubMed] [Google Scholar]
- Loppin B., Lepetit D., Dorus S., Couble P., Karr T. L., 2005. Origin and neofunctionalization of a Drosophila paternal effect gene essential for zygote viability. Curr. Biol. 15: 87–93 [DOI] [PubMed] [Google Scholar]
- Orsi G. A., Joyce E. F., Couble P., McKim K. S., Loppin B., 2010. Drosophila I-R hybrid dysgenesis is associated with catastrophic meiosis and abnormal zygote formation. J. Cell Sci. 123: 3515–3524 [DOI] [PubMed] [Google Scholar]
- Rodrigue S., Malmstrom R. R., Berlin A. M., Birren B. W., Henn M. R., et al. , 2009. Whole genome amplification and de novo assembly of single bacterial cells. PLoS ONE 4: e6864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothwell W. F., Sullivan W., 2007a. Drosophila embryo collection. CSH Protoc. pdb.prot4825 [DOI] [PubMed] [Google Scholar]
- Rothwell W. F., Sullivan W., 2007b. Drosophila embryo dechorionation. CSH Protoc. pdb.prot4826 [DOI] [PubMed] [Google Scholar]
- Sullivan W., Daily D., Fogarty P., Yook K., Pimpinelli S., 1993. Delays in anaphase initiation occur in individual nuclei of the syncytial Drosophila embryo. Mol. Biol. Cell 4: 885–896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinson J. P., Jaffe D. B., O’Neill K., Karlsson E. K., Stange-Thomann N., et al. , 2005. Assembly of polymorphic genomes: algorithms and application to Ciona savignyi. Genome Res. 15: 1127–1135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasuda G. K., Schubiger G., Wakimoto B. T., 1995. Genetic characterization of ms(3)K81, a paternal effect gene of Drosophila melanogaster. Genetics 140: 219–229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang K., Martiny A. C., Reppas N. B., Barry K. W., Malek J., et al. , 2006. Sequencing genomes from single cells by polymerase cloning. Nat. Biotechnol. 24: 680–686 [DOI] [PubMed] [Google Scholar]