Abstract
Strand misalignments at DNA repeats during replication are implicated in mutational hotspots. To study these events, we have generated strains carrying mutations in the Escherichia coli chromosomal lacZ gene that revert via deletion of a short duplicated sequence or by template switching within imperfect inverted repeat (quasipalindrome, QP) sequences. Using these strains, we demonstrate that mutation of the distal repeat of a quasipalindrome, with respect to replication fork movement, is about 10-fold higher than the proximal repeat, consistent with more common template switching on the leading strand. The leading strand bias was lost in the absence of exonucleases I and VII, suggesting that it results from more efficient suppression of template switching by 3′ exonucleases targeted to the lagging strand. The loss of 3′ exonucleases has no effect on strand misalignment at direct repeats to produce deletion. To compare these events to other mutations, we have reengineered reporters (designed by Cupples and Miller 1989) that detect specific base substitutions or frameshifts in lacZ with the reverting lacZ locus on the chromosome rather than an F′ element. This set allows rapid screening of potential mutagens, environmental conditions, or genetic loci for effects on a broad set of mutational events. We found that hydroxyurea (HU), which depletes dNTP pools, slightly elevated templated mutations at inverted repeats but had no effect on deletions, simple frameshifts, or base substitutions. Mutations in nucleotide diphosphate kinase, ndk, significantly elevated simple mutations but had little effect on the templated class. Zebularine, a cytosine analog, elevated all classes.
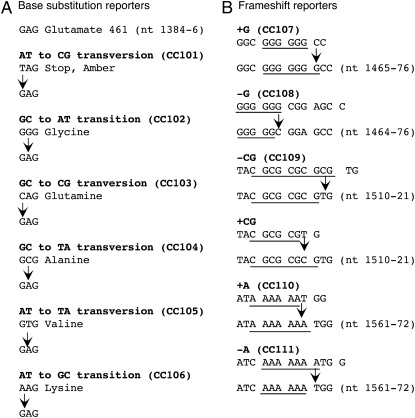
THE lacZ gene of Escherichia coli, which encodes the enzyme β-galactosidase, has been used widely as a reporter gene in a number of applications because of the ease of enzymatic and genetic assays for its function (for example, Schofield et al. 1992; Lopez et al. 2007). Cupples and Miller (1989) pioneered the use of lacZ to report specific mutational events, designing a set of F′ lac strains that can revert to Lac+ only by one of the six specific base substitution mutations. This specificity was possible because only glutamate can be tolerated at amino acid position 461 of the β-galactosidase; this set of strains includes mutations at codon 461 that can restore function by one and only one base substitution (Figure 1A). Cupples et al. (1990) later extended this set to include strains that revert to Lac+ by frameshift mutations in nucleotide runs, including ±1A, or ±1G, or −2CG (Figure 1B). These strains are widely used for determination of the specificity of mutagens and were also key to the isolation of mutator strains that affect specific mutational classes. One example was isolation of mutY, whose mutant phenotype was the specific elevation of GC to TA transversions (Nghiem et al. 1988). MutY glycosylase specifically removes the adenine in G:A mispairs, explaining this specificity (Au et al. 1988).
Figure 1.—
Mutational reporter design. Design is based on Cupples and Miller (1989) and Cupples et al. (1990) for detection of specific base substitutions (A) and frameshift mutations (B) by lacZ reversion.
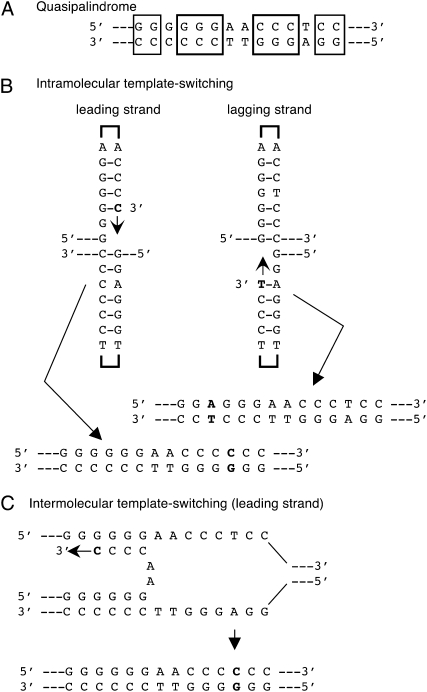
Base substitution and simple frameshift mutations do not account for all potential mutational events that may inactivate genes. An important class of mutational hotspots includes short imperfect inverted repeat sequences, also known as quasipalindromes (QPs) (Figure 2A). These can be seen in mutational spectra in bacteriophage, bacteria, yeast, and in mutations responsible for several human genetic diseases (Stewart and Sherman 1974; Ripley 1982; De Boer and Ripley 1984; Mo et al. 1991; Demarini et al. 1993; Cebula 1995; Greenblatt et al. 1996; Bissler 1998; Yoshiyama and Maki 2003; Schultz et al. 2006). Genetic analysis of the mechanism of mutagenesis suggests that these hotspot mutations, which always improve the perfection of the inverted repeat sequence, occur by template-switch reactions during normal replication (Viswanathan et al. 2000; Dutra and Lovett 2006), as proposed initially by Ripley (1982). The formation of an intrastrand hairpin structure on the nascent strand produces an alternative template for synthesis leading to mutations (Figure 2B). Alternatively, strand displacement and annealing across the replication fork at the site of the inverted repeats provides a template for synthesis of the hotspot mutation (Figure 2C) (Ripley 1982; De Boer and Ripley 1984; Rosche et al. 1995, 1997, 1998). These template-switch reactions have been postulated to occur after stalled replication since mutations in E. coli SOS-induced translesion polymerases elevate hotspot mutagenesis in the thyA gene (Dutra and Lovett 2006). Mutations in E. coli single-strand exonucleases I and VII also elevate thyA hotspot mutagenesis 30- to 50-fold (Viswanathan et al. 2000; Dutra and Lovett 2006), suggesting that the exonucleases suppress mutagenesis by degradation of displaced 3′ ends that occur during the template switch. Mutagens that stimulate such template-switch events have not been well characterized and the generality of genetic effects on the thyA hotspot has not been established for other quasipalindrome-associated mutations.
Figure 2.—
Mechanisms for mutagenesis in quasipalindromes involving replication template switching. (A) Sequence of an imperfect inverted repeat (boxed nucleotides will form base pairs in the quasipalindrome.) (B) Intramolecular template switching. Hairpin formation and templated mutation can occur by a template switch to copy one arm of the repeat, leading to perfection of the inverted repeat. A second template switch to resume normal replication produces the mutational event (shown in boldface type). Note that hairpin formation on leading and lagging strands leads to mutations on different sides of the inverted repeat.
A second class of mutational hotspots, as revealed by mutational spectra, are additions or deletions at short directly repeated DNA sequences. In the Lac repressor, lacI, gene, the repeat of (CTGG)3 accounts for two-thirds of the mutations that inactivate the gene, by deletion or addition of the four-nucleotide repeat (Farabaugh et al. 1978; Schaaper et al. 1986). Deletion hotspots also occur between more dispersed repeats, including a 17-bp nucleotide sequence with 14 bases of homology separated by 759 bases, that accounted for 60% of deletions in a lacI-lacZ gene fusion (Albertini et al. 1982). The mechanism of these events appears to be misalignment (or “slippage”) of the nascent DNA chain at the site of the repeats, as proposed first by Streisinger et al. (1966) (see review in Lovett 2004). Cellular functions that inhibit these mutations have been characterized and include single-strand DNA specific exonucleases that may act to degrade displaced DNA strands that are intermediates in the reaction (Allgood and Silhavy 1991; Bzymek et al. 1999; Feschenko et al. 2003). In addition, impairments in many replication functions in E. coli can predispose to deletion/expansion at direct repeats including mutations in the polymerase itself and proofreading subunit (Bierne et al. 1997; Saveson and Lovett 1997). There is evidence that slipped alignment mutational events occur during DNA repair reactions, as defined by a DnaK-dependent pathway in E. coli (Goldfless et al. 2006) and a Rad5, -6, -18–dependent pathway in yeast (Torres-Ramos et al. 2002), since mutations in these repair functions decrease the rate of deletion at short direct repeats.
In the set of strains described here, we have used direct or inverted repeats in the E. coli chromosomal lacZ gene to engineer reporters that specifically detect mutations by strand misalignment in the context of inverted or direct repeats. These reporter strains should be useful to ascertain whether mutagens or genetic factors enhance mutations of these important classes. We show that exonuclease I and VII deficiency elevates templated mutations in lacZ inverted repeats; this is consistent with our previously published results that show that exonuclease I and VII deficiency elevates templated mutations at inverted repeats within the thyA gene (Dutra and Lovett 2006). Template-switch mutagenesis at inverted repeats is more prevalent at the distal repeat than the proximal repeat, with respect to the direction of the leading strand of replication and the replication fork. This preference may indicate that template-switch events associated with inverted repeats occur predominantly on the leading strand of replication.
To allow comparison of these templated mutational events to simple polymerase error mutations, we have reengineered the frameshift and base substitution reporters of Cupples and Miller (1989) and Cupples et al. (1990), so that the lacZ reporter gene resides on the E. coli chromosome rather than on a plasmid. A similar set of base substitution reporters was previously engineered in the lambda attachment site by Fijalkowska et al. (1998). The chromosomal location not only improves the stability of the reporter, but also eliminates potential contributions of F plasmid-specific DNA metabolism. These may include DNA metabolism genes expressed by the plasmid, such as dinB (Kim et al. 2001), from the fact that the majority of F plasmid replication does not occur during the same phases of bacterial growth where chromosomal replication occurs (Pritchard et al. 1975), and from the rolling circle mechanism of plasmid DNA replication, which may increase both the rate of −1 frameshifts (Radicella et al. 1995) and may promote recombination (Slechta et al. 2002). Using a linked tetracycline-resistance genetic marker, the chromosomal lacZ alleles can be easily moved to other strains of interest. Using this set, we investigate the effect of nucleotide pool alterations caused by hydroxyurea (HU), an inhibitor of ribonucleotide reductase, or by inactivation of nucleoside diphosphate kinase, ndk, and by the cytosine analog zebularine.
MATERIALS AND METHODS
Growth media:
E. coli K-12 strains were routinely grown in LB broth (Miller 1992). Tetracycline, chloramphenicol, and ampicillin, when required, were added to 15, 30, and 100 μg/ml, respectively. Minimal medium employed 56/2 salts (Willetts et al. 1969), glucose, or lactose at 0.4% and 0.001% thiamine. To visualize the Lac phenotype, X-gal (40 μg/ml) and IPTG (0.1 mM) were added to plate medium on which Lac+ colonies appeared blue and Lac− were white.
Strain construction:
E. coli K-12 strains used in this study are available by request from the E. coli Genetic Stock Center (http://cgsc.biology.yale.edu/). We constructed all strains (Table 1) in the MG1655 background, using “recombineering” by lambda recombination and single-strand DNA oligonucleotides (Table 2), as described previously (Datta et al. 2006). To facilitate recovery of recombinants, recombineering was performed in an MG1655 mutS background, strain STL13725 or STL13726, that carries a Tn10 insertion near lac, mhpC281::Tn10 (zah-281::Tn10) (Singer et al. 1989; Nichols et al. 1998). We designed oligonucleotides of ∼70 nt that contained the desired mutations, transforming these by electroporation (Dower et al. 1988) into cells containing heat inducible recombineering plasmids pSIM5 or pSIM6. After heat activation and recovery, cells were plated on LB plates containing X-gal and IPTG. Colonies whose lacZ gene had been modified were identified as white colonies. Mutations (DNA sequences of sites and appropriate PCR primers are given in supporting information, Figure S1) were confirmed by sequencing, and then the altered lacZ genes were transferred to wild-type (wt) MG1655 or mutS strain STL13726 by P1 viral transduction (Miller 1992) with mhpC281::Tn10 (zah-281::Tn10), selecting tetracycline resistance. The LacZ− phenotype was confirmed by white color on X-gal IPTG plates. Deletion and quasipalindrome-associated mutational reporters were also introduced by similar P1 transduction into a strain carrying deletion mutations in exonuclease I and exonuclease VII, STL12325.
TABLE 1.
MG1655-derived strains used in this study
| Strain number | Genotypea | Origin or reference |
| MG1655 | F- rph-1 | Bachmann (1972) |
| CAG12080 | zah-281::Tn10 rph-1 | Singer et al. (1989) |
| STL13133 | zah-281::Tn10 mutS::FRT kan | Kms Transductant STL13726 × CAG12080 |
| STL13137 | zah-281::Tn10 mutS::FRT | Kms derivative of STL13133 induced by pCP1 |
| STL13726 | mutS::FRT kan | Kmr transductant of JW2703 (Baba et al. 2006) × MG1655 |
| STL13727 | mutS::FRT kan | Lab collection |
| STL12325 | xonA::FRT xseA::FRT | Lab collection |
| STL12326 | xonA::FRT xseA::FRT | Lab collection |
| STL13146 | mutS::FRT | Transformation of pSIM5 (CmR) into STL 13137 |
| STL13149 | mutS::FRT | Transformation of pSim6 (ApR) into STL13137 |
| STL 13219 | lacZ(T1384G) zah-281::Tn10 mutS::FRT | Original transformant |
| STL13222 | lacZ(G1385A) zah-281::Tn10 mutS::FRT | Original transformant |
| STL13223 | lacZ(C1384G) zah-281::Tn10 mutS::FRT | Original transformant |
| STL13898 | lacZ(C1385A) zah-281::Tn10 mutS::FRT | Original transformant |
| STL13226 | lacZ(T1385A) zah-281::Tn10 mutS::FRT | Original transformant |
| STL13228 | lacZ(A1384G) zah-281::Tn10 mutS::FRT | Original transformant |
| STL13230 | lacZ(+1G1473) zah-281::Tn10 mutS::FRT | Original transformant |
| STL14046 | lacZ(−1G1474) zah-281::Tn10 mutS::FRT | Original transformant |
| STL13177 | lacZ(−2CG1520) zah-281::Tn10 mutS::FRT | Original transformant |
| STL13181 | lacZ(+1A1569) zah-281::Tn10 mutS::FRT | Original transformant |
| STL13186 | lacZ(−1A1570) zah-281::Tn10 mutS::FRT | Original transformant |
| STL13233 | lacZ(+2CG1518) zah-281::Tn10 mutS::FRT | Original transformant |
| STL13235 | lacZ(QP3) zah-281::Tn10 mutS::FRT | Original transformant |
| STL13237 | lacZ(QP4) zah-281::Tn10 mutS::FRT | Original transformant |
| STL14454 | lacZ(QP5) zah-281::Tn10 mutS::FRT | Original transformant |
| STL15779 | lacZ(QP6) zah-281::Tn10 mutS::FRT | Original transformant |
| STL14022 | lacZ(+11) zah-281::Tn10 mutS::FRT | Original transformant |
| STL 13719 | lacZ(T1384G) zah-281::Tn10 | Tcr transductant STL13219 × MG1655 |
| STL14842 | lacZ(G1385A) zah-281::Tn10 | Tcr transductant STL13222 × MG1655 |
| STL13814 | lacZ(C1384G) zah-281::Tn10 | Tcr transductant STL13223 × MG1655 |
| STL13997 | lacZ(C1385A) zah-281::Tn10 | Tcr transductant STL13898 × MG1655 |
| STL13722 | lacZ(T1385A) zah-281::Tn10 | Tcr transductant STL13226 × MG1655 |
| STL13724 | lacZ(A1384G) zah-281::Tn10 | Tcr transductant STL13228 × MG1655 |
| STL13663 | lacZ(+1G1473) zah-281::Tn10 | Tcr transductant STL13230 × MG1655 |
| STL14095 | lacZ(−1G1474) zah-281::Tn10 | Tcr transductant STL14046 × MG1655 |
| STL13817 | lacZ(−2CG1520) zah-281::Tn10 | Tcr transductant STL13177 × MG1655 |
| STL13812 | lacZ(+1A1569) zah-281::Tn10 | Tcr transductant STL13181 × MG1655 |
| STL14172 | lacZ(−1A1570) zah-281::Tn10 | Tcr transductant STL13186 × MG1655 |
| STL14173 | lacZ(−1A1570) zah-281::Tn10 | Tcr transductant STL13186 × MG1655 |
| STL13764 | lacZ(+2CG1518) zah-281::Tn10 | Tcr transductant STL13233 × MG1655 |
| STL13766 | lacZ(QP3) zah-281::Tn10 | Tcr transductant STL13235 × MG1655 |
| STL14051 | lacZ(QP4) zah-281::Tn10 | Tcr transductant STL13237 × MG1655 |
| STL14553 | lacZ(QP5) zah-281::Tn10 | Tcr transductant STL14454 × MG1655 |
| STL15654 | lacZ(QP6) zah-281::Tn10 | Tcr transductant STL15779 × MG1655 |
| STL14025 | lacZ(+11) zah-281::Tn10 | Tcr transductant STL14022 × MG1655 |
| STL13773 | lacZ(T1384G) mutS::FRT kan zah-281::Tn10 | Tcr transductant STL13219 × STL13726 |
| STL14844 | lacZ(G1385A) mutS::FRT kan zah-281::Tn10 | Tcr transductant STL13222 × STL13726 |
| STL13777 | lacZ(C1384G) mutS::FRT kan zah-281::Tn10 | Tcr transductant STL13223 × STL13726 |
| STL14078 | lacZ(C1385A) mutS::FRT kan zah-281::Tn10 | Tcr transductant STL13898 × STL13726 |
| STL13778 | lacZ(T1385A) mutS::FRT kan zah-281::Tn10 | Tcr transductant STL13226 × STL13726 |
| STL14081 | lacZ(A1384G) mutS::FRT kan zah-281::Tn10 | Tcr transductant STL13228 × STL13726 |
| STL13779 | lacZ(+1G1473) mutS::FRT kan zah-281::Tn10 | Tcr transductant STL13230 × STL13726 |
| STL14097 | lacZ(−1G1474) mutS::FRT kan zah-281::Tn10 | Tcr transductant STL14046 × STL13726 |
| STL13770 | lacZ(−2CG1520) mutS::FRT kan zah-281::Tn10 | Tcr transductant STL13177 × STL13726 |
| STL14048 | lacZ(+1A1569) mutS::FRT kan zah-281::Tn10 | Tcr transductant STL13181 x STL13726 |
| STL14049 | lacZ(+1A1569) mutS::FRT kan zah-281::Tn10 | Tcr transductant STL13181 × STL13726 |
| STL13771 | lacZ(−1A1570) mutS::FRT kan zah-281::Tn10 | Tcr transductant STL13186 × STL13726 |
| STL13780 | lacZ(+2CG1518) mutS::FRT kan zah-281::Tn10 | Tcr transductant STL13233 × STL13726 |
| STL13781 | lacZ(QP3) mutS::FRT kan zah-281::Tn10 | Tcr transductant STL13235 × STL13726 |
| STL13783 | lacZ(QP4) mutS::FRT kan zah-281::Tn10 | Tcr transductant STL13237 × STL13726 |
| STL14778 | lacZ(QP5) mutS::FRT kan zah-281::Tn10 | Tcr transductant STL14454 × STL13726 |
| STL15655 | lacZ(QP6) mutS::FRT kan zah-281::Tn10 | Tcr transductant STL15779 × STL13726 |
| STL14027 | lacZ(+11) mutS::FRT kan zah-281::Tn10 | Tcr transductant STL14022 × STL13727 |
| STL14785 | lacZ(−2CG1520) xonA::FRT xseA::FRT zah-281::Tn10 | Tcr transductant STL13177 × STL12325 |
| STL14166 | lacZ(QP3) xonA::FRT xseA::FRT zah-281::Tn10 | Tcr transductant STL13235 × STL12326 |
| STL14167 | lacZ(QP3) xonA::FRT xseA::FRT zah-281::Tn10 | Tcr transductant STL13235 × STL12326 |
| STL14170 | lacZ(QP4) xonA::FRT xseA::FRT zah-281::Tn10 | Tcr transductant STL13237 × STL12326 |
| STL14171 | lacZ(QP4) xonA::FRT xseA::FRT zah-281::Tn10 | Tcr transductant STL13237 × STL12326 |
| STL14776 | lacZ(QP5) xonA::FRT xseA::FRT zah-281::Tn10 | Tcr transductant STL14454 × STL12326 |
| STL14187 | lacZ(+11) xonA::FRT xseA::FRT zah-281::Tn10 | Tcr transductant STL14022 × STL12325 |
| STL15823 | lacZ(QP6) xonA::FRT xseA::FRT zah-281::Tn10 | Tcr transductant STL15779 × STL12325 |
| STL15952 | lacZ(T1385A) ndk∆::FRT kan zah-281::Tn10 | Kmr transductant of JW2502 (Baba et al. 2006) × STL13722 |
| STL15954 | lacZ(+1G1473) ndk∆::FRT kan zah-281::Tn10 | Kmr transductant of JW2502 (Baba et al. 2006) × STL13663 |
| STL15956 | lacZ(QP5) ndk∆::FRT kan zah-281::Tn10 | Kmr transductant of JW2502 (Baba et al. 2006) × STL14553 |
| STL15958 | lacZ(+11) ndk∆::FRT kan zah-281::Tn10 | Kmr transductant of JW2502 (Baba et al. 2006) × STL14025 |
Unless otherwise noted, all strains are derived from MG1655 and contain the added genotype of F- rph-1 not listed.
Figure 1 gives a more comprehensive review of the mutations created in lacZ gene.
TABLE 2.
Oligonucleotides used in this study
| Oligo name | Purpose | Sequence (5′->3′) |
| lacZCC101A | Recombineering A→C primer | cacccgagtgtgatcatctggtcgctggggaattagtcaggccacggcgctaatcacgacgcgctgtatc |
| lacZCC102A | Recombineering G→A primer | cacccgagtgtgatcatctggtcgctggggaatgggtcaggccacggcgctaatcacgacgcgctgtatc |
| lacZCC103A | Recombineering G→C primer | cacccgagtgtgatcatctggtcgctggggaatcagtcaggccacggcgctaatcacgacgcgctgtatc |
| lacZCC104A | Recombineering G→T primer | cacccgagtgtgatcatctggtcgctggggaatgcgtcaggccacggcgctaatcacgacgcgctgtatc |
| lacZCC105A | Recombineering A→T primer | cacccgagtgtgatcatctggtcgctggggaatgtgtcaggccacggcgctaatcacgacgcgctgtatc |
| lacZCC106A | Recombineering A→G primer | cacccgagtgtgatcatctggtcgctggggaataagtcaggccacggcgctaatcacgacgcgctgtatc |
| lacZCC107A | Recombineering +1G primer | ctcgatccttcccgcccggtgcagtatgaaggcggggggccgacaccacggccaccgatattatttgc |
| lacZCC108A | Recombineering −1G primer | ctcgatccttcccgcccggtgcagtatgaaggggggcggagccgacaccacggccaccgatattatttgc |
| lacZCCnewA | Recombineering +2CG primer | cacggccaccgatattatttgcccgatgtacgcgcgtggatgaagaccagcccttcccggctgtgc |
| lacZCC109A | Recombineering −2CG primer | cacggccaccgatattatttgcccgatgtacgcgcgcgcgtggatgaagaccagcccttcccggctgtgc |
| lacZCC110A | Recombineering +1A primer | agcccttcccggctgtgccgaaatggtccatcaaaaatggctttcgctacctggagagacgcgcccgctg |
| lacZCC111A | Recombineering −1A primer | agcccttcccggctgtgccgaaatggtccatcaaaaaaatggctttcgctacctggagagacgcgcccgctg |
| lacZCCCon1 | PCR for cc strains forward primer | atggtgccaatgaatcgtctg |
| lacZCCCon2 | PCR for cc strains reverse primer | ccgccctgtaaacggggatactgac |
| lacZccseq1 | Sequencing for cc strains forward primer | tgatccgcgctggctaccggc |
| lacZccseq2 | Sequencing for cc strains reverse primer | acgaaacgcctgccagtattt |
| lacZdup11A | Recombineering +11 dup | gttgcagtgcacggcagatacacttgctgatgcggtgctgatgcggtgctgattacgaccgctcacgcgtggcagcatcag |
| lacZdup11Con1 | PCR for +11 dup forward primer | ctttcacagatgtggattggc |
| lacZdup11Con2 | PCR for +11 dup reverse primer | cctaatccgagccagtttaccc |
| lacZdup11seq1 | Sequencing for +11 dup forward primer | gataaaaaacaactgctgacgccgc |
| lacZdup11seq2 | Sequencing for +11dup reverse primer | cagtttacccgctctgctac |
| lacZshpQP3A | Recombineering QP3 primer A | gaatggcgaatggcgctttgcctggtttccgcggcaccggaagcggtgccggaaagctggctggagtgcg |
| lacZshpQP4A | Recombineering QP4 | gaatggcgctttgcctggtttccggcaccggaagcggtgccgcggaaagctggctggagtgcgatcttcctg |
| newQP5oligo | Recombineering QP5 | cgcagcctgaatggcgaatggcgctttgccagcttctcttccggcaccggaagcggtgccggaaagctggctgga |
| QP6ultra | Recombineering QP6 | gcgaatggcgctttgccagctttccggcaccggaagcggtgccggaatct |
| lacZQPcon1 | PCR for QP3/4/5/6 | cctggcgttacccaacttaatcgcc |
| lacZQPcon2 | PCR for QP3/4/5/6 | gtgggaacaaacggcggattgaccg |
| lacZQPseq1 | Sequencing primers QP3/4/5/6 | ttgcagcacatccccctttcgccag |
| lacZQPseq2 | Sequencing primers QP3/4/5/6 | cgtaatgggataggtcacgttggtg |
Mutation rate determination:
Reversion rates were determined for each strain using fluctuation analysis as follows. Cultures inoculated from entire single colonies in 1.5 ml LB medium were grown overnight at 37° with aeration. For hydroxyurea experiments, test tubes containing LB with 0, 3, or 4 mM hydroxyurea were inoculated with <200 cells and grown without aeration overnight at 37°. For zebularine experiments, test tubes containing LB with 0, 15, or 50 μg/ml zebularine were inoculated with <200 cells. Cells from the entire culture were recovered by microcentrifugation, washed twice with 1 ml of 56/2 buffer, and resuspended in ∼150 μl of 1 × 56/2 buffer. A small fraction <1% of the cells was subjected to serial dilution and plating on LB plates to determine number of colony forming units (cfu). The remainder were plated on minimal lactose medium containing X-gal and IPTG (LacMinXI) and incubated for 2 days at 37°. (The X-gal and IPTG in the medium are not necessary for the selection but aid in visualization of the colonies). For those strains with high reversion rates (>0.3 reversions per 108 cell divisions), serial dilutions of cultures in 56/2 were plated on LacMinXI plates. For those data sets where only a fraction of the culture was plated, the following equation was used: (where actual mutation rate = mact, observed mutation rate = mobs, and fraction plated = z) mact = mobs*(z − 1)/(z*ln(z)) as described previously (Rosche and Foster 2000). Ninety-five percent confidence intervals were also calculated (Rosche and Foster 2000). Calculation of mutation rate was performed using the Ma-Sandri-Sarkar (MSS) method (Sarkar et al. 1992) as described in Rosche and Foster (2000). We wrote a VB.net program (Table S1, K. Seier) that performs the recursive calculations necessary to determine the M value by the MSS. The program runs on Windows computers and requires a .Net 3.5 platform and OS, XP, Vista, Windows 7 or better and is also available for download at http://www.bio.brandeis.edu/msscalc/. We calculated all mutation rates to two significant figures. To verify the results of our program, we also calculated by the method of the median by Lea and Coulson (1949) and/or by the p0 method (Rosche and Foster 2000). All three calculation methods gave nearly identical results (Table S1). Confidence intervals were calculated as described by Rosche and Foster (2000).
DNA sequence determination of revertants:
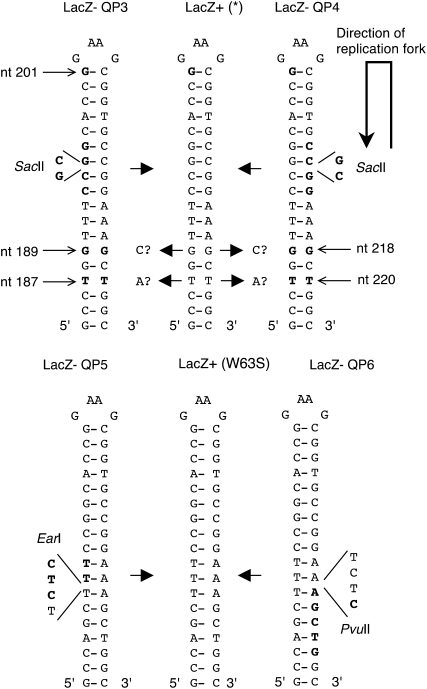
Polymerase chain reaction was performed with Taq polymerase (GoTaq Green Master Mix; Promega), with hot start at 95°, annealing 56° for 45 sec, extension 72° for 1 min, and 29–35 cycles. To confirm the specificity of the chromosomally introduced mutational reporters, the lacZ gene from four revertants from each of the strains was recovered by PCR using one of three sets of PCR primers described in Table 2. One set of primers covers all nucleotide substitutions, ±1 nt, and ±2 nt frameshifts. One set covers the quasipalindromes. One set is specific for the 11-bp duplication. Sequence analysis was performed (Molecular Biology Core Facility at the Dana Farber Cancer Institute, Boston, MA) with one of three sets of sequencing primers described in Table 2. See Figures 1, 3, and 6 for an illustration of these mutations. For the new reporters detecting deletion and quasipalindrome mutations, additional revertants were sequenced. For QP3/4/5/6 (quasipalindrome) mutation reporter strains, the inverted repeats made sequencing difficult and sequencing was performed using “high GC” conditions. For these strains, template-switch mutations were characterized by loss of the SacII (QP3 and QP4), EarI (QP5), or PvuII restriction sites (QP6). PCR products were generated from primers (Table 2) and digested for 1 hr at 37° with the restriction endonuclease (New England Biolabs), followed by resolution on 2% agarose gels and ethidium bromide staining. For QP5 and QP6, the PCR fragments include a natural EarI or PvuII site, in addition to the one affected by mutation in the quasipalindromes, such that digestion distinguishes between one or two cleavage sites.
Figure 3.—
Quasipalindrome (QP) mutational reporter design. Shown are hairpin structures for the 5′ sense strand. The orientation of lacZ is such as the replication fork proceeds in the antisense direction relative to lacZ, as shown by the arrow. Reporters QP3 and QP4 have a silent mutation, nucleotide A201G (G shown in boldface type), that strengthens the hairpin by 1 bp. A GC dinucleotide has been inserted at the 5′ side (QP3) or 3′ side (QP4) that shifts lacZ out of frame and generates a unique SacII site. Template-switch mutagenesis will remove the GC, generating an intact lacZ gene (LacZ*) and may also produce comutations G to C and T to A at nucleotides 189 and 187 for QP3 and nucleotides 218 and 220 for QP4. Second generation reporters QP5 and QP6 incorporate nucleotide T187A and G189C mutations (that produce a W63S amino acid change that does not interfere with lacZ function) and are mutated to Lac− by insertion of a TCTC sequence, generating either an EarI site (QP5) or PvuII site (QP6), shown in boldface type. Templated reversion restores Lac+, with concomitant loss of the EarI or PvuII site.
Figure 6.—
Deletion mutational reporter. An eleven nucleotide sequence was duplicated, inactivating lacZ. A deletion event, produced by “slippage” of the nascent strand on its template, gives rise to lacZ reversion.
Papillation assay:
Reversion of lacZ reporters was detected on special medium including 1% Bacto-Tryptone (BD Biosciences), 0.1% yeast extract, 0.5% NaCl, 1% lactose, and 1.5% agar to which X-gal (40 μg/ml) and IPTG (0.1 mM) had been added.
RESULTS
Quasipalindrome-associated template-switch mutational reporters:
Mutational hotspots are frequently found in imperfect inverted repeats, also known as quasipalindromes. These sequences promote templated mutations in which, during replication, one side of the quasipalindrome serves as a template for replicating the other side, resulting in an increase in symmetry for the quasipalindrome. This can occur in an intramolecular hairpin structure formed by the nascent strand or by intermolecular mispairing of the nascent strand on an alternative template consisting of the parental strand of the sister chromosome, across the replication fork (Figure 2, B and C).
To design a reporter for this type of templated mutation, we searched the lacZ sequence for inverted repeat sequences and found at nucleotides 184–223, relative to the lacZ ATG start site, a 15-bp palindrome with two imperfections and a intervening sequence of 6 nucleotides (Figure 3). This sequence was identified in a previous study of potential secondary structures in lacZ (Burkala et al. 2007). Without alteration of the amino acid sequence, we could add one additional base pair by mutation of A to G at nucleotide 201, which reduced the intervening region to 4 nucleotides. To compare potential effects of the direction of replication on mutation rates, we inserted a +GC frameshift mutation either on the 5′ strand relative to the coding arm (just after nucleotide 194) or on the 3′ side (after nucleotide 212). The +GC insertions create SacII restriction sites (5′ CCGCGG). A templated mutation would remove the GC insertion and restore the wild-type LacZ coding sequence (Figure 4). In theory, +1 or −2 frameshifts at other sites could potentially contribute to reversion in these reporters. Templated removal of GC can be distinguished from these other events by the loss of the SacII restriction site. We named these first-generation quasipalindrome reporters QP3 and QP4.
Figure 4.—
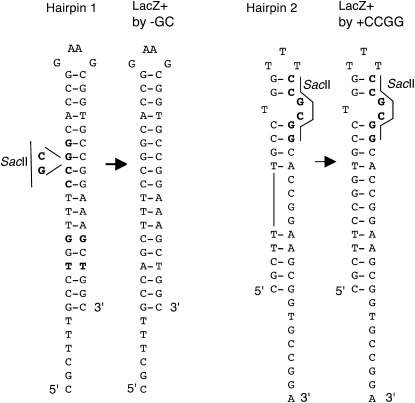
Alternative hairpin structures that template reversion in the lacZ–QP3 reporter. Hairpin 1 represents the most frequent reversion event for QP3, generating a −GC frameshift (and loss of the SacII site) with comutations, nucleotides T187A and G189C. More infrequently, hairpin 2 formation templates a +CCGG mutation, restoring LacZ function without loss of the SacII site.
The orientation of these reporters is such that the QP3 reporter templates the reversion by a simple intramolecular template switch on the nascent leading strand, whereas QP4 reports an intramolecular hairpin-associated mutation on the nascent lagging strand. On the basis of our past experience (Dutra and Lovett 2006), we expected that template switching might also produce, in addition to the −GC mutation, comutations at nucleotides 187 and 189 (T to A and G to C, respectively) for QP3 and nucleotides 218 and 220 (G to C and T to A, respectively) for QP4, which further perfect the palindrome (Figure 3). If these changes did not disturb β-galactosidase function, these might be recovered among the revertants. Comutations in QP3 generate a tryptophan to serine mutation at amino acid 63 in the coding sequence and in QP4 generate asparagine to serine and arginine to tryptophan mutations at amino acids 73 and 74.
In wild-type strains, Lac+ reversion of the QP3 reporter was detected at a rate of 1.6 × 10−9 and of the QP4 reporter somewhat lower at 3.3 × 10−10. To determine the proportion of template switch-derived events that contribute to reversion in these strains, we isolated PCR fragments encompassing the mutational region from independent revertants and subjected them to SacII digestion. For QP3, 13/15 of independently isolated revertants had lost the SacII site, indicative of a templated mutation. For QP4, 6/15 were SacII−, indicating that the majority of events that restored the Lac+ phenotype were not template-switch events. The rate of QP4 templated reversion is so low that it is similar to that detected for insertion of a nonsense codon (A214T) in the otherwise wild-type lacZ sequence (Burkala et al. 2007).
The QP3 and QP4 lacZ loci were moved to the mutS strain background to determine the effects of mismatch repair on reversion rates. Because 2-bp frameshift loops formed between nascent and template strand are potential substrates for mismatch repair, we expected an increase in the mutS background. We observed a seven- to eightfold stimulation of reversion rates of QP3 and QP4 in mutS strains, confirming this expectation (Table 3). Sequence analysis of mutS Lac+ revertants of QP3 showed that 12/17 had a templated −GC mutation, with the remainder +1 nucleotide additions upstream or downstream of the +GC. Of the 12 templated mutations, most (9/12) had comutated nucleotides 187 and 189; 3/12 had only the −GC mutation. For QP4, 10/14 had +1 frameshifts elsewhere and only 4/14 had templated the −GC, without comutation at 218 and 220. These data, combined with the reversion rates, show that templated reversion at lacZ–QP3 is about 13-fold intrinsically higher than that at lacZ QP4, with a rate of 9.2 × 10−9 for QP3 and 6.9 × 10−10 for QP4. This may be because of the orientation of the mutation relative to the replication fork (leading vs. lagging strand synthesis) or because of other unknown sequence context effects, such as the sequence in the hairpin loop region.
TABLE 3.
Reversion rates for deletion and quasipalindrome-associated mutations
| Rate of reversion per 108 cells |
|||||
| Mutation | Strain background | Rate | 95th per. (+) | 95th per. (−) | N |
| 11-bp deletion | (wt) | 6.2 | 6.6 | 5.90 | 34 |
| mutS | 4.9 | 5.4 | 4.4 | 14 | |
| xonA xseA | 7.4 | 8.1 | 6.8 | 18 | |
| QP3 | (wt) | 0.16a | 0.26 | 0.10 | 12 |
| mutS | 1.3a | 1.4 | 1.1 | 19 | |
| xonA xseA | 0.51b | 0.64 | 0.41 | 17 | |
| QP4 | (wt) | 0.033c | 0.058 | 0.019 | 19 |
| mutS | 0.24c | 0.31 | 0.19 | 15 | |
| xonA xseA | 0.37b | 0.47 | 0.29 | 19 | |
| QP5 | (wt) | 9.7b | 11 | 8.6 | 10 |
| mutS | 8.3b | 9.8 | 7.0 | 13 | |
| xonA xseA | 50b | 52 | 47 | 15 | |
| QP6 | (wt) | 1.9b | 2.2 | 1.6 | 12 |
| mutS | 2.0b | 2.4 | 1.7 | 12 | |
| xonA xseA | 76b | 80 | 73 | 16 | |
| −CG | (wt) | 0.86 | 1.08 | 0.69 | 12 |
| mutS | 130 | 130 | 120 | 24 | |
| xonA xseA | 0.60 | 0.72 | 0.50 | 18 | |
Ninety-fifth percentile (per.) determined by methd described in equations 24 and 25 in Rosche and Foster (2000).
Using the frequency of SacII− revertants (13/15) for the wild-type background and sequence analysis for the mutS background (12/17 templated), the rate of templated mutation at QP3 is calculated to be 1.4 × 10−9 for wt and 9.2 × 10−9 for the mutS background.
More than 90% reversions appear to be templated, as determined by sequence analysis and restriction assays.
Using the frequency of SacII− revertants (6/15) for the wild-type background and sequence analysis for the mutS background (4/14 templated), the rate of templated mutation at QP4 is calculated to be 1.3 × 10−10 for wt and 6.9 × 10−10 for the mutS background.
E. coli 3′ ssDNA exonucleases ExoI and ExoVII redundantly repress template-switch-generated mutation at the quasipalindrome-associated mutational hotspot in the thyA gene (Viswanathan et al. 2000; Dutra and Lovett 2006). This effect is largely specific to the hotspot; ExoI and ExoVII do not significantly affect base substitutions and have only slight effects on frameshift mutations (Viswanathan and Lovett 1998). To confirm a similar effect of the exonucleases on templated mutational reporters in the lacZ gene, we moved the QP3 and QP4 lacZ alleles to a strain with the xonA and xseA genes deleted and measured mutation rates (Table 3). Deficiency in these ssDNA exonucleases elevated LacZ reversion rates 3- and 11-fold, respectively, for the QP3 and QP4 reporters, confirming a universal role for these exonucleases in the avoidance of template-switch mutations. Furthermore, SacII digests and sequence analysis revealed that proportionately more reversions in the ssDNA Exo− background occurred by template-switch events. Not all of the template-switch events involved perfection of the quasipalindrome; some involved other, unexpected, sequence misalignments. For QP3, 15/15 isolates lacked the SacII site. Sequence analysis of an additional set of 12 isolates showed that 9/12 of the sequenced isolates had a templated −GC with comutations at 187 and 189. Two sequenced revertants had a templated +CCGG insertion after nucleotide 192, which can be produced by an alternative hairpin structure (Figure 4). The remaining revertant was produced by a triplication of a seven nucleotide 5′ GAATGGC 3′ duplicated sequence at nucleotide 165–178 (1/12). These events, although templated, restore reading frame without loss of the SacII restriction site. For QP4, 13/13 independent isolates lacked the SacII site. Seven sequenced independent isolates, different from the latter set, showed that 4 are −GC with comutations at nucleotides 218 and 220. Two are simple +1 frameshifts (+T after nucleotide 187, +C after nucleotide 189); these are not in nucleotide runs and we suspect they are templated from sequences nearby. One is a seven-nucleotide duplication 5′ GGAAGCG of nucleotides 201–207. Unlike the templated −GC frameshift detected by QP3 and QP4, the simple −CG frameshift mutation (equivalent to that detected by CC109, see construction below) was not affected by exonuclease deficiency (Table 3), confirming that the effect of exonucleases I and VII are exclusive to templated events.
Given that the QP3 and QP4 reporters did not exclusively revert via templated mutational events, we created second-generation reporters, QP5 and QP6, with features that would strengthen the hairpin and promote template switching. The Lac+ phenotype of W63S revertants (the comutated class of QP3) suggested that the hairpin could be strengthened by 2 bp by mutation to A at 187 and C at 189, without loss of LacZ function. We inserted 4 nucleotides, 5′ TCTC after nucleotide 188, generating a frameshift mutation and an EarI restriction site for QP5 or after nucleotide 216, generating a PvuII site for QP6 (Figure 3). The mutable site in these second-generation constructs was moved closer to the base of the predicted hairpin structure. The predicted hairpin structure therefore contains 11 bp before the site of the templated reversion site and 7 bp after the site (compared to 7 premutation and 9 postmutation bp for QP3 and QP4 structures). Furthermore, the four nucleotide frameshift intermediate should be refractory to the mismatch repair system (Parker and Marinus 1992), allowing a higher frequency of detection in mismatch repair-proficient, wild-type strains.
The reversion rate of the second-generation quasipalindrome reporters in the wild-type strain was indeed higher, at 9.7 per 108 cells for QP5 and 1.9 per 108 cells for QP6; reversion rates for both reporters, as expected, were unaffected by deficiency in mutS (Table 3). As was seen with the first-generation reporters, the construct that could revert via an intramolecular template switch on the leading strand (QP5) gave higher rates than for the lagging strand (QP6). Exonuclease I and VII deficiency elevated rates 5-fold for QP5 and 40-fold for QP6. A total of 24/24 independent isolates from QP5 revertants in the wild-type background had lost the EarI site, indicating a templated event. A total of 12/12 independent isolates from QP6 revertants in each of three genetic backgrounds (wt, MutS−, and Exo−) had lost the PvuII site, indicative of a templated event. Therefore both QP5 and QP6 strains specifically report quasipalindrome-associated template-switching events. We also note that, in both cases, increased reversion of the leading strand reporters (QP3 and QP5) relative to lagging strand reporters (QP4 and QP6) is lost in the Exo− genetic background.
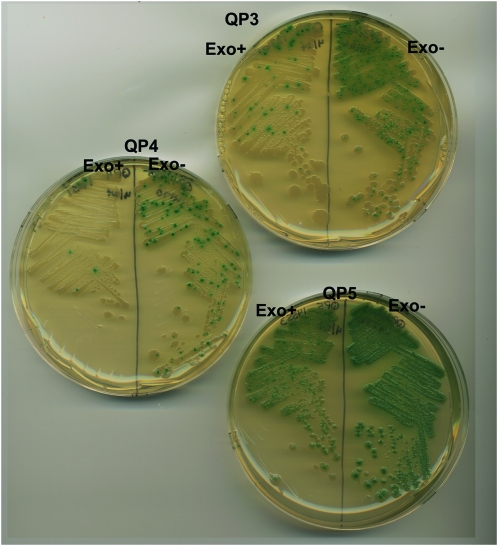
These higher reversion rates of the second generation quasipalindrome reporters are detected easily by a colony papillation assay, where Lac+ revertants within a colony are seen as blue pimples on an otherwise white colony (Figure 5). This provides a convenient visual assay for reversion in multiple independent populations and should prove useful for genetic mutator and mutagen screens for effects on this specific class of mutations.
Figure 5.—
Colony papillation assay for reversion. Supplementation of growth medium with lactose allows revertants that occur during the growth of the colony to outgrow as blue papillae, in the presence of X-gal and IPTG. Shown are wild-type and Exo− (xseA xonA) strains carrying reporters QP3, QP4, and QP5, where differences in reversion rate are clearly visible.
Deletion mutational reporter:
Deletions at short tandem repeats are also associated with mutational hotspots (Farabaugh et al. 1978; Albertini et al. 1982; Schaaper et al. 1986). To design a lacZ reporter for this type of event, we created an 11-bp tandem duplication of lacZ from nucleotides 2500–2510, which produces a frameshift in lacZ and a premature stop codon (Figure 6). Deletion of this repeat restores the wild-type reading frame and Lac+ phenotype. Like the previous reporters, this was produced by oligonucleotide recombineering and then backcrossed into wild-type and mutS strain background by P1 transduction.
Deletion of the 11-bp tandem repeat occurs at high rates, between 5 and 6 × 10−8 per cell (Table 3). Mismatch repair was not expected to change deletion rates, because the 11-base loop structures formed by slipped misalignment of template and nascent strand is too large to be detected by MutS (Parker and Marinus 1992). Indeed, mutS has no effect on reversion rate in this construct. Sequence analysis of 12 revertants confirmed that all are produced by precise loss of the 11-bp repeat. Unlike the mutations templated at inverted repeats, deletion of the 11-bp direct repeat was not significantly affected by loss of exonucleases I and VII.
Reengineering base substitution and frameshift reporters:
Using the design of Cupples and Miller (1989), we constructed a set of six lacZ mutant strains, which revert by one specific base substitution mutation. Unlike the Cupples and Miller reporters on plasmid F lac, our set employs mutations in the natural chromosomal lac locus, which would allow them to be compared to the reporters developed above. Fijalkowska et al. (1998) had previously engineered four base substitution reporters (equivalent to CC102, CC104, CC105, and CC106) into the chromosome by insertion of the lac operon in two orientations at attB (Makiela-Dzbenska et al. 2009).
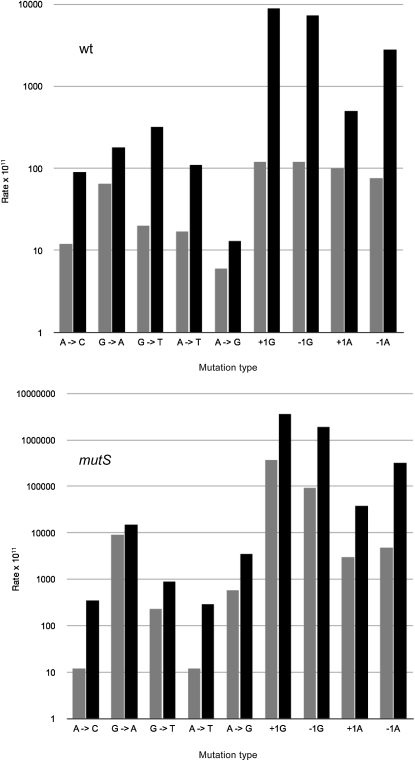
By Lac+ reversion assays, we calculated rates of each base substitution mutation in both wild-type and mutS mismatch repair-defective strains (Table 4). Reversion rates for all events were low, between 10−11 to 10−9 per cell in wild-type strains. Deficiency in mismatch repair led to a large increase in transition mutations A to G and G to A, as expected (140-fold and 197-fold, respectively), consistent with the known efficiency of mismatch repair on A:C and G:T mispairs (reviewed in Modrich and Lahue 1996). A lesser effect of mismatch repair was seen on the rates of transversion mutations, ranging from no effect to 12-fold elevation in the mutS strain background. Again, this is consistent with the relative inefficiency of mismatch repair on purine:purine or pyrimidine:pyrimidine mispairs, which are the intermediates of these mutational events (reviewed in Modrich and Lahue 1996).
TABLE 4.
Reversion rates for base substitutions and frameshift mutations
| Rate of reversion per 108 cells |
||||||||
| Wild type |
mutS |
|||||||
| Mutation | Rate | 95th per. (+) | 95th per. (−) | N | Rate | 95th per. (+) | 95th per. (−) | N |
| A→C | 0.012 | 0.033 | 0.005 | 11 | 0.012 | 0.023 | 0.006 | 14 |
| G→A | 0.065 | 0.119 | 0.036 | 12 | 9.1 | 10.0 | 8.3 | 12 |
| G→C | 0.013 | 0.029 | 0.006 | 11 | 0.055 | 0.091 | 0.033 | 9 |
| G→T | 0.020 | 0.048 | 0.008 | 14 | 0.23 | 0.36 | 0.15 | 14 |
| A→T | 0.017 | 0.027 | 0.011 | 15 | 0.012 | 0.025 | 0.006 | 10 |
| A→G | 0.006 | 0.015 | 0.002 | 11 | 0.58 | 0.70 | 0.48 | 16 |
| +1G | 0.12 | 0.17 | 0.09 | 18 | 370 | 380 | 360 | 19 |
| −1G | 0.12 | 0.16 | 0.09 | 16 | 93 | 96 | 89 | 16 |
| +2CG | < 0.006 | 0.027 | 0.001 | 14 | 0.020 | 0.055 | 0.007 | 14 |
| −2CG | 0.86 | 1.08 | 0.69 | 12 | 130 | 130 | 120 | 24 |
| +1A | 0.10 | 0.13 | 0.08 | 30 | 3.0 | 3.4 | 2.7 | 13 |
| −1A | 0.076 | 0.114 | 0.051 | 18 | 4.8 | 5.3 | 4.3 | 20 |
Ninety-fifth percentile (per.) determined by method described in equations 24 and 25 in Rosche and Foster (2000).
In addition to the base substitution reporters, we reengineered a set of frameshift mutation reporters at the chromosomal lacZ locus. Nucleotide repeats are sites of frameshift hotspot mutations, which are believed to be produced by slipped alignment of the nascent strand and its template at the site of the repeats (Streisinger et al. 1966). Cupples et al. (1990) designed a set of frameshift reporter strains that measure single nucleotide additions or deletion in runs of six guanines or six or seven adenine residues. A reporter for a dinucleotide frameshift was also generated that reports −CG in a run of five CG dinucleotide repeats. We recreated these reporter mutations in chromosomal lacZ and designed an additional reporter for a +CG in a three CG repeat run (Figure 1B).
The observed rates of single frameshift mutations in wild-type strains were higher than the base substitution mutations, in the range of 10−9 to 10−8 per cell (Table 4). The −CG mutation rate was also higher, at ∼10−8. The +CG mutation was undetectable (<10−10 per cell) in wild-type cells. The difference between the rates of addition or loss of CG in these reporters likely reflects the probability of slippage as a function of the size of the repeat run in the reporter strains, five repeats for the −CG, and three repeats for the +CG, rather than any intrinsic difference between +2 or −2 frameshift mutations.
Loss of mismatch repair capacity led to a dramatic increase in G or C frameshifts: 3100-fold elevation of +G and 780-fold elevation of −G reversion. A or T frameshifts were also enhanced by mutS but to a lesser extent: 33-fold for +A and 63-fold for −A. The dinucleotide frameshift was also enhanced by mismatch repair defects: at least 3-fold for +CG and 150-fold for −CG.
Comparison of mutant reversion rates measured in our chromosomal lac reversion assays vs. those measured by Miller et al. (2002) for those resident on F′ lac (Figure 7) indicate that, in all nine cases for which there are data in both studies, chromosomal reversion is more infrequent than reversion of the same lac allele on the F′ plasmid. This effect can be as high as a factor of 74 and is especially apparent for frameshift mutations. In mutS, mismatch repair-defective strains the effect of plasmid vs. chromosome location persisted, indicating that it likely represents differences in the occurrence of the mutation, rather than its repair. We did note, however, that mutS ameliorated somewhat the difference between plasmid and chromosome lacZ loci for the G to T transversion and the ±1G frameshifts, suggesting that some premutations are refractory to mismatch repair in the plasmid, but not in the chromosomal, context.
Figure 7.—
Comparison of lacZ reversion rates on the chromosome vs. F′ lac plasmid. Reversion rates of specific mutation events measured on the chromosome (shaded bars) compared to the same alleles carried on F′ lac (solid bars, data from Miller et al. 2002) in wild-type strains (top) or mismatch repair defective, mutS strains (bottom).
Nucleotide effects on templated mutations vs. simple mutations:
Using members of the constructed and validated set above, we assayed two conditions that might perturb DNA replication by alterations in deoxynucleotide pools. In addition, we assayed effects of a mutagenic cytosine analog.
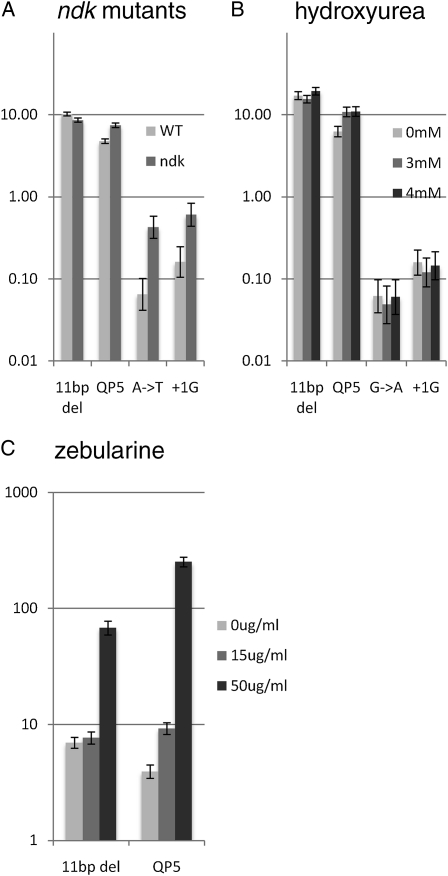
The first treatment was with hydroxyurea, an inhibitor of ribonucleotide reductase that converts ribonucleoside diphosphates to deoxynucleotide diphosphates, thereby depleting precursor pools for DNA synthesis (Timson 1975). We used sublethal doses of HU during expansion of the cultures and measured Lac+ reversion rates in the deletion and QP5 reporters, as well as a transition mutation (AT to GC) and a frameshift (+1G) reporter. Because we have observed that HU loses potency in aerated cultures, these cultures were prepared in test tubes that were capped and unshaken. We observed that HU significantly stimulated reversion, about twofold, of the QP5 reporter, at both 3 mM and 4 mM HU; no other reporter showed significant difference (Figure 8).
Figure 8.—
Reversion rates for selected reporter strains by alterations in nucleotide pools. (A) Rates of an 11-bp deletion, quasipalindrome-associated mutation (QP5), AT to TA transversion, and +1G frameshift mutations in wild-type cells (bars with light shading) compared to ndk-deficient strains (bars with dark shading). (B) Effects of hydroxyurea on deletion of an 11-bp tandem repeat and reversion rates for the quasipalindrome reporter QP5 and GC→AT transition mutation and +1G frameshift. Mock-treated cells (bars with light shading) were compared to strains treated with 3 mM (bars with medium shading) and 4 mM HU (bars with dark shading). (C) Effects of zebularine on deletion of an 11-bp tandem repeat and reversion rates for the quasipalindrome reporter QP5. Mock-treated cells (bars with light shading) were compared to strains treated with 15 μg/ml (bars with medium shading) or 50 μg/ml zebularine (bars with dark shading).
In addition, we tested the effects of nucleotide pool alterations by loss of nucleotide diphosphate kinase (ndk) activity. Such strains are viable but exhibit a strong mutator phenotype (Lu et al. 1995; Miller et al. 2002), possibly due to perturbations of nucleotide pools, including a modest elevation of dCTP and diminishment of dATP (Shen et al. 2006) or accumulation of dUTP (Nordman and Wright 2008). We transduced an ndk knockout allele into our AT to TA and +1G reporter strains [which are strongly affected by ndk on F′ lac (CC105 and CC107); Miller et al. 2002], as well as into the −11 deletion and QP5 reporters. Although we verified effects of ndk on the AT to TA transversion and +1G frameshift reversions (Figure 8), ndk had no detectable effect on either the deletion or quasipalindrome-associated mutational events.
Zebularine is a cytosine analog, lacking its amino group at C4, and has previously shown to be strongly mutagenic, especially GC to AT transitions, as assayed by mutagenesis of the rpoB gene to rifampicin resistance and using the CC reversion assay strains (Lee et al. 2004). Zebularine also stimulates mutation to rifampicin resistance in mutS strains, with the mutational spectrum resembling that without treatment, suggesting that zebularine stimulates polymerase errors of many types. We tested two doses of zebularine, 15 and 50 μg/ml, (the 15 μg/ml dose was used in Lee et al. (2004) and was found to be mutagenic and compatible with growth), on reversion of the deletion and QP5 templated mutation reporters (Figure 8). Zebularine at 15 μg/ml was detectably mutagenic for QP5 and at 50 μg/ml was strongly mutagenic for both the −11 deletion and the quasipalindrome-associated QP5 mutation, with >10-fold stimulation of the latter reporter. Although the mechanism of zebularine mutagenicity is not known, this supports the idea that zebularine increases errors during replication, including the templated class.
DISCUSSION
Using lambda Red recombination between the chromosome and transformed oligonucleotides, we created a series of lacZ reversion reporter strains that are more complete than any set previously available. Our strains detect two classes of templated events associated with mutational hotspots and include specific reporters for all six base substitutions and short frameshift mutations.
Our reporters that revert to lacZ+ by a templated mutation within a quasipalindromic sequence cause concomitant loss of a restriction site that can be conveniently assayed. These reporters were designed by adding 1 or 3 additional base pairs to a naturally occurring inverted repeat at nucleotides 184–223 of the lacZ coding region and by the insertion of 2 or 4 nucleotides to one of the repeats, moving the remainder of the lacZ coding region out of frame. Template switching, such that one side of the repeat templates the other, removes the insertion and gives rise to a Lac+ revertant (Figure 2). Two of these, QP5 and QP6, appear to revert exclusively by the template switch-generated mutation, at high frequency in the population, comparable to the rate for the natural quasipalindromic hotspot in the thyA gene. The use of these new generation reporters should facilitate identification of the genes and mutagens that specifically affect this class of mutations.
Using a reversion assay for these lacZ reporters, we confirmed that deficiency in the 3′ single-strand DNA exonucleases, ExoI and ExoVII, elevate template-switch mutations, as it does for the mutational hotspot discovered in the thyA gene of E. coli. In Exo− strains carrying the first-generation QP3 and QP4 reporters, we saw evidence of multiple types of templated events involving novel misalignments, including two hairpin structures (Figure 4) and a seven nucleotide duplication and triplication. Increasing the number of potential base pairs in the inverted repeat from 16 to 18 in the second generation reporters QP5 and QP6, elevates mutation rate one to two orders of magnitude over that of the first-generation reporters and increases the specificity of the reversion event. Our experiments with the thyA quasipalindrome mutational hotspot suggest that base-pairing potential both before and after a templated mutation in a hairpin structure is important (Dutra and Lovett 2006), although it is not clear what rules govern the efficiency of this class of mutations. Because the amino acid sequence in this region of lacZ appears to not be important for function, we hope to use these reporters in the future to dissect systemically the parameters that govern the frequency of template switching.
Other studies of quasipalindrome-associated mutation have established a strand bias with respect to replication (Rosche et al. 1997, 1998; Yoshiyama et al. 2001; Yoshiyama and Maki 2003). In most cases, it is not possible to distinguish a simple intramolecular template switch in a hairpin structure from the more complex intermolecular template switch to the sister strand across the replication fork (Figure 3). However, an intermolecular template switch during replication of the first repeat can lead to an inversion of the unpaired intervening sequence between the repeats; such events are indeed more common on the leading strand (Rosche et al. 1997). In no cases in this study did we observe such inversion. We did, however, observe higher reversion of a mutation present on the repeat distal to the replication origin, relative to that in the proximal repeat; this bias was 11-fold for the QP3/QP4 reporter pair and 5.2-fold for the QP5/QP6 reporter pair. Such a bias is consistent with a stronger propensity of mutation on the leading strand, which has been cited as a reason to support an intermolecular template-switching mechanism for all hotspots of this type (Rosche et al. 1997). Interestingly, this bias disappeared when reversion was assayed in strains lacking the 3′ ssDNA exonucleases; exonuclease deficiency elevates templated reversion of leading strand reporters QP3 and QP5 weakly (3.6- and 5.1-fold, respectively) and lagging strand reporters QP4 and QP6 more strongly (28- and 40-fold, respectively). Therefore, strand bias could reflect the higher probability that 3′ exonucleases will abort template switching on the lagging strand than on the leading strand. A potential explanation for this may be the recruitment of the exonucleases through interactions with single-strand DNA binding protein, SSB, which is more prevalent on the lagging strand. (ExoI is recruited and stimulated by an interaction with the C-terminal domain of SSB (Shereda et al. 2008); an interaction between SSB and ExoVII has not been reported).
In addition to the reporters of template switches at quasipalindromes, we designed another reversion reporter to detect deletion between 11-bp tandem directly repeated sequences. Such events were strikingly higher than base substitutions and frameshift mutations in wild-type strains, with rates approaching 10−7 per cell per generation. Although both deletions and quasipalindrome-associated mutations occur at elevated frequencies and involve misalignments of template and nascent strands, they appear to be affected differentially by genetic background and mutagens.
Because blocks to replication might stimulate the templated class of mutations, we investigated the effects of agents and genetic backgrounds that alter nucleotide pools. HU, which lowers deoxynucleotide pools via inhibition of ribonucleotide reductase, might be expected to promote nascent strand:template misalignment by polymerase dissociation or processing of stalled replication forks. We saw a modest, twofold, stimulation by HU on quasipalindrome-associated mutagenesis but not deletion at 11-bp repeats, +1G frameshift, or G to A base substitution mutations.
Loss of nucleotide diphosphate kinase activity encoded by the ndk gene has been shown to cause a strong mutator phenotype (Lu et al. 1995; Miller et al. 2002), especially for AT to TA transversions (CC105) and +1G frameshifts (CC107), which we confirm here with chromosomal lacZ reversion reporters. The source of mutagenic effect is unclear but there is modest elevation of dCTP and reduction of dATP pools in ndk mutants (Shen et al. 2006); another study correlated ndk mutagenicity with dUTP incorporation into DNA (Nordman and Wright 2008). In addition, there is evidence that ndk mutations may slow the replication fork; ndk was isolated as suppressor of the lethality associated with overinitiation of replication (conferred by the allele dnaAcos), despite a lack of effect on initiation rate (Nordman et al. 2007). Rather surprisingly, ndk did not affect the templated class of mutations, neither deletion nor quasipalindrome-associated mutations.
Zebularine, a mutagenic cytidine analog, strongly stimulates GC to AT mutations, shown by mutational spectrum analysis of rifampicin-resistant mutants in rpoB (Lee et al. 2004). Zebularine also stimulated trimethoprim-resistance mutations in thyA in this study, although sequence determination was not done to determine whether stimulation occurred at the quasipalindromic hotspot, which normally accounts for 60% of trimethoprim-resistant mutants in wild-type strains (Viswanathan et al. 2000). We show here a strong stimulation of quasipalindrome-associated templated mutations and deletions in lacZ, suggesting that zebularine may interfere with the progression of replication.
Although both deletions and quasipalindrome-associated mutations occur at elevated frequencies and involve misalignments of template and nascent strands, they appear to be affected differentially by genetic background and mutagens. Reversion of the quasipalindromic reporter QP5 was affected by zebularine and HU more strongly than the 11-bp deletion reporter. Likewise, exonuclease I and VII deficiency elevated quasipalindrome-associated mutagenesis but not deletion formation.
Using lambda Red recombination between the chromosome and transformed oligonucleotides, we recreated a set of lacZ reporters, initially designed by Cupples et al. (1990) and Cupples and Miller (1989), to reside on the chromosomal lacZ locus rather than on a F′ lac plasmid. These revert to Lac+ by specific base substitutions or by frameshift mutations in nucleotide runs and can be compared to the reporters of templated mutation, described above. Comparison of the mutation rates of F′ or chromosomal loci shows that the F′ lacZ locus is more mutable in all assays relative to the chromosomal lacZ, ranging from 2-fold to almost 80-fold. The frameshift mutations, in particular, were especially elevated in the plasmid reporter. F′ rolling circle replication during the transfer process has been implicated in the mutagenic process, known as “adaptive mutation,” where −1 frameshift mutations accumulate in nongrowing cells under lactose selection (Foster and Trimarchi 1995; Radicella et al. 1995). Rolling circle replication, even under nonselective conditions such as used in our experiments, may constitute a mutagenic process, especially for frameshift mutations. Difference between mutant frequencies of chromosomal vs. plasmid lacZ was still apparent in mismatch-deficient cells, relative to those proficient in mismatch repair, suggesting that plasmid replication may generate more mutations. Another advantage of this chromosomal set of reporters, in addition to the lower background of spontaneous mutagenesis, includes stability of the reporter construct.
This set, constructed in the genetic background of MG1655, can be used directly to screen potential mutagens. We included in the strain design a tetracycline-resistance marker tightly linked to lacZ so that individual lacZ mutant reporters can be moved to any recombination-proficient E. coli genetic background by P1 transduction to test genetic effects on specific mutations.
Acknowledgments
We thank Ken Seier for assistance in the development of the MSS Calc 2.0 program. We also thank the Advanced Bacterial Genetics Course at Cold Spring Harbor, June 2008 (John Guillinger, Michael Bednarz, Jennifer Brigati, Javier Buceta, Maureen Coleman, Linda de Vooght, Jennifer Gaines, Ivana Ivancic Bace, Kristina Jonas Anna Konings, Carey Nadell, Udi Qimron, Desmond Lun, Anne Meyer, Venugopal Sathyamoorthy, Bo Shopsin, and Bhupender Singh) and the Brandeis University BIOL155a Project Laboratory in Genetics and Genomics course, fall 2009 (Honey Nagakura, Julie Brooks, William Collins, Sandy Danh, Michelle Faits, Rebecca Gologorsky, Jawad Mirzai, Katherine Mucci, Taehyun Ryu, Claudia Vasquez, Dylan Wolman, Hannah Worchel, and Felix Ye) for work on the development of strains and methods. This work was supported by grants from the National Science Foundation (NSF) Molecular and Cellular Biosciences (MCB) 0645850 and National Institutes of Health RO1 GM051753 to S.T.L. and T32 007122 to T.S. Coursework on this project was supported in part by NSF Course, Curriculum and Laboratory Improvement grant DUE-0736995 (Brandeis University) and NSF Genes and Genomes Systems grant MCB-0919304 (Cold Spring Harbor Laboratory).
LITERATURE CITED
- Albertini A. M., Hofer M., Calos M. P., Miller J. H., 1982. On the formation of spontaneous deletion: the importance of short sequence homologies in the generation of large deletions. Cell 29: 319–328 [DOI] [PubMed] [Google Scholar]
- Allgood N. D., Silhavy T. J., 1991. Escherichia coli xonA (sbcB) mutants enhance illegitimate recombination. Genetics 127: 671–680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Au K. G., Cabrera M., Miller J. H., Modrich P., 1988. Escherichia coli mutY gene product is required for specific A-G—C.G mismatch correction. Proc. Natl. Acad. Sci. USA 85: 9163–9166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baba T., Ara T., Hasegawa M., Takai Y., Okumura Y., et al. , 2006. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol. Syst. Biol. 2: 1–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachmann B. J., 1972. Pedigrees of some mutant strains of Escherichia coli K-12. Bacteriol. Rev. 36: 525–557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bierne H., Vilette D., Ehrlich S. D., Michel B., 1997. Isolation of a dnaE mutation which enhances RecA-independent homologous recombination in the Escherichia coli chromosome. Mol. Microbiol. 24: 1225–1234 [DOI] [PubMed] [Google Scholar]
- Bissler J. J., 1998. DNA inverted repeats and human disease. Front Biosci. 3: d408–d418 [DOI] [PubMed] [Google Scholar]
- Burkala E., Reimers J. M., Schmidt K. H., Davis N., Wei P., et al. , 2007. Secondary structures as predictors of mutation potential in the lacZ gene of Escherichia coli. Microbiology 153: 2180–2189 [DOI] [PubMed] [Google Scholar]
- Bzymek M., Saveson C. J., Feschenko V. V., Lovett S. T., 1999. Slipped misalignment mechanisms of deletion formation: in vivo susceptibility to nucleases. J. Bacteriol. 181: 477–482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cebula T., 1995. Allele-specific hybridization and polymerase chain reaction in mutation analysis: the Salmonella typhimurium his paradigm, pp. 11–33 Molecular Biological Methods, Environmental Chemistry and Biological Engineering. CRC Press, Boca Raton, FL [Google Scholar]
- Cupples C. G., Cabrera M., Cruz C., Miller J. H., 1990. A set of lacZ mutations in Escherichia coli that allow rapid detection of specific frameshift mutations. Genetics 125: 275–280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cupples C. G., Miller J. H., 1989. A set of lacZ strains in Escherichia coli that allow rapid detection of each of the six base substitutions. Proc. Natl. Acad. Sci. USA 86: 5345–5349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datta S., Costantino N., Court D., 2006. A set of recombineering plasmids for gram-negative bacteria. Gene 379: 109–115 [DOI] [PubMed] [Google Scholar]
- de Boer J. G., Ripley L. S., 1984. Demonstration of the production of frameshift and base-substitution mutations by quasipalindromic DNA sequences. Proc. Natl. Acad. Sci. USA 81: 5528–5531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeMarini D. M., Bell D. A., Levine J. G., Shelton M. L., Abu-Shakra A., 1993. Molecular analysis of mutations induced at the hisD3052 allele of Salmonella by single chemicals and complex mixtures. Environ. Health Perspect. 101(Suppl 3): 207–212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dower W. J., Miller J. F., Ragsdale C. W., 1988. High efficiency transformation of E. coli by high voltage electroporation. Nucleic Acids Res. 16: 6127–6145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutra B. E., Lovett S. T., 2006. Cis and trans-acting effects on a mutational hotspot involving a replication template-switch. J. Mol. Biol. 356: 300–311 [DOI] [PubMed] [Google Scholar]
- Farabaugh P. J., Schmeissner U., Hofer M., Miller J. H., 1978. Genetic studies of the lac repressor. VII. On the molecular nature of spontaneous hotspots in the lacI gene of Escherichia coli. J. Mol. Biol. 126: 847–857 [DOI] [PubMed] [Google Scholar]
- Feschenko V. V., Rajman L. A., Lovett S. T., 2003. Stabilization of perfect and imperfect tandem repeats by single-strand DNA exonucleases. Proc. Natl. Acad. Sci. USA 100: 1134–1139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fijalkowska I. J., Jonczyk P., Tkaczyk M. M., Bialoskorska M., Schaaper R. M., 1998. Unequal fidelity of leading strand and lagging strand DNA replication on the Escherichia coli chromosome. Proc. Natl. Acad. Sci. USA 95: 10020–10025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster P. L., Trimarchi J. M., 1995. Adaptive reversion of an episomal frameshift mutation in Escherichia coli requires conjugal functions but not actual conjugation. Proc. Natl. Acad. Sci. USA 92: 5487–5490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldfless S. J., Morag A. S., Belisle K. A., Sutera V. A., Jr., Lovett S. T., 2006. DNA repeat rearrangements mediated by DnaK-dependent replication fork repair. Mol. Cell 21: 595–604 [DOI] [PubMed] [Google Scholar]
- Greenblatt M. S., Grollman A. P., Harris C. C., 1996. Deletions and insertions in the p53 tumor suppressor gene in human cancers: confirmation of the DNA polymerase slippage/misalignment model. Cancer Res. 56: 2130–2136 [PubMed] [Google Scholar]
- Kim S. R., Matsui K., Yamada M., Gruz P., Nohmi T., 2001. Roles of chromosomal and episomal dinB genes encoding DNA pol IV in targeted and untargeted mutagenesis in Escherichia coli. Mol. Genet. Genomics 266: 207–215 [DOI] [PubMed] [Google Scholar]
- Lea D. E., Coulson C. A., 1949. The distribution of the numbers of mutants in bacterial populations. J. Genet. 49: 264–285 [DOI] [PubMed] [Google Scholar]
- Lee G., Wolff E., Miller J. H., 2004. Mutagenicity of the cytidine analog zebularine in Escherichia coli. DNA Repair (Amst.) 3: 155–161 [DOI] [PubMed] [Google Scholar]
- Lopez E., Elez M., Matic I., Blazquez J., 2007. Antibiotic-mediated recombination: ciprofloxacin stimulates SOS-independent recombination of divergent sequences in Escherichia coli. Mol. Microbiol. 64: 83–93 [DOI] [PubMed] [Google Scholar]
- Lovett S. T., 2004. Encoded errors: mutations and rearrangements mediated by misalignment at repetitive DNA sequences. Mol. Microbiol. 52: 1243–1253 [DOI] [PubMed] [Google Scholar]
- Lu Q., Zhang X., Almaula N., Mathews C. K., Inouye M., 1995. The gene for nucleoside diphosphate kinase functions as a mutator gene in Escherichia coli. J. Mol. Biol. 254: 337–341 [DOI] [PubMed] [Google Scholar]
- Makiela-Dzbenska K., Jaszczur M., Banach-Orlowska M., Jonczyk P., Schaaper R. M., et al. , 2009. Role of Escherichia coli DNA polymerase I in chromosomal DNA replication fidelity. Mol. Microbiol. 74: 1114–1127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller J. H., 1992. A Short Course in Bacterial Genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- Miller J. H., Funchain P., Clendenin W., Huang T., Nguyen A., et al. , 2002. Escherichia coli strains (ndk) lacking nucleoside diphosphate kinase are powerful mutators for base substitutions and frameshifts in mismatch-repair-deficient strains. Genetics 162: 5–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mo J., Maki H., Sekiguchi M., 1991. Mutational specificity of the dnaE173 mutator associated with a defect in the catalytic subunit of DNA polymerase III of Escherichia coli. J Mol Biol 222: 925–936 [DOI] [PubMed] [Google Scholar]
- Modrich P., Lahue R., 1996. Mismatch repair in replication fidelity, genetic recombination, and cancer biology. Annu. Rev. Biochem. 65: 101–133 [DOI] [PubMed] [Google Scholar]
- Nghiem Y., Cabrera M., Cupples C. G., Miller J. H., 1988. The mutY gene: a mutator locus in Escherichia coli that generates G.C—-T.A transversions. Proc. Natl. Acad. Sci. USA 85: 2709–2713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nichols B. P., Shafiq O., Meiners V., 1998. Sequence analysis of Tn10 insertion sites in a collection of Escherichia coli strains used for genetic mapping and strain construction. J. Bacteriol. 180: 6408–6411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nordman J., Wright A., 2008. The relationship between dNTP pool levels and mutagenesis in an Escherichia coli NDP kinase mutant. Proc. Natl. Acad. Sci. USA 105: 10197–10202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nordman J., Skovgaard O., Wright A., 2007. A novel class of mutations that affect DNA replication in E. coli. Mol. Microbiol. 64: 125–138 [DOI] [PubMed] [Google Scholar]
- Parker B. O., Marinus M. G., 1992. Repair of DNA heteroduplexes containing small heterologous sequences in Escherichia coli. Proc. Natl. Acad. Sci. USA 89: 1730–1734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pritchard R. H., Chandler M. G., Collins J., 1975. Independence of F replication and chromosome replication in Escherichia coli. Mol. Gen. Genet. 138: 143–155 [DOI] [PubMed] [Google Scholar]
- Radicella J. P., Park P. U., Fox M. S., 1995. Adaptive mutation in Escherichia coli: a role for conjugation. Science 268: 418–420 [DOI] [PubMed] [Google Scholar]
- Ripley L. S., 1982. Model for the participation of quasi-palindromic DNA sequences in frameshift mutation. Proc. Natl. Acad. Sci. USA 79: 4128–4132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosche W. A., Foster P. L., 2000. Determining mutation rates in bacterial populations. Methods 20: 4–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosche W. A., Trinh T. Q., Sinden R. R., 1995. Differential DNA secondary structure-mediated deletion mutation in the leading and lagging strands. J. Bacteriol. 177: 4385–4391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosche W. A., Trinh T. Q., Sinden R. R., 1997. Leading strand specific spontaneous mutation corrects a quasipalindrome by an intermolecular strand switch mechanism. J. Mol. Biol. 269: 176–187 [DOI] [PubMed] [Google Scholar]
- Rosche W. A., Ripley L. S., Sinden R. R., 1998. Primer-template misalignments during leading strand DNA synthesis account for the most frequent spontaneous mutations in a quasipalindromic region in Escherichia coli. J. Mol. Biol. 284: 633–646 [DOI] [PubMed] [Google Scholar]
- Sarkar S., Ma W. T., Sandri G. H., 1992. On fluctuation analysis: a new, simple and efficient method for computing the expected number of mutants. Genetica 85: 173–179 [DOI] [PubMed] [Google Scholar]
- Saveson C. J., Lovett S. T., 1997. Enhanced deletion formation by aberrant DNA replication in Escherichia coli. Genetics 146: 457–470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaaper R. M., Danforth B. N., Glickman B. W., 1986. Mechanisms of spontaneous mutagenesis: an analysis of the spectrum of spontaneous mutation in the Escherichia coli lacI gene. J Mol. Biol. 189: 273–284 [DOI] [PubMed] [Google Scholar]
- Schofield M. A., Agbunag R., Miller J. H., 1992. DNA inversions between short inverted repeats in Escherichia coli. Genetics 132: 295–302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz G., Carver G., Drake J., 2006. A role for replication repair in the genesis of templated mutations. J. Mol. Biol. 358: 963–973 [DOI] [PubMed] [Google Scholar]
- Shen R., Wheeler L. J., Mathews C. K., 2006. Molecular interactions involving Escherichia coli nucleoside diphosphate kinase. J. Bioenerg. Biomembr. 38: 255–259 [DOI] [PubMed] [Google Scholar]
- Shereda R. D., Kozlov A. G., Lohman T. M., Cox M. M., Keck J. L., 2008. SSB as an organizer/mobilizer of genome maintenance complexes. Crit. Rev. Biochem. Mol. Biol. 43: 289–318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singer M., Baker T. A., Schnitzler G., Deischel S. M., Goel M., et al. , 1989. A collection of strains containing genetically linked alternating antibiotic resistance elements for genetic mapping of Escherichia coli. Microbiol. Rev. 53: 1–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slechta E. S., Harold J., Andersson D. I., Roth J. R., 2002. The effect of genomic position on reversion of a lac frameshift mutation (lacIZ33) during non-lethal selection (adaptive mutation). Mol. Microbiol. 44: 1017–1032 [DOI] [PubMed] [Google Scholar]
- Stewart J. W., Sherman F., 1974. Yeast frameshift mutants identified by sequence changes in iso-1-cytochrome C., pp. 102–127 in Molecular and Environmental Aspects of Mutagenesis, edited by Miller M. W., Charles C. Thomas Publishing, Springfield, IL [Google Scholar]
- Streisinger G., Okada Y., Emrich J., Newton J., Tsugita A., et al. , 1966. Frameshift mutations and the genetic code. Cold Spring Harbor Symp. Quant. Biol. 31: 77–86 [DOI] [PubMed] [Google Scholar]
- Timson J., 1975. Hydroxyurea. Mutat. Res. 32: 115–132 [DOI] [PubMed] [Google Scholar]
- Torres-Ramos C. A., Prakash S., Prakash L., 2002. Requirement of RAD5 and MMS2 for postreplication repair of UV-damaged DNA in Saccharomyces cerevisiae. Mol. Cell. Biol. 22: 2419–2426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viswanathan M., Lacirignola J. J., Hurley R. L., Lovett S. T., 2000. A novel mutational hotspot in a natural quasipalindrome in Escherichia coli. J. Mol. Biol. 302: 553–564 [DOI] [PubMed] [Google Scholar]
- Viswanathan M., Lovett S. T., 1998. Single-strand DNA-specific exonucleases in Escherichia coli. Roles in repair and mutation avoidance. Genetics 149: 7–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willetts N. S., Clark A. J., Low B., 1969. Genetic location of certain mutations conferring recombination deficiency in Escherichia coli. J. Bacteriol. 97: 244–249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshiyama K., Higuchi K., Matsumura H., Maki H., 2001. Directionality of DNA replication fork movement strongly affects the generation of spontaneous mutations in Escherichia coli. J. Mol. Biol. 307: 1195–1206 [DOI] [PubMed] [Google Scholar]
- Yoshiyama K., Maki H., 2003. Spontaneous hotspot mutations resistant to mismatch correction in Escherichia coli: transcription-dependent mutagenesis involving template-switching mechanisms. J. Mol. Biol. 327: 7–18 [DOI] [PubMed] [Google Scholar]