Abstract
We have studied hypoxic induction of transcription by studying the seripauperin (PAU) genes of Saccharomyces cerevisiae. Previous studies showed that PAU induction requires the depletion of heme and is dependent upon the transcription factor Upc2. We have now identified additional factors required for PAU induction during hypoxia, including Hog1, a mitogen-activated protein kinase (MAPK) whose signaling pathway originates at the membrane. Our results have led to a model in which heme and ergosterol depletion alters membrane fluidity, thereby activating Hog1 for hypoxic induction. Hypoxic activation of Hog1 is distinct from its previously characterized response to osmotic stress, as the two conditions cause different transcriptional consequences. Furthermore, Hog1-dependent hypoxic activation is independent of the S. cerevisiae general stress response. In addition to Hog1, specific components of the SAGA coactivator complex, including Spt20 and Sgf73, are also required for PAU induction. Interestingly, the mammalian ortholog of Spt20, p38IP, has been previously shown to interact with the mammalian ortholog of Hog1, p38. Taken together, our results have uncovered a previously unknown hypoxic-response pathway that may be conserved throughout eukaryotes.
CHANGES in the environmental level of molecular oxygen can have profound effects on the growth of most organisms. Oxygen is required as an electron receptor in aerobic respiration, as well as for the biosynthesis of sterols, unsaturated fatty acids (UFAs), and heme, all of which are essential cellular components (Rosenfeld and Beauvoit 2003). In contrast, oxygen can also have negative consequences when it is metabolized into reactive oxygen species that can damage cellular components (Jamieson 1998). To adapt to altered levels of oxygen in the environment, most organisms, from bacteria to humans, respond to changes in oxygen levels by extensive changes in transcription (Bunn and Poyton 1996).
Several distinct mechanisms govern how cells respond to low levels of oxygen (hypoxia) in both metazoans and microorganisms, and four have been well described. First, in metazoans, many genes are induced during hypoxia by the transcription factor, hypoxia-inducible factor (HIF) (Kaelin 2005). In the presence of oxygen, HIF is inhibited by an oxygen-dependent hydroxylation that targets HIF for degradation; however, when oxygen levels are low, this hydroxylation cannot occur and HIF accumulates to activate transcription. Second, in Rhizobia and other bacterial species, the FixJ transcription factor promotes transcription during hypoxia (Rodgers 1999; Delgado-Nixon et al. 2000). Oxygen, when present, binds to a heme molecule attached to FixL, a histidine kinase, thereby preventing it from phosphorylating and activating FixJ. Third, in the yeast Schizosaccharomyces pombe, the transcription factor, sterol regulatory element-binding protein (SREBP) is activated during hypoxia (Hughes et al. 2005). This activation is caused by a decreased level of sterols, whose biosynthesis is oxygen dependent. Like its human ortholog, SREBP is tethered to a cellular membrane and is released to the nucleus when sterol levels are low (Brown and Goldstein 1997; Delgado-Nixon et al. 2000). While there is no SREBP ortholog in Saccharomyces cerevisiae, the Upc2 transcription factor is considered a functional homolog (Vik and Rine 2001) and is one focus of this work. Finally, S. cerevisiae contains an oxygen-responsive transcription factor, Hap1, that is described below.
In S. cerevisiae, ∼400 genes respond to changes in oxygen levels. Several studies have shown that Hap1 plays a prominent role in this regulation, under both hypoxic and aerobic conditions (Zitomer and Lowry 1992; Ter Linde et al. 1999; Becerra et al. 2002; Kwast et al. 2002; Lai et al. 2005, 2006; Hickman and Winston 2007). Hap1 directly regulates many aerobic genes through activation in the presence of oxygen and repression in hypoxia (Zitomer and Lowry 1992; Hickman and Winston 2007). This switch between repression and activation is regulated through the binding of heme, which, like sterols, requires oxygen for its biosynthesis and is thus absent from hypoxic cells (Hon et al. 2003; Hickman and Winston 2007). Hap1 also regulates several hypoxic genes and does so indirectly through aerobic induction of MOT3 and ROX1, which encode transcriptional repressors of these hypoxic genes (Sertil et al. 2003; Lai et al. 2006). However, there is likely at least one other oxygen-sensing pathway in S. cerevisiae since many genes respond to oxygen levels in the absence of Hap1 (Becerra et al. 2002; Hickman and Winston 2007).
In this work, we have studied hypoxic induction in S. cerevisiae by analysis of the hypoxia-induced seripauperin (PAU) genes. The PAU genes comprise a family of 24 genes that encode proteins related to fungal cell wall proteins and are thought to be important in remodeling the cell wall during certain types of stress, including hypoxic stress (Viswanathan et al. 1994; Rachidi et al. 2000; Abramova et al. 2001; Ai et al. 2002; Luo and van Vuuren 2009). Most PAU genes are subtelomeric and previous studies have shown that they are subject to regulation both by the Sir complex and by stresses that phosphorylate Sir3 (Ai et al. 2002; Radman-Livaja et al. 2011). With respect to hypoxic induction, the focus of our studies, one study has shown that adding heme to hypoxic cells prevents PAU induction (Rachidi et al. 2000), suggesting that the PAU genes are regulated through the known heme sensor, Hap1. However, the finding that PAU induction is Rox1 independent (Rachidi et al. 2000) and data from our previous microarray experiments (Hickman and Winston 2007) suggested that the hypoxic induction of PAU genes is Hap1 independent, and we confirm that in this study. Other work has shown that the PAU genes are induced by the transcription factor, Upc2, known to activate many other hypoxic genes through an unknown mechanism (Abramova et al. 2001; Kwast et al. 2002; Wilcox et al. 2002; Lai et al. 2006; Luo and van Vuuren 2009).
Here, we have identified additional factors and conditions required for the Upc2-dependent hypoxic induction of PAU genes, including the depletion of both sterols and heme, and the Hog1 mitogen-activated protein kinase (MAPK) pathway. Microarray analysis shows that, in addition to PAU genes, several other genes involved in maintaining the cell membrane or cell wall are induced by this pathway, demonstrating that the Hog1 pathway contributes to the maintenance of cell integrity during hypoxic growth. Extensive work has shown that Hog1 is required for the response to osmotic stress (Hohmann et al. 2007; de Nadal and Posas 2010); our results are the first demonstration that Hog1 is required as part of the response to hypoxic growth. Finally, we show that the SAGA transcriptional coactivator complex is also required for the expression of PAU genes. Our data, taken together, suggest the existence of a hypoxic-response regulatory system that may be conserved throughout eukaryotes.
MATERIALS AND METHODS
Yeast strains:
All S. cerevisiae strains are listed in Table 1 and are isogenic with a GAL2+ derivative of S288C containing a repaired HAP1 allele (Winston et al. 1995; Hickman and Winston 2007). Strains were constructed by standard methods, either by crosses or by transformation (Ausubel et al. 1991). The deletion alleles were created by replacing the respective ORF with the KanMX, URA3, LEU2, HIS3 (Brachmann et al. 1998), or NatMX marker (Goldstein and McCusker 1999). All primers used for strain construction are listed in supporting information, Table S1. Strains containing the hem1Δ::KanMX allele were grown on 200 μg/ml δ-ala, except where indicated. The KanMX::GAL1pr-ERG25 allele was constructed by placing the KanMX marker and the GAL1 promoter upstream of the ERG25 ORF (Longtine et al. 1998); strains containing this allele of the essential ERG25 gene were maintained on YP galactose medium, except where indicated. The HOG1-13X-myc::KanMX allele was created by inserting 13 copies of the myc epitope tag at the C-terminal end while adding a KanMX marker (Longtine et al. 1998); this allele did not affect hypoxic PAU induction (data not shown).
TABLE 1.
Saccharomyces cerevisiae strains
| Strain | Genotype | Reference |
| FY2609 | MATaura3Δ0 his3Δ200 leu2Δ0 lys2-128δ HAP1 | Hickman and Winston (2007) |
| FY2637 | MATaura3Δ0 his3Δ200 leu2Δ0 lys2-128δ HAP1 hem1Δ0::KanMX | Hickman and Winston (2007) |
| FY2611 | MATaura3Δ0 his3Δ200 leu2Δ0 lys2-128δ hap1Δ0::KanMX | Hickman and Winston (2007) |
| FY2867 | MATaura3Δ0 his3Δ200 leu2Δ0 lys2-128δ HAP1 hap2Δ0::LEU2 | This study |
| FY2868 | MATaura3Δ0 his3Δ200 leu2Δ0 lys2-128δ hap1Δ0::KanMX hap2Δ0::LEU2 | This study |
| FY2869 | MATaura3Δ0 his3Δ200 leu2Δ0 lys2-128δ HAP1 upc2Δ0::HIS3 | This study |
| FY2870 | MATaura3Δ0 his3Δ200 leu2Δ0 lys2-128δ HAP1 KanMX-GAL1pr-ERG25 | This study |
| FY2871 | MATaura3Δ0 his3Δ200 leu2Δ0 lys2-128δ HAP1 KanMX-GAL1pr-ERG25 upc2Δ0::HIS3 | This study |
| FY2872 | MATaura3Δ0 his3Δ200 leu2Δ0 lys2-128δ HAP1 hem1Δ0::KanMX upc2Δ0::HIS3 | This study |
| FY2873 | MATaura3Δ0 his3Δ200 leu2Δ0 lys2-128δ HAP1 hog1Δ0::LEU2 | This study |
| FY2874 | MATaura3Δ0 his3Δ200 leu2Δ0 lys2-128δ HAP1 HOG1-13X-myc::KanMX | This study |
| FY2875 | MATaura3Δ0 his3Δ200 leu2Δ0 lys2-128δ HAP1 ste11Δ0::KanMX | This study |
| FY2876 | MATaura3Δ0 his3Δ200 leu2Δ0 lys2-128δ HAP1 hog1Δ0::LEU2 ste11Δ0::KanMX | This study |
| FY2877 | MATaura3Δ0 his3Δ200 leu2Δ0 lys2-128δ HAP1 pbs2Δ0::HIS3 | This study |
| FY2878 | MATaura3Δ0 his3Δ200 leu2Δ0 lys2-128δ HAP1 ssk1Δ1::HIS3 | This study |
| FY2879 | MATaura3Δ0 his3Δ200 leu2Δ0 lys2-128δ HAP1 ssk1Δ1::HIS3 ste11Δ0::KanMX | This study |
| FY2880 | MATaura3Δ0 his3Δ200 leu2Δ0 lys2-128δ HAP1 mga2Δ0::NatMX | This study |
| FY2881 | MATaura3Δ0 his3Δ200 leu2Δ0 lys2-128δ HAP1 spt23Δ0::URA3 | This study |
| FY2882 | MATaura3Δ0 his3Δ200 leu2Δ0 lys2-128δ HAP1 mga2Δ0::NatMX spt23Δ0::URA3 | This study |
| FY2883 | MATaura3Δ0 his3Δ200 leu2Δ0 lys2-128δ HAP1 spt20Δ0::URA3 | This study |
| FY2884 | MATaura3Δ0 his3Δ200 leu2Δ0 lys2-128δ HAP1 gcn5Δ::URA3 | Roberts and Winston (1997) and this study |
| FY2885 | MATaura3-52 HAP1 spt3Δ-202 | Winston and Minehart (1986) and this study |
| FY2886 | MATaura3Δ0 his3Δ200 leu2Δ0 lys2-128δ HAP1 sgf73Δ0::KanMX | Martens et al. (2005) and this study |
| FY2887 | MATaura3Δ0 his3Δ200 leu2Δ0 lys2-128δ HAP1 sus1Δ::HIS3 | This study |
| FY2888 | MATaura3Δ0 his3Δ200 leu2Δ0 lys2-128δ HAP1 sgf11Δ::KanMX | This study |
| FY2889 | MATaura3Δ0 his3Δ200 leu2Δ0 lys2-128δ HAP1 ubp8Δ::URA3 | This study |
| FY2890 | MATaura3Δ0 his3Δ200 leu2Δ0 lys2-128δ HAP1 sus1Δ::HIS3 sgf11Δ::KanMX ubp8Δ::URA3 | This study |
| FY2891 | MATaura3Δ0 his3Δ200 leu2Δ0 lys2-128δ HAP1 sgf73Δ::KanMX sus1Δ::HIS3 sgf11Δ::KanMX ubp8Δ::URA3 | This study |
| FY2892 | MATaura3Δ0 his3Δ200 leu2Δ0 lys2-128δ HAP1 nsg1Δ::HIS3 | This study |
| FY2893 | MATaura3Δ0 his3Δ200 leu2Δ0 lys2-128δ HAP1 nsg2Δ::LEU2 | This study |
| FY2894 | MATaura3Δ0 his3Δ200 leu2Δ0 lys2-128δ HAP1 nsg1Δ::HIS3 nsg2Δ::LEU2 | This study |
| FY2895 | MATaura3Δ0 his3Δ200 leu2Δ0 lys2-128δ HAP1 msn2Δ::URA3 | This study |
| FY2896 | MATaura3Δ0 his3Δ200 leu2Δ0 lys2-128δ HAP1 msn4Δ::LEU2 | This study |
| FY2897 | MATaura3Δ0 his3Δ200 leu2Δ0 lys2-128δ HAP1 msn2Δ::URA3 msn4Δ::LEU2 | This study |
| FY2898 | MATα ura3Δ0 his3Δ200 leu2Δ0 lys2-128δ HAP1 PAU5-URA3::NatMX can1Δ::MFA1pr-HIS3 | Tong et al. (2001) and this study |
Media and growth conditions:
Culture and hypoxic methods were as described (Hickman and Winston 2007), except where noted below. For all experiments, cells were grown at 30° in YP (1% yeast extract and 2% peptone) supplemented with 2% glucose or, where indicated, 2% galactose. For RNA and protein analyses, overnight saturated cultures were diluted and grown aerobically for approximately four generations to mid-log (1–2 × 107 cells/ml). For hypoxic growth, these aerobic mid-log cultures were diluted appropriately and grown again in 250-ml flasks continuously sparged with ultra-high-purity nitrogen gas at ∼3 liters/min. For analysis of mRNA levels or proteins during hypoxic growth, cells were grown for 5 hr after the shift to nitrogen unless otherwise noted. Cells were chilled on ice for 5 min before removing from nitrogen gas. Heme, in the form of hemin (BioChemika), was made up at 50 mg/ml in 1:1 (ethanol):(water with 0.1 m NaOH). Ergosterol (>95%) (Sigma) was made up at 2 mg/ml in 1:1 Tween 80:ethanol. δ-Aminolevulinate (δ-ala; Sigma) was dissolved in water at 20 mg/ml and added to the media at the indicated concentration. Importantly, except where indicated, ergosterol was not included in the media for our hypoxic experiments because it has an effect on expression of some genes, as described elsewhere in this report. For the short time points used, there was no decrease in cell viability even though the ergosterol solution is essential for long-term growth in hypoxia. The appropriate amount of the stock solution was added to cells to achieve the desired final concentration, and an equal volume of the solvent was used as a solvent-only control. For the heme and ergosterol depletion experiments in Figure 1, cells were grown for 12 hr.
Figure 1.—
Factors and conditions that control PAU induction. PAU mRNA levels were measured by Northern analysis, with SNR190 serving as the loading control. The experiments were done as described in materials and methods. (A) Upc2 is required for PAU induction. Wild-type (FY2609), upc2Δ (FY2869), hap1Δ (FY2611), hap2Δ (FY2867), and hap1Δ hap2Δ (FY2868) strains were grown in the presence (+) or absence (−) of oxygen for 5 hr. (B) Depletion of heme or ergosterol during aerobic growth leads to PAU induction. To test the effect of heme depletion, wild-type (lanes 1 and 2, FY2609) and hem1Δ (lanes 3 and 4, FY2637) strains were grown with or without 280 µg/ml δ-ala for 12 hr and PAU mRNA levels were measured. To test the effect of ergosterol depletion, wild-type (lanes 5 and 6, FY2609) and GAL1pr-ERG25 (lanes 7 and 8, FY2870) strains were grown for 12 hr with 2% galactose or 2% glucose and PAU mRNA levels were measured. (C) Addition of heme or ergosterol represses the hypoxic induction of PAU genes. A wild-type (FY2609) strain was grown aerobically (lanes 1 and 4), for 4 hr hypoxically (lanes 2 and 3), or for 8 hr hypoxically (lanes 5 and 6). Heme (at 500 µg/ml, lane 3) or ergosterol (at 20 µg/ml, lane 6) was added to the media 40 min before shifting to hypoxia. For the −heme (lane 2) and −erg (lane 5) controls, an equal volume of the solvent was added as described in materials and methods. (D) Upc2 is required for PAU induction during heme or ergosterol depletion. To test whether Upc2 is required during heme depletion, hem1Δ (FY2637, lane 1) and hem1Δ upc2Δ (FY2872, lane 2) strains were grown in the absence of δ-ala for 12 hr and PAU mRNA levels were measured. To test whether Upc2 is required during ergosterol depletion, GAL1pr-ERG25 (FY2870, lane 3) and GAL1pr-ERG25 upc2Δ (FY2871, lane 4) were each grown in glucose for 12 hr and PAU mRNA levels were measured. (E) Manipulating membrane fluidity with DMSO or glycerol affects hypoxic PAU induction. Northern analysis is shown of wild-type cells (FY2609) grown in the presence (+O2) or absence (−O2) of oxygen for 4 hr. DMSO (5%, 10%, or 15% v/v; lanes 3–5) or glycerol (5%, 10%, or 15% v/v; lanes 7–9) was added to the medium immediately before shifting cells to hypoxic growth.
Expression microarray, Northern blot analysis, and real-time PCR:
The mRNA expression of all yeast genes was analyzed by hybridization to an Agilent yeast array as described (Brauer et al. 2008). The 24 PAU genes are >95% identical in nucleotide sequence (Luo and van Vuuren 2009), and hence probes designed for one PAU gene will cross-hybridize with all the other genes. Thus, we averaged all the expression values for PAU genes into one value that represents the average PAU expression. The microarray data (Table S2) have been deposited in the NCBI Gene Expression Omnibus (GEO) (http://www.ncbi.nlm.nih.gov/geo/) database and are accessible through GEO series accession no. GSE26593.
Northern blot analysis was performed as described (Ausubel et al. 1991), using PCR-amplified and random-primed 32P-labeled probes. The primers for PCR are listed in Table S1. The Northern probe for PAU genes also detected a weak band and we showed that it was the homologous DAN1 gene by Northern analysis of a dan1Δ strain (data not shown). For all panels in all figures, a representative of at least three independent experiments is shown. SNR190 encodes a small nucleolar RNA and serves as a loading control.
For real-time PCR, RNA was isolated from cells as described above and then reverse transcribed using a poly(T) primer. The resulting cDNA was quantitated on the Stratagene (La Jolla, CA) MX3000P. Primers used are listed in Table S1. Shown are the mean and standard deviation for at least three independent experiments. Detection of UPC2 mRNA by Northern analysis was hampered because the UPC2 mRNA overlaps with a ribosomal band. Thus, real-time PCR was used to quantify UPC2 mRNA levels.
Western analysis:
Whole-cell extracts were prepared by bead lysis. Protein concentrations were measured by Bradford assay and equal amounts of extract were separated by SDS–PAGE and transferred to a nitrocellulose membrane (Invitrogen, Carlsbad, CA). The membrane was incubated overnight at 4° with primary antibodies to myc (1:1000; clone 9E10; BD Pharmingen no. 51-1485GR), total Hog1 (1:1000; polyclonal yC-20; Santa Cruz no. sc-6815), phospho-Hog1 (1:1000; clone 3D7; Cell Signaling no. 9215S), and Mbp1 [1:5000; polyclonal; New England BioLabs (Beverly, MA) no. E8030S]. The membrane was then incubated for 1 hr at room temperature with HRP-conjugated secondary antibody (1:10,000; Jackson ImmunoResearch Laboratories) and treated with Lumigen HRP substrate (GE Healthcare).
Deletion set screen for mutants unable to induce PAU genes:
To screen for mutants with altered regulation of PAU genes, we constructed a strain in which the PAU5 ORF was replaced with that of URA3, and a NatMX cassette was integrated 152 nucleotides downstream of the PAU5 stop codon to provide a closely linked selectable marker. This strain has a Ura− phenotype during aerobic growth and a Ura+ phenotype during hypoxic growth; it also showed the expected URA3 expression by Northern analysis (data not shown). The strain also contains the can1Δ::MFA1pr-HIS3 marker, to allow a screen of the S. cerevisiae deletion set (Tong et al. 2001). After crossing by the deletion set on YPD plates, diploids were selected on YPD plates containing the drugs nourseothricin (to select for the NatMX marker) and G418 (to select for the KanMX marker). The diploids were sporulated and the MATa haploids were selected on synthetic complete media lacking histidine and arginine and containing the drug canavanine. The haploids were then tested for hypoxic growth in a BBL GasPak chamber (Becton, Dickinson) on the same media containing nourseothricin, G418, and 5-fluoroorotic acid (5FOA). Under these conditions, most strains exhibit very weak growth on 5FOA due to the hypoxic expression of PAU5. Those strains that grew better were considered to be candidates for defective PAU5 expression and were further tested by making fresh deletions and testing PAU expression by Northern analysis.
RESULTS
PAU genes are strongly regulated by Upc2 but not by Hap1 or Hap2:
Previous studies have demonstrated that PAU genes are tightly regulated by oxygen levels. During aerobic growth, PAU transcription is undetectable; however, upon a shift to hypoxic conditions, PAU transcription is strongly induced (Rachidi et al. 2000; Abramova et al. 2001). Our previous microarray experiments (Hickman and Winston 2007) suggested that hypoxic induction of PAU genes can occur independently of Hap1 and other studies have shown that the induction is dependent upon Upc2 (Abramova et al. 2001; Kwast et al. 2002). To test these results in our strains, we performed Northern analysis, also testing the requirement for Hap2, another heme-responsive transcription factor that regulates many of the same genes as Hap1 (Pinkham and Guarente 1985; Lai et al. 2006). Our results show a modest requirement for Hap1 and no requirement for Hap2 for PAU activation during hypoxic growth (Figure 1A). The modest requirement for Hap1 is likely due to its requirement as a repressor of heme and ergosterol biosynthetic genes under hypoxic conditions (Hickman and Winston 2007 and see below). Further, Hap1 and Hap2 play no role in the aerobic repression of PAU genes (Figure 1A). In contrast to Hap1 and Hap2 independence, our results show a strong dependence upon Upc2 for the hypoxic induction of PAU (Figure 1A). This induction is likely direct, as there is a Upc2 DNA-binding site found in almost all PAU gene promoters (Cohen et al. 2001; Kwast et al. 2002; Luo and van Vuuren 2009). Thus, hypoxic induction of PAU expression is controlled by a Upc2-dependent pathway that is independent of Hap1 and Hap2.
PAU transcription is regulated by heme and ergosterol levels:
A previous study suggested that aerobic repression of PAU transcription requires heme (Rachidi et al. 2000). As heme is required for ergosterol biosynthesis in S. cerevisiae (Gollub et al. 1977; Lorenz and Parks 1991), we tested the roles of both heme and ergosterol in PAU regulation. First, we depleted either heme or ergosterol during aerobic growth to test the effect on aerobic repression of PAU transcription. To deplete heme during aerobic growth, we used a strain with a deletion of HEM1, required for the production of δ-ala in the first committed step of heme biosynthesis (Gollub et al. 1977). When hem1Δ cells were grown without δ-ala (and thus could not produce heme), the PAU genes were induced (Figure 1B). To deplete cells of ergosterol, we constructed a strain containing the essential ERG25 gene, required for ergosterol biosynthesis, under the control of the GAL1 promoter and shifted it from galactose, which induces the gene, to glucose, which represses the gene. Similar to heme depletion, when ERG25 is repressed, PAU transcription is induced (Figure 1B). Thus, depletion of either heme or ergosterol during aerobic growth abolishes repression of PAU transcription. As heme is required for ergosterol biosynthesis (Gollub et al. 1977; Lorenz and Parks 1991), heme depletion might induce PAU genes because it leads to ergosterol depletion. To address this possibility, we added heme to ergosterol-depleted cells during aerobic growth and found that the addition of heme blocked PAU induction (Figure S1A). Similarly, adding ergosterol to heme-depleted cells dramatically reduces PAU induction (Figure S1B). These results suggest that heme and ergosterol each are required to regulate PAU transcription. As the depletion experiments suggest that decreased levels of heme and ergosterol are necessary during hypoxic growth for PAU induction, we added exogenous heme or ergosterol to hypoxic cells. In both cases, we found that they repressed PAU induction (Figure 1C). Finally, to determine the relationship of heme or ergosterol depletion to activation by Upc2, we repeated each depletion in a upc2Δ mutant background. Our results show that neither heme nor ergosterol depletion leads to PAU induction in a upc2Δ mutant (Figure 1D), suggesting that Upc2 activation occurs downstream of the signal from heme or ergosterol depletion. Taken together, these results show that heme and ergosterol levels each play critical roles downstream of oxygen levels and upstream of Upc2 in mediating the regulation of PAU.
Membrane fluidity, but not sterol sensing, is important for PAU regulation:
Our finding that ergosterol depletion mimics hypoxic induction of PAU genes suggested that S. cerevisiae might employ a sterol-sensing system, similar to what has been described in S. pombe and mammals (Yang et al. 2002; Hughes et al. 2005). However, the results described in the previous section suggest that this is not the case as heme itself plays a role. Second, we did not find a significant role for putative sterol sensors in hypoxic PAU induction. While S. cerevisiae does not have an SREBP ortholog, it does contain two orthologs of the S. pombe and human INSIG protein, required for sterol-sensing and SREBP regulation (Yang et al. 2002; Hughes et al. 2005). We found that neither of the S. cerevisiae orthologs, Nsg1 and Nsg2 (Flury et al. 2005), plays a significant role in PAU regulation (Figure S2). In addition, Ncr1, predicted to be a sterol sensor (Malathi et al. 2004), is not required for PAU regulation (data not shown). These results suggest that direct sterol sensing, as occurs in S. pombe and humans, does not play a role in PAU induction in S. cerevisiae.
On the basis of these results, we speculated that the PAU genes may be induced in response to a more general effect on membranes, as both heme and sterols are known to decrease membrane fluidity (Lees et al. 1979; Shviro et al. 1982; Ginsburg and Demel 1984; Schmitt et al. 1993; Berg et al. 2002; Abe et al. 2009). To test directly whether changing membrane fluidity contributes to PAU induction, we employed dimethyl sulfoxide (DMSO) and glycerol, both of which are known to decrease membrane fluidity (Surewicz 1984; Lewis et al. 1994; Gurtovenko and Anwar 2007). Indeed, we found that increased levels of DMSO or glycerol repressed PAU transcription during hypoxia (Figure 1E) or sterol depletion (data not shown). This result suggests that during the hypoxic response, the increased membrane fluidity that occurs as a consequence of decreased heme and ergosterol levels is required for PAU induction.
The Hog1 MAP kinase is activated by hypoxia and is required for PAU induction:
The Hog1 MAP kinase is important for the response to osmotic stress, as well as other stresses that may affect membranes (Hohmann et al. 2007). We wondered whether Hog1 is also involved in the response to hypoxia, given our evidence that membrane changes are a key aspect of PAU induction. Indeed, we found that Hog1 plays a major role in the hypoxic induction of PAU expression, as a hog1Δ mutant exhibited little induction compared to wild type (Figure 2A). Hog1 is activated in response to osmotic stress by phosphorylation of two threonine residues (Brewster et al. 1993; Saito and Tatebayashi 2004). Using a phospho-specific antisera and Western blot analysis, we found that Hog1 is also phosphorylated under hypoxic conditions (Figure 2B). We verified the identity of Hog1 in our Western blots by showing that a myc epitope tag added to Hog1 caused a band shift to a higher molecular weight (Figure 2B).
Figure 2.—
The role of the Hog1 MAP kinase in PAU and UPC2 induction. (A) PAU hypoxic induction is impaired in a hog1Δ mutant. PAU expression was monitored by Northern blot analysis of wild-type (FY2609) or hog1Δ (FY2873) cells grown in the presence (+) or absence (−) of O2 for 5 hr. (B) Hog1 becomes phosphorylated after a shift to hypoxic growth. The expression and phosphorylation of Hog1 protein were determined by Western blot analysis of wild-type (FY2609) or HOG1-myc (FY2874) cells grown in the presence (+) or absence (−) of O2 for 5 hr, using the indicated antibodies. (C) UPC2 mRNA levels are not affected by hog1Δ. UPC2 mRNA levels were monitored by real-time PCR in wild-type (FY2609) or hog1Δ (FY2873) cells in the presence (aerobic) or absence (hypoxic) of oxygen for 5 hr.
The UPC2 gene is transcriptionally induced under hypoxia (Abramova et al. 2001) and we wanted to test whether this induction is Hog1 dependent. To do this, we measured UPC2 mRNA levels by real-time PCR. While we did confirm that UPC2 is induced by hypoxia, we found that Hog1 is not required for the induction (Figure 2C). Thus, Hog1 does not induce the PAU genes through regulation of UPC2 mRNA levels.
Other members of the Hog1 MAPK pathway are required for the hypoxic response:
The Hog1 pathway contains two independent branches upstream of Hog1, both of which are used during osmotic stress (Saito and Tatebayashi 2004; de Nadal and Posas 2010). One pathway uses two redundant MAPKKKs, Ssk2 and Ssk22, while the other pathway uses a different MAPKKK, Ste11. Both of these pathways activate the MAPKK, Pbs2, which then phosphorylates Hog1 (Brewster et al. 1993; Saito and Tatebayashi 2004), although some evidence suggests that the Ssk2/Ssk22 pathway plays a more prominent role (O’Rourke and Herskowitz 2004). To determine whether either of these pathways is required during the hypoxic response, we measured PAU induction in mutants that are defective for each pathway. To test the Ssk2/Ssk22 pathway, we used an ssk1Δ mutant. Ssk1 is an upstream kinase that phosphorylates and activates both Ssk2 and Ssk22; an ssk1Δ allele thus abrogates activation of both of these MAPKKKs (Maeda et al. 1994). Our results show that an ssk1Δ mutant exhibited the same low level of PAU induction as a hog1Δ mutant (Figure 3A), suggesting that this pathway is activated during hypoxia. In contrast, a ste11Δ mutation did not have any effect on hypoxic PAU induction (Figure 3A and Figure S3), suggesting that this pathway does not play a role in this hypoxic response. As Ste11 is also the MAPKKK for the filamentous/invasive growth and pheromone response pathways (Harris et al. 2001), our results suggest that these other pathways that signal from the cell membrane do not participate in hypoxic PAU induction. In addition, an ssk1Δste11Δ double mutant had the same defect as an ssk1Δ single mutant, again suggesting that Ste11 plays no role in the hypoxic activation of Hog1 (Figure 3A). As expected, a pbs2Δ mutant had the same defect in PAU induction as a hog1Δ strain (Figure 3B). These data show that the Ssk2/Ssk22 branch of the Hog1 pathway, but not the Ste11 branch, is required for PAU induction.
Figure 3.—
Role of the Hog1 MAPK cascade in PAU induction. (A) PAU hypoxic induction requires Ssk1 but not Ste11. Northern analysis is shown of PAU expression in wild-type (FY2609), hog1Δ (FY2873), ste11Δ (FY2875), ssk1Δ (FY2878), and ssk1Δ ste11Δ (FY2879) strains grown in the presence (+) or absence (−) of O2 for 5 hr. (B) PAU hypoxic induction requires Pbs2. Northern analysis is shown of PAU expression in wild-type (FY2609), hog1Δ (FY2873), and pbs2Δ (FY2877) strains grown in the presence (+) or absence (−) of O2 for 5 hr. (C) PAU hypoxic induction does not require Msn2 or Msn4. Northern analysis is shown of wild-type (FY2609), hog1Δ (FY2873), msn2Δ (FY2895), msn4Δ (FY2896), and msn2Δ msn4Δ (FY2897) strains grown in the presence (+) or absence (−) of O2 for 5 hr. (D) PAU hypoxic induction requires Mga2 but not Spt23. Northern analysis is shown of wild-type (FY2609), hog1Δ (FY2873), mga2Δ (FY2880), and spt23Δ (FY2881) strains grown in the presence (+) or absence (−) of O2 for 5 hr.
The Hog1-mediated hypoxic response does not require the general stress response factors Msn2 and Msn4:
Previous studies have suggested that the Hog1 pathway interacts with the general stress response pathway (Martinez-Pastor et al. 1996; Gasch et al. 2000; Rep et al. 2000; Capaldi et al. 2008). The general stress response pathway is activated by a number of environmental changes, including osmotic shock, oxidative stress, heat shock, and nutritional starvation (Martinez-Pastor et al. 1996; Schmitt and McEntee 1996; Gorner et al. 1998). Upon any one of these events, the transcription factors Msn2/Msn4 are activated and induce a transcriptional response. We tested whether Msn2 and Msn4 are required for the hypoxic induction of PAU transcription and found that in msn2Δ, msn4Δ, and msn2Δmsn4Δ mutants the hypoxic induction of PAU transcription was normal (Figure 3C). Thus, the Hog1-mediated hypoxic response is independent of the general stress response.
The ER-membrane protein, Mga2, is required for PAU induction:
The transcription factor Mga2 has been previously shown to be required for the hypoxic induction of the OLE1 gene (Zhang et al. 1999; Jiang et al. 2001, 2002). Additional experiments have shown that Mga2 is normally associated with the endoplasmic reticulum, but can be activated by ubiquitin-dependent proteolysis, under a set of conditions that includes low levels of unsaturated fatty acids (UFAs) and hypoxia (Hoppe et al. 2000; Jiang et al. 2002). UFA biosynthesis, like that of heme and sterols, is dependent on oxygen and is therefore impaired in hypoxic cells. We wanted to test whether the Mga2 pathway plays a role in hypoxic induction of PAU genes. Indeed, we have found that in an mga2Δ mutant, PAU induction is defective, equivalent to the defect in a hog1Δ mutant (Figure 3D). As MGA2 is partially redundant with the SPT23 gene, we also tested an spt23Δ mutant, but found that it did not impair PAU induction (Figure 3D). These results are similar to induction of OLE1, which requires Mga2 but not Spt23 (Jiang et al. 2001).
Hog1 distinguishes between the hypoxic and osmotic stress signals:
The demonstration that Hog1 is required for a normal hypoxic response and that this pathway requires Pbs2 and Ssk1 raised the question of whether the hypoxic pathway is distinct from that for osmotic stress, the pathway best characterized for Hog1. To examine this, we assayed the regulation of GRE2, a gene that responds to osmotic stress (Garay-Arroyo and Covarrubias 1999), and PAU genes, which respond to hypoxic conditions. Our results show that each response is specific, as GRE2 is induced only upon osmotic stress, and PAU genes are induced only upon hypoxic induction, all occurring in a Hog1-dependent manner (Figure 4A).
Figure 4.—
Hog1 distinguishes between the hypoxic and osmotic stress signals. (A) Expression of PAU mRNA during hypoxic growth and during osmotic stress. Northern analysis is shown of PAU and GRE2 mRNA levels in wild-type (FY2609) and hog1Δ (FY2873) strains. Cells were grown for the indicated times in hypoxia (lanes 1–6) or 1 m sorbitol (lanes 7–12). (B) Hog1 is activated with different kinetics under osmotic stress and during hypoxic induction. Western analysis is shown of Hog1 phosphorylation in a wild-type (FY2609) strain grown in 1 m sorbitol (lanes 1–5) or hypoxia (lanes 6–12) for the indicated times.
We then compared the kinetics of Hog1 phosphorylation during osmotic stress and hypoxic induction. We found that osmotic stress causes a dramatic peak of Hog1 phosphorylation within minutes of treatment (Figure 4B, lanes 1–5), as previously reported (Maeda et al. 1995). We also found a second peak of Hog1 that reproducibly appeared 2 hr after treatment (Figure 4B). In contrast, hypoxia led to a gradual increase in Hog1 phosphorylation, reaching a peak at 5 hr (Figure 4B, lanes 6–12 and data not shown). The kinetics of Hog1 phosphorylation in response to each treatment are consistent with the kinetics of gene induction seen in Figure 4A, strongly supporting the idea that Hog1 responds differently to hypoxia and osmotic stress.
Hog1 plays a role in the induction of several hypoxic genes:
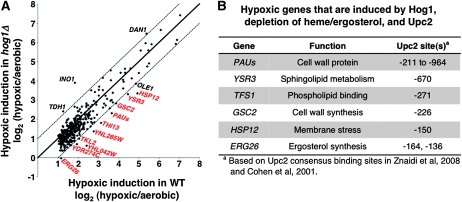
As Hog1 regulates the transcription of 50–200 genes in response to osmotic stress (Posas et al. 2000; Rep et al. 2000), we wanted to determine how many genes are regulated by Hog1 in response to hypoxia. To test this, we used expression microarrays to compare the hypoxic induction of genes in wild-type and hog1Δ strains. In addition to PAU genes, there were 9 other hypoxic genes that exhibited a more than twofold decrease in mRNA levels in a hog1Δ mutant compared to wild type under hypoxic conditions (Figure 5A, gene names in red). There were also three hypoxic genes (DAN1, INO1, and TDH1) that had increased mRNA levels in a hog1Δ mutant under hypoxic conditions; these have not yet been analyzed further.
Figure 5.—
Several hypoxic genes are dependent upon Hog1. (A) Expression microarray analysis showing hypoxic induction of genes in wild-type (FY2609, x-axis) and hog1Δ (FY2873, y-axis) strains. Only the genes hypoxically induced more than twofold in two of two experiments are shown on the plot. The solid line represents the same induction in wild-type and hog1Δ strains, while the dotted lines represent a twofold difference in induction. Genes shown in red have mRNA levels decreased by twofold or more in the hog1Δ mutant. The data point labeled “PAUs” represents the average expression of all 24 PAU genes. (B) Genes in the sterol/heme/Hog1/Upc2 pathway are listed with their function and conserved Upc2 binding site(s). The list of genes includes only those that meet three criteria: (1) regulated by O2 and Hog1 (Figure 5A), (2) induced by depletion of sterols and depletion of heme (Figure S4), and (3) contain a conserved Upc2 binding site in the promoter, on the basis of the previously determined consensus (TCGTATA or TCGTTYAG) (Cohen et al. 2001; Znaidi et al. 2008).
To test if the nine Hog1-dependent, hypoxia-induced genes that we identified are also regulated by ergosterol and heme levels, we examined a previous data set of gene expression changes upon heme and ergosterol depletion (Mnaimneh et al. 2004). Of the nine genes, five were induced in both heme and ergosterol depletion, similar to PAU genes (Figure S4). All five of these genes contain at least one Upc2 consensus binding site in the promoter region (TCGTATA or TCGTTYAG) (Cohen et al. 2001; Znaidi et al. 2008). These five genes, listed in Figure 5B, encode proteins that are involved in either cell membrane (YSR3, TFS1, HSP12, and ERG26) or cell wall (PAU and GSC2) functions, and three of them have been previously shown to be hypoxia induced: PAU (Rachidi et al. 2000; Abramova et al. 2001; Hickman and Winston 2007), GSC2 (Lai et al. 2005), and HSP12 (Becerra et al. 2002; Hickman and Winston 2007). Thus, there are several genes regulated by the Hog1-mediated hypoxic response that are likely to be important in maintaining the cell wall and cell membrane when oxygen is not present. Interestingly, part of the osmotic stress response involves decreased expression of some ERG genes (Montanes et al. 2011), perhaps to maintain proper membrane fluidity during this different form of membrane stress.
The SAGA coactivator complex is required for PAU and UPC2 induction:
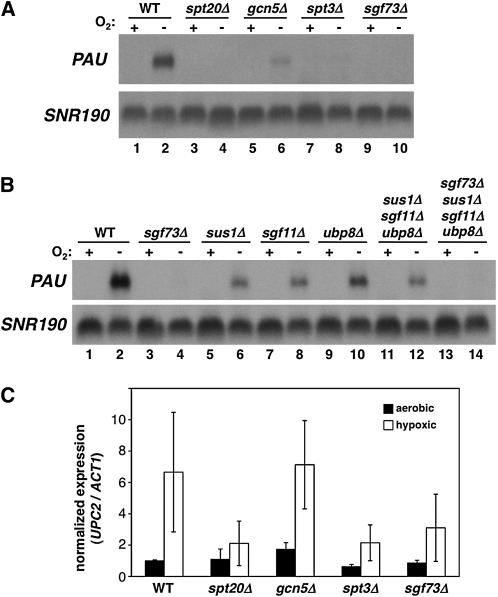
To identify other regulators of PAU expression, we screened the S. cerevisiae deletion set for mutants unable to express a PAU reporter under hypoxic conditions. This screen identified several components of the SAGA transcriptional coactivator complex, including Ada2, Gcn5, and Sgf29. We then directly tested the requirement for most nonessential SAGA genes by Northern analysis and found that the SAGA mutants with the strongest defects in PAU induction occurred in four classes of SAGA genes (Figure 6A and data not shown): those encoding core components (ADA1, SPT7, and SPT20), those required for histone acetyltransferase activity (GCN5, ADA2, and ADA3), a component required for TBP recruitment (SPT3), and a component required for the assembly of the histone deubiquitination (DUB) module of SAGA (SGF73) (see Koutelou et al. 2010 for a review).
Figure 6.—
The SAGA coactivator complex is required for PAU induction. (A) Northern analysis of PAU mRNA levels in wild-type (FY2609), spt20Δ (FY2883), gcn5Δ (FY2884), spt3Δ (FY2885), and sgf73Δ (FY2886) strains grown in the presence (+) or absence (−) of O2 for 5 hr. (B) Northern analysis of PAU mRNA levels in wild-type (FY2609), sgf73Δ (FY2886), sus1Δ (FY2887), sgf11Δ (FY2888), ubp8Δ (FY2889), sus1Δ sgf11Δ ubp8Δ (FY2890), and sgf73Δ sus1Δ sgf11Δ ubp8Δ (FY2891) strains grown in the presence (+) or absence (−) of O2 for 5 hr. (C) Real-time PCR analysis of UPC2 mRNA levels in wild-type (FY2609), spt20Δ (FY2883), gcn5Δ (FY2884), spt3Δ (FY2885), and sgf73Δ (FY2886) strains grown in the presence (aerobic) or absence (hypoxic) of oxygen for 5 hr. The high standard deviation for some samples is due to an outlier experiment that showed the same relative levels for all of the mutants.
We were particularly interested in the strong requirement for Sgf73 in PAU induction. While Sgf73 is clearly important in humans (Helmlinger et al. 2004), only a few studies have identified phenotypes for sgf73 mutants in S. cerevisiae (Shukla et al. 2006; Jordan et al. 2007; Gresham et al. 2008). Sgf73 was recently found to anchor the DUB module of SAGA within the complex (Lee et al. 2009; Rodriguez-Navarro 2009; Kohler et al. 2010; Samara et al. 2010). The DUB module contains three proteins that are required for histone deubiquitination: (1) Ubp8, the deubiquitinase; (2) Sus1, also involved in SAGA-mediated mRNA export; and (3) Sgf11, a possible structural protein. To determine whether Sgf73 plays a role in PAU expression via the DUB module, we deleted the genes encoding each of the DUB components and compared their effects, singly and in combination, to sgf73Δ’s. Our results show that even in a sus1Δsgf11Δubp8Δ triple mutant, there is still significant PAU induction, albeit reduced (Figure 6B, lanes 11 and 12). In contrast, in the sgf73Δ single mutant, or when sgf73Δ is combined with the other three mutations, there is no detectable PAU induction. These results show that Sgf73 plays a prominent role in the hypoxic induction of PAU genes and does so, at least partly, in a DUB-independent manner.
To further define the role of SAGA in the hypoxic response, we tested whether SAGA is required for UPC2 transcription. Our results show that most of the SAGA mutants tested that impair the hypoxic expression of PAU also impair the hypoxic expression of UPC2 (Figure 6C). One notable exception is gcn5Δ, which significantly reduces PAU mRNA levels but not those for UPC2. Thus, SAGA may contribute to PAU expression in two ways: (1) by regulating transcription of UPC2 in an Sgf73- and Spt3-dependent fashion and (2) by directly regulating PAU expression via Gcn5.
DISCUSSION
In this work, we have identified several factors that control the hypoxic induction of the PAU genes of S. cerevisiae. Combined with previous results concerning PAU regulation (Rachidi et al. 2000; Abramova et al. 2001; Kwast et al. 2002), our studies suggest the existence of a previously unknown hypoxic-response regulatory system that is distinct from the well-studied Hap1-dependent pathway and that requires the Hog1 MAPK pathway. Our microarray analysis has shown that Hog1 is required for the hypoxic induction of several genes in addition to the PAU genes, showing that it plays a significant role in the S. cerevisiae hypoxic response. As the mammalian Hog1 ortholog, p38, is also activated by hypoxia (Seko et al. 1997; Jin et al. 2000; Kacimi et al. 2000; Blaschke et al. 2002; Kulisz et al. 2002; Zhu et al. 2002; Emerling et al. 2005), this role may be conserved throughout eukaryotes.
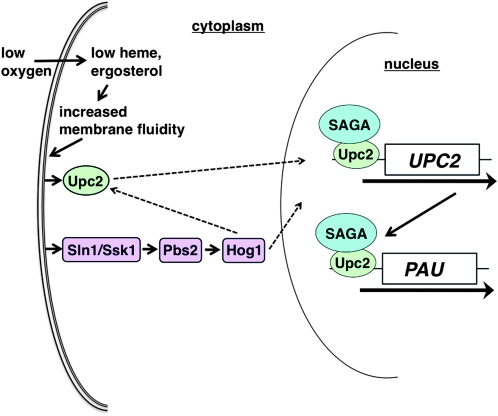
Our results suggest a regulatory framework for most of the factors now known to be required for the hypoxic induction of PAU genes (Figure 7). In this regulatory pathway, the direct consequence of decreased oxygen levels is the depletion of heme and sterols, which causes an increase in membrane fluidity. This step is supported by our result that chemicals that decrease membrane fluidity (DMSO and glycerol) block the hypoxic induction of PAU transcription and that the levels of both ergosterol (Lees et al. 1979; Berg et al. 2002; Abe et al. 2009) and heme (Kirschner-Zilber et al. 1982; Shviro et al. 1982; Ginsburg and Demel 1984; Shaklai et al. 1985; Light and Olson 1990; Balla et al. 1991; Schmitt et al. 1993) affect membrane fluidity.
Figure 7.—
A model for hypoxic induction of PAU gene expression. During hypoxic growth, when heme and ergosterol levels are low, membrane fluidity is altered. This change is proposed to activate the Hog1 MAPK cascade via the Sln1/Ssk1 pathway. In turn, activated Hog1 may induce PAU transcription directly, by translocating to the nucleus and activating transcription factors, or indirectly, by influencing signaling events in the cytoplasm, such as activation of Upc2. In addition, a SAGA-dependent, Hog1-independent pathway induces UPC2 transcription. The induction of PAU and other genes by this pathway is important in maintaining the cell wall and membrane in the absence of oxygen. Solid arrows indicate parts of the model strongly supported by our results; dashed arrows represent speculative consequences of Hog1 activation. We have left Mga2 out of the current model, pending further information on its relationship to the factors shown.
Our results showing that PAU activation requires Ssk1 and Pbs2 suggest that the change in membrane fluidity then activates the Hog1 pathway via Sln1, a transmembrane protein that responds to osmotic stress and that is part of the Sln1-Ypd1-Ssk1 phospho-relay complex that activates the Hog1 pathway (Maeda et al. 1994). Sln1 has been proposed to be a sensor of membrane turgor pressure (Reiser et al. 2003), but Sln1 may more directly be a sensor of membrane fluidity, since membrane turgor influences membrane fluidity (Yamazaki et al. 1989; Laroche et al. 2001; Hayashi and Maeda 2006; Panadero et al. 2006). Indeed, studies have shown that treatments that affect membrane fluidity, such as DMSO and low temperature, influence Hog1 signaling via the Sln1 pathway (Hayashi and Maeda 2006; Panadero et al. 2006). Once activated by changing membrane fluidity, Sln1 would initiate the MAPK cascade that leads to activation of Pbs2 and Hog1. Our results have also shown that another pathway to activate Hog1, via Sho1 and Ste11, is not involved in the hypoxic response. Past results have suggested that the Sln1 branch plays a more prominent role in Hog1 activation under certain conditions, including low-temperature activation, and the Sho1/Ste11 branch plays a distinct role in pseudohyphal growth (O’Rourke and Herskowitz 1998, 2004; Maeta et al. 2005; Hayashi and Maeda 2006; Panadero et al. 2006). Our results have provided a further distinction between these two pathways.
Once activated, Hog1 may have multiple functions in hypoxic induction and our results have not yet distinguished among these possibilities. In the osmotic stress response, Hog1 functions in several ways, with both transcriptional and cytoplasmic roles (Proft and Struhl 2004; Mettetal et al. 2008; Westfall et al. 2008; de Nadal and Posas 2010). Interestingly, the cytoplasmic roles of Hog1 are sufficient for the osmotic stress response (Westfall et al. 2008). In the hypoxic response, one obvious possibility is that Hog1 activates Upc2. While we have shown that Hog1 is not required for UPC2 transcriptional induction under hypoxic conditions, previous studies have suggested that Upc2 requires an activation step beyond transcriptional induction (Kwast et al. 2002; Davies and Rine 2006) and that it relocalizes from intracellular membranes to the nucleus upon sterol depletion (Marie et al. 2008). Thus, Hog1 may control Upc2 localization or activity by phosphorylation, the latter possibility similar to its role in activation of Sko1 during osmotic stress (Proft et al. 2001). Chromatin immunoprecipitation experiments have not yet been able to detect either Hog1 or Upc2 association with PAU promoters. However, all PAU promoters have consensus Upc2 binding sites (Kwast et al. 2002; Luo and van Vuuren 2009), suggesting that Upc2 acts directly on PAU induction and that the lack of a ChIP signal is due to the technical complication of multiple copies of PAU promoters. Future experiments will address the levels at which Hog1 controls the hypoxic response.
The role of the SAGA transcriptional coactivator in hypoxic induction may involve both Hog1-independent and Hog1-dependent activities. One role of SAGA must be independent of Hog1 activity, as SAGA is required for UPC2 transcriptional induction, while Hog1 is not. However, SAGA may also function directly at PAU promoters, possibly recruited by Upc2, where SAGA components could be regulated more directly by Hog1 in response to changing membrane fluidity. Indeed, there is strong evidence for connections between stress, Hog1, and SAGA: (1) SAGA-dependent genes are mainly those regulated by stress (Huisinga and Pugh 2004), (2) SAGA is required for Hog1-dependent gene expression in response to osmotic stress (Proft and Struhl 2002; Zapater et al. 2007), and (3) during mouse development, the SAGA subunit Spt20 interacts with p38, the mammalian Hog1 ortholog (Zohn et al. 2006; Wang et al. 2008; Nagy et al. 2009). Whatever the exact role of SAGA in hypoxic induction, it is of interest that its activity during hypoxic induction is strongly dependent upon the SAGA component, Sgf73. This role of Sgf73 is at least partially independent of its well-characterized requirement for the assembly and activity of the histone deubiquitylase activity of SAGA (Lee et al. 2009; Rodriguez-Navarro 2009; Kohler et al. 2010; Samara et al. 2010). Other studies have shown that Sgf73 is also required for preinitiation complex formation at some SAGA-dependent promoters (Shukla et al. 2006) and that Sgf73 can bind to nucleosomes (Bonnet et al. 2010). Further analysis of Sgf73 in the hypoxic response may reveal whether these or additional Sgf73 functions are important during the hypoxic response.
While both osmotic stress and hypoxic induction require Hog1 activation, there are clear differences in these responses. There is a large difference in the kinetics of Hog1 activation with respect to osmotic stress and hypoxic induction. This is likely due to the distinct ways that these environmental changes affect membrane fluidity. Osmotic stress rapidly changes the membrane turgor pressure and therefore likely changes the fluidity almost immediately. In contrast, hypoxia causes sterol and heme depletion, with a change in membrane fluidity probably occurring more slowly, as the dilution of heme and ergosterol is required to change membrane fluidity. The kinetics of Hog1 activation correlate with the induction of the respective transcriptional programs. In addition, although both pathways depend upon Hog1, they have distinct transcriptional outputs, as the PAU genes respond solely to hypoxia while GRE2 responds solely to osmotic stress. This result strongly suggests that there are additional components that are distinctly required for each response.
To conclude, our results have shown that the Hog1 MAPK pathway, SAGA, and other factors are required for the hypoxic induction of several genes in S. cerevisiae. These genes likely help cells adapt to an environment with little or no oxygen, where they are unable to make heme and sterols and hence unable to properly maintain the membrane. Most of these genes were previously shown to be induced by different stresses, such as changing temperature, that cause damage to membranes and are all involved in protecting and maintaining cell membrane and cell wall components (Viswanathan et al. 1994; Mazur et al. 1995; Mao et al. 1997; Mandala et al. 1998; Rachidi et al. 2000; Sales et al. 2000; Abramova et al. 2001; Swain et al. 2002; Caesar and Blomberg 2004; Serrano et al. 2006; Pacheco et al. 2009). In addition to conservation of heme, sterols, the Hog1 pathway, and SAGA, at least three of the induced genes (YSR3, ERG26, and TFS1) are conserved in humans and in most eukaryotes. We thus speculate that parts of this pathway also function in metazoans and play important roles in the adaptation of cells and tissues to changing oxygen levels.
Acknowledgments
We are grateful to David Botstein for his support of this work. We thank Patrick Gibney, Dominique Helmlinger, and Christine Kiely for helpful comments on the manuscript and Lisa Laprade for technical help with the initial experiments. M.J.H. was funded by a fellowship from the National Institutes of Health (NIH) (F32GM71102). This work was supported by grants from the NIH (GM45720 to F.W., GM046406 to David Botstein, and P50GM071508 to the Center for Quantitative Biology at Princeton University).
LITERATURE CITED
- Abe F., Usui K., Hiraki T., 2009. Fluconazole modulates membrane rigidity, heterogeneity, and water penetration into the plasma membrane in Saccharomyces cerevisiae. Biochemistry 48: 8494–8504 [DOI] [PubMed] [Google Scholar]
- Abramova N. E., Cohen B. D., Sertil O., Kapoor R., Davies K. J., et al. , 2001. Regulatory mechanisms controlling expression of the DAN/TIR mannoprotein genes during anaerobic remodeling of the cell wall in Saccharomyces cerevisiae. Genetics 157: 1169–1177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ai W., Bertram P. G., Tsang C. K., Chan T. F., Zheng X. F., 2002. Regulation of subtelomeric silencing during stress response. Mol. Cell 10: 1295–1305 [DOI] [PubMed] [Google Scholar]
- Ausubel F. M., Brent R., Kingston R. E., Moore D. D., Seidman J. G., et al. , 1991. Current Protocols in Molecular Biology. Greene Publishing and Wiley-Interscience, New York [Google Scholar]
- Balla G., Vercellotti G. M., Muller-Eberhard U., Eaton J., Jacob H. S., 1991. Exposure of endothelial cells to free heme potentiates damage mediated by granulocytes and toxic oxygen species. Lab. Invest. 64: 648–655 [PubMed] [Google Scholar]
- Becerra M., Lombardia-Ferreira L. J., Hauser N. C., Hoheisel J. D., Tizon B., et al. , 2002. The yeast transcriptome in aerobic and hypoxic conditions: effects of hap1, rox1, rox3 and srb10 deletions. Mol. Microbiol. 43: 545–555 [DOI] [PubMed] [Google Scholar]
- Berg J. M., Tymoczko J. L., Stryer L., 2002. Biochemistry. W. H. Freeman, New York [Google Scholar]
- Blaschke F., Stawowy P., Goetze S., Hintz O., Grafe M., et al. , 2002. Hypoxia activates beta(1)-integrin via ERK 1/2 and p38 MAP kinase in human vascular smooth muscle cells. Biochem. Biophys. Res. Commun. 296: 890–896 [DOI] [PubMed] [Google Scholar]
- Bonnet J., Wang Y. H., Spedale G., Atkinson R. A., Romier C., et al. , 2010. The structural plasticity of SCA7 domains defines their differential nucleosome-binding properties. EMBO Rep. 11: 612–618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brachmann C. B., Davies A., Cost G. J., Caputo E., Li J., et al. , 1998. Designer deletion strains derived from Saccharomyces cerevisiae S288C: a useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast 14: 115–132 [DOI] [PubMed] [Google Scholar]
- Brauer M. J., Huttenhower C., Airoldi E. M., Rosenstein R., Matese J. C., et al. , 2008. Coordination of growth rate, cell cycle, stress response, and metabolic activity in yeast. Mol. Biol. Cell 19: 352–367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brewster J. L., de Valoir T., Dwyer N. D., Winter E., Gustin M. C., 1993. An osmosensing signal transduction pathway in yeast. Science 259: 1760–1763 [DOI] [PubMed] [Google Scholar]
- Brown M. S., Goldstein J. L., 1997. The SREBP pathway: regulation of cholesterol metabolism by proteolysis of a membrane-bound transcription factor. Cell 89: 331–340 [DOI] [PubMed] [Google Scholar]
- Bunn H. F., Poyton R. O., 1996. Oxygen sensing and molecular adaptation to hypoxia. Physiol. Rev. 76: 839–885 [DOI] [PubMed] [Google Scholar]
- Caesar R., Blomberg A., 2004. The stress-induced Tfs1p requires NatB-mediated acetylation to inhibit carboxypeptidase Y and to regulate the protein kinase A pathway. J. Biol. Chem. 279: 38532–38543 [DOI] [PubMed] [Google Scholar]
- Capaldi A. P., Kaplan T., Liu Y., Habib N., Regev A., et al. , 2008. Structure and function of a transcriptional network activated by the MAPK Hog1. Nat. Genet. 40: 1300–1306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen B. D., Sertil O., Abramova N. E., Davies K. J., Lowry C. V., 2001. Induction and repression of DAN1 and the family of anaerobic mannoprotein genes in Saccharomyces cerevisiae occurs through a complex array of regulatory sites. Nucleic Acids Res. 29: 799–808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies B. S., Rine J., 2006. A role for sterol levels in oxygen sensing in Saccharomyces cerevisiae. Genetics 174: 191–201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delgado-Nixon V. M., Gonzalez G., Gilles-Gonzalez M.-A., 2000. Dos, a heme-binding PAS protein from Escherichia coli, is a direct oxygen sensor. Biochemistry 39: 2685–2691 [DOI] [PubMed] [Google Scholar]
- de Nadal E., Posas F., 2010. Multilayered control of gene expression by stress-activated protein kinases. EMBO J. 29: 4–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emerling B. M., Platanias L. C., Black E., Nebreda A. R., Davis R. J., et al. , 2005. Mitochondrial reactive oxygen species activation of p38 mitogen-activated protein kinase is required for hypoxia signaling. Mol. Cell. Biol. 25: 4853–4862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flury I., Garza R., Shearer A., Rosen J., Cronin S., et al. , 2005. INSIG: a broadly conserved transmembrane chaperone for sterol-sensing domain proteins. EMBO J. 24: 3917–3926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garay-Arroyo A., Covarrubias A. A., 1999. Three genes whose expression is induced by stress in Saccharomyces cerevisiae. Yeast 15: 879–892 [DOI] [PubMed] [Google Scholar]
- Gasch A. P., Spellman P. T., Kao C. M., Carmel-Harel O., Eisen M. B., et al. , 2000. Genomic expression programs in the response of yeast cells to environmental changes. Mol. Biol. Cell 11: 4241–4257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ginsburg H., Demel R. A., 1984. Interactions of hemin, antimalarial drugs and hemin-antimalarial complexes with phospholipid monolayers. Chem. Phys. Lipids 35: 331–347 [DOI] [PubMed] [Google Scholar]
- Goldstein A. L., McCusker J. H., 1999. Three new dominant drug resistance cassettes for gene disruption in Saccharomyces cerevisiae. Yeast 15: 1541–1553 [DOI] [PubMed] [Google Scholar]
- Gollub E. G., Liu K. P., Dayan J., Adlersberg M., Sprinson D. B., 1977. Yeast mutants deficient in heme biosynthesis and a heme mutant additionally blocked in cyclization of 2,3-oxidosqualene. J. Biol. Chem. 252: 2846–2854 [PubMed] [Google Scholar]
- Gorner W., Durchschlag E., Martinez-Pastor M. T., Estruch F., Ammerer G., et al. , 1998. Nuclear localization of the C2H2 zinc finger protein Msn2p is regulated by stress and protein kinase A activity. Genes Dev. 12: 586–597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gresham D., Desai M. M., Tucker C. M., Jenq H. T., Pai D. A., et al. , 2008. The repertoire and dynamics of evolutionary adaptations to controlled nutrient-limited environments in yeast. PLoS Genet. 4: e1000303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurtovenko A. A., Anwar J., 2007. Modulating the structure and properties of cell membranes: the molecular mechanism of action of dimethyl sulfoxide. J. Phys. Chem. B 111: 10453–10460 [DOI] [PubMed] [Google Scholar]
- Harris K., Lamson R. E., Nelson B., Hughes T. R., Marton M. J., et al. , 2001. Role of scaffolds in MAP kinase pathway specificity revealed by custom design of pathway-dedicated signaling proteins. Curr. Biol. 11: 1815–1824 [PubMed] [Google Scholar]
- Hayashi M., Maeda T., 2006. Activation of the HOG pathway upon cold stress in Saccharomyces cerevisiae. J. Biochem. 139: 797–803 [DOI] [PubMed] [Google Scholar]
- Helmlinger D., Hardy S., Sasorith S., Klein F., Robert F., et al. , 2004. Ataxin-7 is a subunit of GCN5 histone acetyltransferase-containing complexes. Hum. Mol. Genet. 13: 1257–1265 [DOI] [PubMed] [Google Scholar]
- Hickman M. J., Winston F., 2007. Heme levels switch the function of Hap1 of Saccharomyces cerevisiae between transcriptional activator and transcriptional repressor. Mol. Cell. Biol. 27: 7414–7424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hohmann S., Krantz M., Nordlander B., 2007. Yeast osmoregulation. Methods Enzymol. 428: 29–45 [DOI] [PubMed] [Google Scholar]
- Hon T., Dodd A., Dirmeier R., Gorman N., Sinclair P. R., et al. , 2003. A mechanism of oxygen sensing in yeast. Multiple oxygen-responsive steps in the heme biosynthetic pathway affect Hap1 activity. J. Biol. Chem. 278: 50771–50780 [DOI] [PubMed] [Google Scholar]
- Hoppe T., Matuschewski K., Rape M., Schlenker S., Ulrich H. D., et al. , 2000. Activation of a membrane-bound transcription factor by regulated ubiquitin/proteasome-dependent processing. Cell 102: 577–586 [DOI] [PubMed] [Google Scholar]
- Hughes A. L., Todd B. L., Espenshade P. J., 2005. SREBP pathway responds to sterols and functions as an oxygen sensor in fission yeast. Cell 120: 831–842 [DOI] [PubMed] [Google Scholar]
- Huisinga K. L., Pugh B. F., 2004. A genome-wide housekeeping role for TFIID and a highly regulated stress-related role for SAGA in Saccharomyces cerevisiae. Mol. Cell 13: 573–585 [DOI] [PubMed] [Google Scholar]
- Jamieson D. J., 1998. Oxidative stress responses of the yeast Saccharomyces cerevisiae. Yeast 14: 1511–1527 [DOI] [PubMed] [Google Scholar]
- Jiang Y., Vasconcelles M. J., Wretzel S., Light A., Martin C. E., et al. , 2001. MGA2 is involved in the low-oxygen response element-dependent hypoxic induction of genes in Saccharomyces cerevisiae. Mol. Cell. Biol. 21: 6161–6169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Y., Vasconcelles M. J., Wretzel S., Light A., Gilooly L., et al. , 2002. Mga2p processing by hypoxia and unsaturated fatty acids in Saccharomyces cerevisiae: impact on LORE-dependent gene expression. Eukaryot. Cell 1: 481–490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin N., Hatton N., Swartz D. R., Xia X., Harrington M. A., et al. , 2000. Hypoxia activates jun-N-terminal kinase, extracellular signal-regulated protein kinase, and p38 kinase in pulmonary arteries. Am. J. Respir. Cell. Mol. Biol. 23: 593–601 [DOI] [PubMed] [Google Scholar]
- Jordan P. W., Klein F., Leach D. R., 2007. Novel roles for selected genes in meiotic DNA processing. PLoS Genet. 3: e222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kacimi R., Chentoufi J., Honbo N., Long C. S., Karliner J. S., 2000. Hypoxia differentially regulates stress proteins in cultured cardiomyocytes: role of the p38 stress-activated kinase signaling cascade, and relation to cytoprotection. Cardiovasc. Res. 46: 139–150 [DOI] [PubMed] [Google Scholar]
- Kaelin W. G., Jr, 2005. The von Hippel-Lindau protein, HIF hydroxylation, and oxygen sensing. Biochem. Biophys. Res. Commun. 338: 627–638 [DOI] [PubMed] [Google Scholar]
- Kirschner-Zilber I., Rabizadeh E., Shaklai N., 1982. The interaction of hemin and bilirubin with the human red cell membrane. Biochim. Biophys. Acta 690: 20–30 [DOI] [PubMed] [Google Scholar]
- Kohler A., Zimmerman E., Schneider M., Hurt E., Zheng N., 2010. Structural basis for assembly and activation of the heterotetrameric SAGA histone H2B deubiquitinase module. Cell 141: 606–617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koutelou E., Hirsch C. L., Dent S. Y., 2010. Multiple faces of the SAGA complex. Curr. Opin. Cell. Biol. 22: 374–382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulisz A., Chen N., Chandel N. S., Shao Z., Schumacker P. T., 2002. Mitochondrial ROS initiate phosphorylation of p38 MAP kinase during hypoxia in cardiomyocytes. Am. J. Physiol. Lung Cell. Mol. Physiol. 282: L1324–L1329 [DOI] [PubMed] [Google Scholar]
- Kwast K. E., Lai L. C., Menda N., James D. T., 3rd, Aref S., et al. , 2002. Genomic analyses of anaerobically induced genes in Saccharomyces cerevisiae: functional roles of Rox1 and other factors in mediating the anoxic response. J. Bacteriol. 184: 250–265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai L. C., Kosorukoff A. L., Burke P. V., Kwast K. E., 2005. Dynamical remodeling of the transcriptome during short-term anaerobiosis in Saccharomyces cerevisiae: differential response and role of Msn2 and/or Msn4 and other factors in galactose and glucose media. Mol. Cell. Biol. 25: 4075–4091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai L. C., Kosorukoff A. L., Burke P. V., Kwast K. E., 2006. Metabolic-state-dependent remodeling of the transcriptome in response to anoxia and subsequent reoxygenation in Saccharomyces cerevisiae. Eukaryot. Cell 5: 1468–1489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laroche C., Beney L., Marechal P. A., Gervais P., 2001. The effect of osmotic pressure on the membrane fluidity of Saccharomyces cerevisiae at different physiological temperatures. Appl. Microbiol. Biotechnol. 56: 249–254 [DOI] [PubMed] [Google Scholar]
- Lee K. K., Swanson S. K., Florens L., Washburn M. P., Workman J. L., 2009. Yeast Sgf73/Ataxin-7 serves to anchor the deubiquitination module into both SAGA and Slik(SALSA) HAT complexes. Epigenet. Chromatin 2: 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lees N. D., Bard M., Kemple M. D., Haak R. A., Kleinhans F. W., 1979. ESR determination of membrane order parameter in yeast sterol mutants. Biochim. Biophys. Acta 553: 469–475 [DOI] [PubMed] [Google Scholar]
- Lewis J. G., Learmonth R. P., Watson K., 1994. Cryoprotection of yeast by alcohols during rapid freezing. Cryobiology 31: 193–198 [DOI] [PubMed] [Google Scholar]
- Light W. R., 3rd, Olson J. S., 1990. The effects of lipid composition on the rate and extent of heme binding to membranes. J. Biol. Chem. 265: 15632–15637 [PubMed] [Google Scholar]
- Longtine M. S., McKenzie A., 3rd, Demarini D. J., Shah N. G., Wach A., et al. , 1998. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14: 953–961 [DOI] [PubMed] [Google Scholar]
- Lorenz R. T., Parks L. W., 1991. Involvement of heme components in sterol metabolism of Saccharomyces cerevisiae. Lipids 26: 598–603 [DOI] [PubMed] [Google Scholar]
- Luo Z., van Vuuren H. J., 2009. Functional analyses of PAU genes in Saccharomyces cerevisiae. Microbiology 155: 4036–4049 [DOI] [PubMed] [Google Scholar]
- Maeda T., Wurgler-Murphy S. M., Saito H., 1994. A two-component system that regulates an osmosensing MAP kinase cascade in yeast. Nature 369: 242–245 [DOI] [PubMed] [Google Scholar]
- Maeda T., Takekawa M., Saito H., 1995. Activation of yeast PBS2 MAPKK by MAPKKKs or by binding of an SH3-containing osmosensor. Science 269: 554–558 [DOI] [PubMed] [Google Scholar]
- Maeta K., Mori K., Takatsume Y., Izawa S., Inoue Y., 2005. Diagnosis of cell death induced by methylglyoxal, a metabolite derived from glycolysis, in Saccharomyces cerevisiae. FEMS Microbiol. Lett. 243: 87–92 [DOI] [PubMed] [Google Scholar]
- Malathi K., Higaki K., Tinkelenberg A. H., Balderes D. A., Almanzar-Paramio D., et al. , 2004. Mutagenesis of the putative sterol-sensing domain of yeast Niemann Pick C-related protein reveals a primordial role in subcellular sphingolipid distribution. J. Cell Biol. 164: 547–556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandala S. M., Thornton R., Tu Z., Kurtz M. B., Nickels J., et al. , 1998. Sphingoid base 1-phosphate phosphatase: a key regulator of sphingolipid metabolism and stress response. Proc. Natl. Acad. Sci. USA 95: 150–155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao C., Wadleigh M., Jenkins G. M., Hannun Y. A., Obeid L. M., 1997. Identification and characterization of Saccharomyces cerevisiae dihydrosphingosine-1-phosphate phosphatase. J. Biol. Chem. 272: 28690–28694 [DOI] [PubMed] [Google Scholar]
- Marie C., Leyde S., White T. C., 2008. Cytoplasmic localization of sterol transcription factors Upc2p and Ecm22p in S. cerevisiae. Fungal Genet. Biol. 45: 1430–1438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martens J. A., Wu P. Y., Winston F., 2005. Regulation of an intergenic transcript controls adjacent gene transcription in Saccharomyces cerevisiae. Genes Dev. 19: 2695–2704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Pastor M. T., Marchler G., Schuller C., Marchler-Bauer A., Ruis H., et al. , 1996. The Saccharomyces cerevisiae zinc finger proteins Msn2p and Msn4p are required for transcriptional induction through the stress response element (STRE). EMBO J. 15: 2227–2235 [PMC free article] [PubMed] [Google Scholar]
- Mazur P., Morin N., Baginsky W., el-Sherbeini M., Clemas J. A., et al. , 1995. Differential expression and function of two homologous subunits of yeast 1,3-beta-D-glucan synthase. Mol. Cell. Biol. 15: 5671–5681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mettetal J. T., Muzzey D., Gomez-Uribe C., van Oudenaarden A., 2008. The frequency dependence of osmo-adaptation in Saccharomyces cerevisiae. Science 319: 482–484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mnaimneh S., Davierwala A. P., Haynes J., Moffat J., Peng W. T., et al. , 2004. Exploration of essential gene functions via titratable promoter alleles. Cell 118: 31–44 [DOI] [PubMed] [Google Scholar]
- Montanes F. M., Pascual-Ahuir A., Proft M., 2011. Repression of ergosterol biosynthesis is essential for stress resistance and is mediated by the Hog1 MAP kinase and the Mot3 and Rox1 transcription factors. Mol. Microbiol. 79: 1008–1023 [DOI] [PubMed] [Google Scholar]
- Nagy Z., Riss A., Romier C., le Guezennec X., Dongre A. R., et al. , 2009. The human SPT20-containing SAGA complex plays a direct role in the regulation of endoplasmic reticulum stress-induced genes. Mol. Cell. Biol. 29: 1649–1660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Rourke S. M., Herskowitz I., 1998. The Hog1 MAPK prevents cross talk between the HOG and pheromone response MAPK pathways in Saccharomyces cerevisiae. Genes Dev. 12: 2874–2886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Rourke S. M., Herskowitz I., 2004. Unique and redundant roles for HOG MAPK pathway components as revealed by whole-genome expression analysis. Mol. Biol. Cell 15: 532–542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pacheco A., Pereira C., Almeida M. J., Sousa M. J., 2009. Small heat-shock protein Hsp12 contributes to yeast tolerance to freezing stress. Microbiology 155: 2021–2028 [DOI] [PubMed] [Google Scholar]
- Panadero J., Pallotti C., Rodriguez-Vargas S., Randez-Gil F., Prieto J. A., 2006. A downshift in temperature activates the high osmolarity glycerol (HOG) pathway, which determines freeze tolerance in Saccharomyces cerevisiae. J. Biol. Chem. 281: 4638–4645 [DOI] [PubMed] [Google Scholar]
- Pinkham J. L., Guarente L., 1985. Cloning and molecular analysis of the HAP2 locus: a global regulator of respiratory genes in Saccharomyces cerevisiae. Mol. Cell. Biol. 5: 3410–3416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Posas F., Chambers J. R., Heyman J. A., Hoeffler J. P., de Nadal E., et al. , 2000. The transcriptional response of yeast to saline stress. J. Biol. Chem. 275: 17249–17255 [DOI] [PubMed] [Google Scholar]
- Proft M., Struhl K., 2002. Hog1 kinase converts the Sko1-Cyc8-Tup1 repressor complex into an activator that recruits SAGA and SWI/SNF in response to osmotic stress. Mol. Cell 9: 1307–1317 [DOI] [PubMed] [Google Scholar]
- Proft M., Struhl K., 2004. MAP kinase-mediated stress relief that precedes and regulates the timing of transcriptional induction. Cell 118: 351–361 [DOI] [PubMed] [Google Scholar]
- Proft M., Pascual-Ahuir A., de Nadal E., Arino J., Serrano R., et al. , 2001. Regulation of the Sko1 transcriptional repressor by the Hog1 MAP kinase in response to osmotic stress. EMBO J. 20: 1123–1133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rachidi N., Martinez M. J., Barre P., Blondin B., 2000. Saccharomyces cerevisiae PAU genes are induced by anaerobiosis. Mol. Microbiol. 35: 1421–1430 [DOI] [PubMed] [Google Scholar]
- Radman-Livaja M., Ruben G., Weiner A., Friedman N., Kamakaka R., et al. , 2011. Dynamics of Sir3 spreading in budding yeast: secondary recruitment sites and euchromatic localization. EMBO J. 30: 1012–1026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiser V., Raitt D. C., Saito H., 2003. Yeast osmosensor Sln1 and plant cytokinin receptor Cre1 respond to changes in turgor pressure. J. Cell Biol. 161: 1035–1040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rep M., Krantz M., Thevelein J. M., Hohmann S., 2000. The transcriptional response of Saccharomyces cerevisiae to osmotic shock. Hot1p and Msn2p/Msn4p are required for the induction of subsets of high osmolarity glycerol pathway-dependent genes. J. Biol. Chem. 275: 8290–8300 [DOI] [PubMed] [Google Scholar]
- Roberts S. M., Winston F., 1997. Essential functional interactions of SAGA, a Saccharomyces cerevisiae complex of Spt, Ada, and Gcn5 proteins, with the Snf/Swi and Srb/mediator complexes. Genetics 147: 451–465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodgers K. R., 1999. Heme-based sensors in biological systems. Curr. Opin. Chem. Biol. 3: 158–167 [DOI] [PubMed] [Google Scholar]
- Rodriguez-Navarro S., 2009. Insights into SAGA function during gene expression. EMBO Rep. 10: 843–850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenfeld E., Beauvoit B., 2003. Role of the non-respiratory pathways in the utilization of molecular oxygen by Saccharomyces cerevisiae. Yeast 20: 1115–1144 [DOI] [PubMed] [Google Scholar]
- Saito H., Tatebayashi K., 2004. Regulation of the osmoregulatory HOG MAPK cascade in yeast. J. Biochem. 136: 267–272 [DOI] [PubMed] [Google Scholar]
- Sales K., Brandt W., Rumbak E., Lindsey G., 2000. The LEA-like protein HSP 12 in Saccharomyces cerevisiae has a plasma membrane location and protects membranes against desiccation and ethanol-induced stress. Biochim. Biophys. Acta 1463: 267–278 [DOI] [PubMed] [Google Scholar]
- Samara N. L., Datta A. B., Berndsen C. E., Zhang X., Yao T., et al. , 2010. Structural insights into the assembly and function of the SAGA deubiquitinating module. Science 328: 1025–1029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt A. P., McEntee K., 1996. Msn2p, a zinc finger DNA-binding protein, is the transcriptional activator of the multistress response in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 93: 5777–5782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt T. H., Frezzatti W. A., Jr, Schreier S., 1993. Hemin-induced lipid membrane disorder and increased permeability: a molecular model for the mechanism of cell lysis. Arch. Biochem. Biophys. 307: 96–103 [DOI] [PubMed] [Google Scholar]
- Seko Y., Takahashi N., Tobe K., Kadowaki T., Yazaki Y., 1997. Hypoxia and hypoxia/reoxygenation activate p65PAK, p38 mitogen-activated protein kinase (MAPK), and stress-activated protein kinase (SAPK) in cultured rat cardiac myocytes. Biochem. Biophys. Res. Commun. 239: 840–844 [DOI] [PubMed] [Google Scholar]
- Serrano R., Martin H., Casamayor A., Arino J., 2006. Signaling alkaline pH stress in the yeast Saccharomyces cerevisiae through the Wsc1 cell surface sensor and the Slt2 MAPK pathway. J. Biol. Chem. 281: 39785–39795 [DOI] [PubMed] [Google Scholar]
- Sertil O., Kapoor R., Cohen B. D., Abramova N., Lowry C. V., 2003. Synergistic repression of anaerobic genes by Mot3 and Rox1 in Saccharomyces cerevisiae. Nucleic Acids Res. 31: 5831–5837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaklai N., Shviro Y., Rabizadeh E., Kirschner-Zilber I., 1985. Accumulation and drainage of hemin in the red cell membrane. Biochim. Biophys. Acta 821: 355–366 [DOI] [PubMed] [Google Scholar]
- Shukla A., Bajwa P., Bhaumik S. R., 2006. SAGA-associated Sgf73p facilitates formation of the preinitiation complex assembly at the promoters either in a HAT-dependent or independent manner in vivo. Nucleic Acids Res. 34: 6225–6232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shviro Y., Zilber I., Shaklai N. 1982. The interaction of hemoglobin with phosphatidylserine vesicles. Biochim. Biophys. Acta 687: 63. [DOI] [PubMed] [Google Scholar]
- Surewicz W. K., 1984. Membrane actions of water-soluble flusogens: effect of demethyl solfoxide, glyerol and sucrose on lipid bilayer order and fluidity. Chem. Phys. Lipids 34: 363–372 [Google Scholar]
- Swain E., Baudry K., Stukey J., McDonough V., Germann M., et al. , 2002. Sterol-dependent regulation of sphingolipid metabolism in Saccharomyces cerevisiae. J. Biol. Chem. 277: 26177–26184 [DOI] [PubMed] [Google Scholar]
- ter Linde J. J., Liang H., Davis R. W., Steensma H. Y., van Dijken J. P., et al. , 1999. Genome-wide transcriptional analysis of aerobic and anaerobic chemostat cultures of Saccharomyces cerevisiae. J. Bacteriol. 181: 7409–7413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong A. H., Evangelista M., Parsons A. B., Xu H., Bader G. D., et al. , 2001. Systematic genetic analysis with ordered arrays of yeast deletion mutants. Science 294: 2364–2368 [DOI] [PubMed] [Google Scholar]
- Vik A., Rine J., 2001. Upc2p and Ecm22p, dual regulators of sterol biosynthesis in Saccharomyces cerevisiae. Mol. Cell. Biol. 21: 6395–6405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viswanathan M., Muthukumar G., Cong Y. S., Lenard J., 1994. Seripauperins of Saccharomyces cerevisiae: a new multigene family encoding serine-poor relatives of serine-rich proteins. Gene 148: 149–153 [DOI] [PubMed] [Google Scholar]
- Wang Y. L., Faiola F., Xu M., Pan S., Martinez E., 2008. Human ATAC is a GCN5/PCAF-containing acetylase complex with a novel NC2-like histone fold module that interacts with the TATA-binding protein. J. Biol. Chem. 283: 33808–33815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westfall P. J., Patterson J. C., Chen R. E., Thorner J., 2008. Stress resistance and signal fidelity independent of nuclear MAPK function. Proc. Natl. Acad. Sci. USA 105: 12212–12217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilcox L. J., Balderes D. A., Wharton B., Tinkelenberg A. H., Rao G., et al. , 2002. Transcriptional profiling identifies two members of the ATP-binding cassette transporter superfamily required for sterol uptake in yeast. J. Biol. Chem. 277: 32466–32472 [DOI] [PubMed] [Google Scholar]
- Winston F., Minehart P. L., 1986. Analysis of the yeast SPT3 gene and identification of its product, a positive regulator of Ty transcription. Nucleic Acids Res. 14: 6885–6900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winston F., Dollard C., Ricupero-Hovasse S. L., 1995. Construction of a set of convenient Saccharomyces cerevisiae strains that are isogenic to S288C. Yeast 11: 53–55 [DOI] [PubMed] [Google Scholar]
- Yamazaki M., Ohnishi S., Ito T., 1989. Osmoelastic coupling in biological structures: decrease in membrane fluidity and osmophobic association of phospholipid vesicles in response to osmotic stress. Biochemistry 28: 3710–3715 [DOI] [PubMed] [Google Scholar]
- Yang T., Espenshade P. J., Wright M. E., Yabe D., Gong Y., et al. , 2002. Crucial step in cholesterol homeostasis: sterols promote binding of SCAP to INSIG-1, a membrane protein that facilitates retention of SREBPs in ER. Cell 110: 489–500 [DOI] [PubMed] [Google Scholar]
- Zapater M., Sohrmann M., Peter M., Posas F., de Nadal E., 2007. Selective requirement for SAGA in Hog1-mediated gene expression depending on the severity of the external osmostress conditions. Mol. Cell. Biol. 27: 3900–3910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S., Skalsky Y., Garfinkel D. J., 1999. MGA2 or SPT23 is required for transcription of the delta9 fatty acid desaturase gene, OLE1, and nuclear membrane integrity in Saccharomyces cerevisiae. Genetics 151: 473–483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y., Mao X. O., Sun Y., Xia Z., Greenberg D. A., 2002. p38 mitogen-activated protein kinase mediates hypoxic regulation of Mdm2 and p53 in neurons. J. Biol. Chem. 277: 22909–22914 [DOI] [PubMed] [Google Scholar]
- Zitomer R. S., Lowry C. V., 1992. Regulation of gene expression by oxygen in Saccharomyces cerevisiae. Microbiol. Rev. 56: 1–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Znaidi S., Weber S., Al-Abdin O. Z., Bomme P., Saidane S., et al. , 2008. Genomewide location analysis of Candida albicans Upc2p, a regulator of sterol metabolism and azole drug resistance. Eukaryot. Cell 7: 836–847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zohn I. E., Li Y., Skolnik E. Y., Anderson K. V., Han J., et al. , 2006. p38 and a p38-interacting protein are critical for downregulation of E-cadherin during mouse gastrulation. Cell 125: 957–969 [DOI] [PubMed] [Google Scholar]