Abstract
The onset of flowering is an important adaptive trait in plants. The small ephemeral species Arabidopsis thaliana grows under a wide range of temperature and day-length conditions across much of the Northern hemisphere, and a number of flowering-time loci that vary between different accessions have been identified before. However, only few studies have addressed the species-wide genetic architecture of flowering-time control. We have taken advantage of a set of 18 distinct accessions that present much of the common genetic diversity of A. thaliana and mapped quantitative trait loci (QTL) for flowering time in 17 F2 populations derived from these parents. We found that the majority of flowering-time QTL cluster in as few as five genomic regions, which include the locations of the entire FLC/MAF clade of transcription factor genes. By comparing effects across shared parents, we conclude that in several cases there might be an allelic series caused by rare alleles. While this finding parallels results obtained for maize, in contrast to maize much of the variation in flowering time in A. thaliana appears to be due to large-effect alleles.
THE correct timing of flower initiation is critical for a variety of reasons. For example, depending on geography, the season during which a plant can successfully complete seed set is more or less limited. Similarly, in outcrossing species, synchronized flowering of conspecifics ensures that pollen can be exchanged, a prerequisite for fertilization. Research aimed at understanding the multiple layers of control in floral initiation has been a very active field over the past 40 years and has followed several complementary directions. First, forward and reverse genetics, primarily in Arabidopsis thaliana and rice, have led to the identification of many genes that promote or repress flowering. These so-called flowering-time genes have since been placed in a number of genetically defined pathways that integrate external stimuli such as photoperiod, ambient temperature, or prolonged exposure to cold, with endogenous signals including phytohormones and plant age (Bäurle and Dean 2006; Kobayashi and Weigel 2007; Turck et al. 2008; Greenup et al. 2009; Amasino 2010).
In parallel, A. thaliana has emerged as a powerful platform from which to study the genetic basis of naturally occurring variation in flowering time. A. thaliana accessions are found across much of the Northern hemisphere and grow under local conditions for which many have presumably become adapted. Usually starting with the mapping of quantitative trait loci (QTL) in crosses derived from two parents, the analysis of naturally occurring alleles has confirmed the importance of several proteins in the control of flowering, among them the photoreceptors CRYPTOCHROME2 (CRY2), PHYTOCHROME B (PHYB), and PHYC (El-Din El-Assal et al. 2001; Balasubramanian et al. 2006; Filiault et al. 2008), HUA2, a likely pre-mRNA processing factor (Wang et al. 2007), the mobile flowering signal FT (Schwartz et al. 2009), and the MADS domain transcription factor FLOWERING LOCUS M (FLM)/MADS AFFECTING FLOWERING 1 (MAF1) (Werner et al. 2005). FLM belongs to a small clade of transcription factors that comprises FLC and the four closely related MAF proteins, MAF2 to MAF5, encoded in a tandem cluster (Ratcliffe et al. 2001; Ratcliffe et al. 2003; Scortecci et al. 2003). This cluster is polymorphic between accessions and has recently been implicated in natural variation of flowering as well (Caicedo et al. 2009; Rosloski et al. 2010).
In addition to these factors, which had been identified already as actual or potential regulators of flowering through forward or reverse genetics studies, the role of FLC and its regulator FRIGIDA (FRI) was revealed only through the analysis of natural accessions, as they are defective in several of the early flowering accessions used commonly for laboratory studies (Michaels and Amasino 1999; Sheldon et al. 1999; Johanson et al. 2000). It has been estimated that the FLC and FRI loci account for almost three-quarters of the flowering-time variation among accessions, when these are not exposed to several weeks of winter-like conditions, known as vernalization. Upon vernalization, the contribution of FLC and FRI becomes markedly reduced (Lempe et al. 2005; Shindo et al. 2005). Notably, the variant alleles identified at CRY2, FLM, FT, HUA2, and PHYB are all rare, and it is therefore unclear how much these genes contribute to the global genetic architecture of flowering-time control in A. thaliana, although functionally distinct PHYC and MAF2-5 alleles appear to be quite common (Balasubramanian et al. 2006; Caicedo et al. 2009).
An alternative to genetic mapping is the use of genome-wide association studies (GWAS) to identify common variants controlling a trait, and this approach has recently been implemented in A. thaliana (Atwell et al. 2010; Brachi et al. 2010). Unfortunately, the analysis of flowering time by GWAS is strongly confounded by population structure, and even the re-identification of FLC and FRI was not straightforward, although this might change in the future with larger populations, or more appropriately chosen collections of accessions. Furthermore, there was essentially no overlap with genes identified in QTL studies.
Here, we took advantage of a set of 18 distinct accessions that present much of the common genetic diversity of A. thaliana (Clark et al. 2007). We generated 17 F2 populations and phenotyped almost 500 plants of each population in a common environment. An integrated analysis of this data set was greatly facilitated by all plants being genotyped with the same intermediate-frequency SNPs chosen to be maximally informative across the 17 populations. The detailed picture of the genetic architecture of flowering-time variation in these F2 populations validates and extends previous studies focused on recombinant inbred lines (RILs), by identifying QTL clusters that have not been described before. Much of the mappable variation in flowering time can be attributed to as few as five genomic regions, mirroring the results of a recent study with a similar design, but growing plants in a variable environment (Brachi et al. 2010). The regions we identified include the locations of the entire FLC/MAF clade of transcription factor genes. By comparing effects across shared parents, we conclude that in several cases there might be an allelic series, which parallels results obtained for maize (Buckler et al. 2009). In contrast to the many but small- to modest-effect QTL in maize, however, much of the variation in flowering time in A. thaliana appears to be due to large-effect alleles.
MATERIALS AND METHODS
Plants and growth conditions:
Seeds of 18 accessions were from the individuals described by Clark and Colleagues (2007). All accessions were crossed to each other in a full diallel. Out of the 306 F1 crosses, 14 were chosen in a simple round-robin design, such that 13 parents were represented twice and two parents once. Three additional crosses represented a triangular design with three parents. The list of parental accessions and the crossing design are provided in Table 1 and supporting information, Figure S1.
TABLE 1.
The 17 Arabidopsis F2 populations generated in this study
| Population | Grandmother | Originb | Grandfather | Originb |
| P35 | Tamm-2 | Finland | Col-0 | (Poland) |
| P66 | Fei-0 | Portugal | Col-0 | (Poland) |
| P145 | Sha | Tajikistan | Fei-0 | Portugal |
| P2 | Lov-5 | Sweden | Sha | Tajikistan |
| P19 | Bay-0 | Germany | Lov-5 | Sweden |
| P3 | Bur-0 | Ireland | Bay-0 | Germany |
| P10 | Bur-0 | Ireland | Cvi-0 | Cape Verde Islands |
| P17 | Cvi-0 | Cape Verde Islands | RRS7 | United States |
| P8 | Est-1 | Estonia | RRS7 | United States |
| P12 | Est-1 | Estonia | Br-0 | Czech Republic |
| P15 | Br-0 | Czech Republic | C24 | (Portugal) |
| P129 | C24 | (Portugal) | RRS10 | United States |
| P9 | Tsu-1 | Japan | RRS10 | United States |
| P169 | Ts-1 | Spain | Tsu-1 | Japan |
| P6a | Van-0 | Canada | Bor-4 | Czech Republic |
| P7a | NFA-8 | United Kingdom | Van-0 | Canada |
| P20a | Bor-4 | Czech Republic | NFA-8 | United Kingdom |
Triangular crossing design.
Parentheses indicate uncertainty of precise geographic origin.
Parents and F1 and F2 progeny were grown under identical conditions. Seeds were stratified in 0.1% top agar for 4 days in the dark at 4°, before being sown on soil. Seeds were first allowed to break dormancy at 16° overnight, before being subjected to 6 weeks vernalization at 4° under 8-hr photoperiods (short days), to reduce differences in flowering time. Upon release from vernalization, all seeds had germinated, and cotyledons were expanded. A single seedling was kept in each pot and allowed to grow in 16-hr-long days, at a constant temperature of 16°. Trays were rotated 180° and moved to a new shelf every other day to minimize position effects within the growth chamber. Humidity inside the chamber was maintained at 65%, and lights were provided with a combination of cool white and warm white fluorescent lights, for a fluence of 125 to 175 μmol m−2 s−1.
For F1 plants and parental accessions, 8 pots were sown for each genotype in a randomized fashion across two 40-pot trays. F2 seeds were sown in 12 40-pot trays, for a total of 480 plants. Because of lack of germination in some pots, the number of F2 plants per population varied between 239 and 462. The 17 populations were analyzed in four overlapping cohorts, grown from July 2007 to January 2008: P2 and P3 (cohort 1); P6, P7, P8, P9, and P10 (cohort 2), P12, P15, P17, P19, and P20 (cohort 3); P35, P66, P129, P145, and P169 (cohort 4). F1 plants and parental accessions were grown immediately following cohort 4, in January and February 2008.
The 96 Nordborg (Nordborg et al. 2005) accessions were grown last in cohort 5; 10 pots per accession were sown. Accessions were divided into two sets of 48, and sown in 12 40-pot trays in a randomized fashion. After 4 weeks of growth at 16°, pots from each accession were grouped together, to decrease shading of smaller accessions.
Phenotyping:
Flowering time, number of rosette, and number of cauline leaves were recorded. Flowering time was first assessed when floral buds became visible in the center of the rosette (DTF1), when the main shoot had elongated to 1 cm (DTF2), and last when the first flower opened (DTF3). The number of rosette leaves was also recorded at DTF2, and the number of cauline leaves was counted 1–2 weeks later. A complete list of traits measured is listed in Table S1.
Genotyping:
A single leaf from each F2 plant and parental accession was collected after plants had flowered and used for DNA extraction using the BioSprint 96 workstation (Qiagen, Hilden, Germany). Quality of genomic DNA was tested on an agarose gel; DNA concentrations were determined on a Nanodrop photometer (Thermo Scientific, Waltham, MA). About 2 μg of genomic DNA was used for genotyping of SNP markers by Sequenom (San Diego, CA), using MassArray technology (Jurinke et al. 2001). Genotypes are available in File S2.
QTL mapping:
Raw genotype data were converted to the appropriate genotype format A, B, H (A being a marker homozygous for parent A; B homozygous for parent B; H a heterozygous marker). Genotype and phenotype data were merged and saved as .csv files. QTL analysis was performed using R/qtl, with simple and composite interval mapping (Broman et al. 2003). The confidence intervals around each significant QTL peak were determined with the baysint function, at 95% confidence levels. Additional information on the extent of variation explained by each QTL, as well as the effect associated with each parental allele, was gathered using the sim.geno, makeqtl, fitqtl, effectplot, and effectscan functions. Epistatic interactions between QTL were identified by using the qb.scantwo function in R/qtlbim (Yandell et al. 2007).
For joint QTL analysis, all F2 plants were combined into a single population, the genotype of each chromosome at any given SNP taking on 1 of the 18 possible identities (from our founding accessions). Evidence for a QTL is given as log P, P being the probability that a QTL is segregating at a given SNP (Kover et al. 2009).
QTL data on flowering time obtained from RIL analysis were taken from published studies (Alonso-Blanco et al. 1998; Loudet et al. 2002; Weinig et al. 2002; El-Lithy et al. 2004, 2006; Werner et al. 2005; O’neill et al. 2008; Simon et al. 2008).
RESULTS
Variation in flowering time among 17 F2 populations:
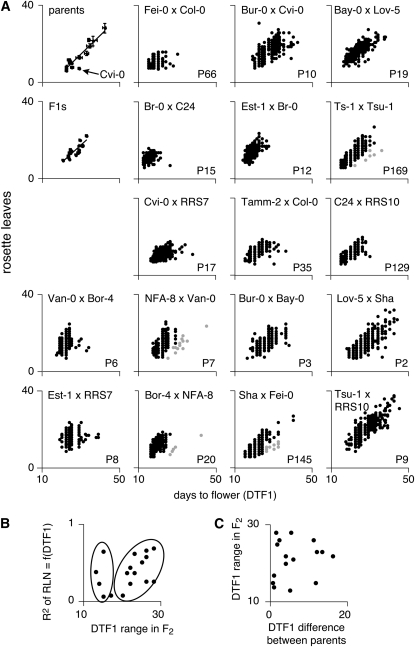
We measured flowering time for 7045 F2 plants, as well as 136 F1 plants and 128 plants from the 18 parental accessions in 5 cohorts as described in materials and methods. A final cohort consisted of 960 plants from a larger group of 96 A. thaliana accessions, chosen over the geographical range of the species and representative of its phenotypic and genetic diversity (Nordborg et al. 2005). Previous studies have found strong correlations between days until flowering and the number of rosette leaves produced on the main shoot (Alonso-Blanco et al. 1998; El-Din El-Assal et al. 2001; Lempe et al. 2005; El-Lithy et al. 2006; Simon et al. 2008), indicating that these two traits are genetically linked in natural accessions. Under our conditions, we observed a similar positive linear relationship between days to flower and leaf number for the parental accessions (r2 = 0.88; see Figure 1A). When we excluded Cvi-0, which grew very slowly, correlation was even higher (r2 = 0.96), similar to what we found with the 96 Nordborg (Nordborg et al. 2005) accessions (r2 = 0.98; Figure S2). The slopes of the regression line between days to flower and leaf number from the founding accessions and the larger set of 96 accessions were close to parallel (the regression coefficient being 0.97 for founding accessions, 0.88 for the full set of 96 accessions, or 0.93 when the latest flowering accessions are excluded).
Figure 1.—
Flowering-time variation in parental accessions, F1 and F2 populations. (A) Correlation between rosette leaf number and days to flower. F2 populations are sorted in order of increasing r2 values. Seedlings with lower leaf initiation rate in the NFA-8 × Van-0 (P7), Bor-4 × NFA-8 (P20), Sha × Fei-0 (P145), and Ts-1 × Tsu-1 (P169) populations are shown as gray dots. (B) Correlation between the r2 values for rosette leaf number and days to flower and flowering-time variance in F2 populations. (C) Flowering-time variance in F2 populations is independent of the phenotypic differences of grandparents.
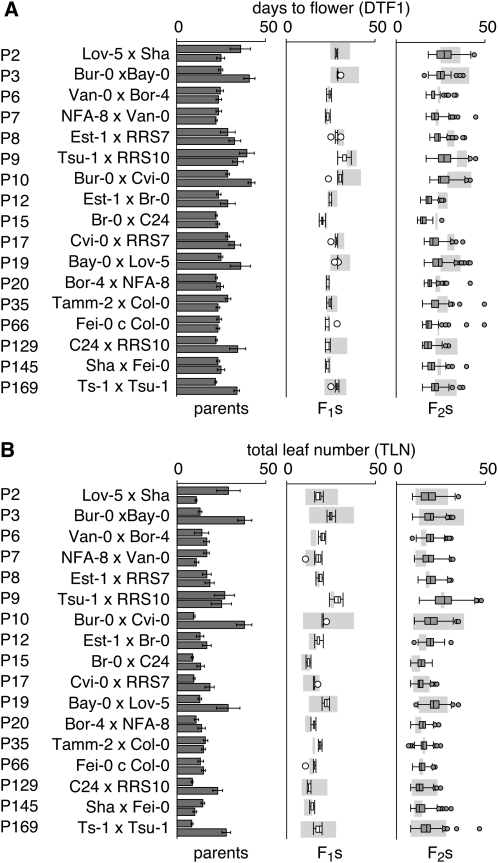
In contrast, the correlation between days to flower and leaf number in F2 populations dramatically decreased, with a maximum of r2 = 0.84 for P9 (Tsu-1 × RRS10). In several instances the correlation was only marginal, as in the P6 (Van-0 × Bor-4: r2 = 0.3), P8 (Est-1 × RRS7: r2 = 0.28), and P66 populations (Fei-0 × Col-0: r2 = 0.26, Figure 1A); this suggests that days to flower and leaf number are canalized in natural accessions, but that the link between the two can be genetically uncoupled. This observation can only partially be explained by the smaller spread of flowering times in each population (Figure 1B). While in most F2 populations r2 values tended to increase with increased variance in flowering time, P12 (Est-1 × Br-0), P15 (Br-0 × C24), P66 (Fei-0 × Col-0), and P129 (C24 × RRS10) formed a distinct group (left-side circle, Figure 1B) with small variances, but differing r2 values. In some populations (for example, NFA-8 × Van-0 [P7], Bor-4 × NFA-8 [P20], Sha × Fei-0 [P145], and Ts-1 × Tsu-1 [P169]), a small group of plants appeared to initiate leaves at a slower rate than their siblings (Figure 1A), which might reflect variation in growth rate. Finally, the range in flowering time measured within each F2 population did not correlate with differences in flowering time between the parental accessions (Figure 1C), reflecting rampant transgressive segregation, which was evident in all populations, even though the founding grandparents had not been selected for differences in flowering time. In fact, 8 of the 17 pairs of grandparents showed no significant differences in their flowering time in our conditions (Figure 2).
Figure 2.—
Transgressive segregation of flowering time. Comparison of days to flower (A) and total leaf number (B) for parental accessions, F1 hybrids, and all F2 populations. Light blue bars indicate the parental difference in flowering time in the F1 and F2 columns.
QTL for flowering time:
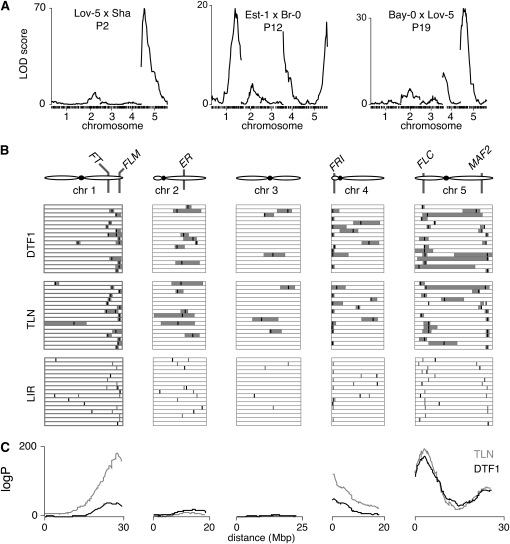
All 7045 F2 plants were genotyped with a common set of SNP markers, with 215 to 257 markers (mean 237) being informative in each population (P. Salomé and D. Weigel, unpublished results). With an average of 400 plants per population, and a mean 237 informative SNPs, the amount of variance explained by a given QTL is very close to the LOD value for that QTL peak (File S1, Figure S3, and not shown). In essence, a QTL with a LOD score of 10 will explain 10% of the observed variation. With a significance threshold of LOD 3–4 for most populations, we therefore have the power to detect QTL with effects as small as 3–4% of the total variance. Population-wide scans revealed two to five flowering-time QTL per population, with an average of 3.2 and a total of 55 QTL (Figure 3, A and B, Table S2, and Table S3). The effects of all but one QTL exceeded 1 day, which is in stark contrast to only 7 of 333 in maize exceeding the 1-day threshold (Figure S4, Buckler et al. 2009). In the vast majority of populations, a single QTL per chromosome could be detected, indicating that measured effects at a given genomic location were not confounded by the local genetic background. When two QTL located to the same chromosome, they mapped to opposite arms and were therefore too distant from each other to influence their colocating QTL and associated effects (Table S2 and Table S3).
Figure 3.—
Location of flowering-time QTL. (A) Examples of QTL maps for DTF1 in three distinct F2 populations from simple interval mapping. (B) Location of QTL for days to flower (DTF1), total leaf number (TLN), and leaf initiation rate (LIR) in all 17 F2 populations. The positions of al DTF1 QTL are shown in gray in the LIR row. (C) Joint QTL analysis, across all 17 F2 populations.
Flowering-time-related traits (such as days to flower [DTF1, DTF2, and DTF3] and leaf number [rosette, cauline, and total leaf number]) were highly correlated and confidence intervals of QTL peaks for these traits usually overlapped in a given population. While variation in leaf initiation rate would provide the simplest explanation for a lack of correlation between days to flower and rosette leaf number, we detected leaf initiation rate QTL irrespective of the degree of correlation (Figure 3B). For the trait DTF1, number of days until the inflorescence first became visible to the unaided eye, QTL could explain between 10 and 64% (mean 39%) of the total phenotypic variation, with individual QTL accounting for 3–54% of variation (Table 2). The remainder is not due to rampant epistatic interactions between these QTL: we detected 11 strong epistatic pairs between QTL from 9 populations, but these accounted for only a mean of 1.6% of the total variance (range 0.35–3.3; see Table S4 and Figure S5).
TABLE 2.
QTL cluster in our study
| Population | FLM | FRI | FLC | MAF2-MAF5 | Total variance (%) |
| Tamm-2 × Col-0 (P35) | Yes | Yes | Yes | Yes | 35.3 |
| Fei-0 × Col-0 (P66) | Yes | Yes | 9.7 | ||
| Sha × Fei-0 (P145) | Yes | Yes | Yes | 31.2 | |
| Lov-5 × Sha (P2) | Yes | 62.4 | |||
| Bay-0 × Lov-5 (P19) | Yes | Yes | 51.7 | ||
| Bur-0 × Bay-0 (P3) | Yes | Yes | Yes | Yes | 34.4 |
| Bur-0 × Cvi-0 (P10) | Yes | Yes | 39.7 | ||
| Cvi-0 × RRS7 (P17) | Yes | Yes | 37.9 | ||
| Est-1 × RRS7 (P8) | Yes | 25.2 | |||
| Est-1 × Br-0 (P12) | Yes | Yes | Yes | 50.9 | |
| Br-0 × C24 (P15) | Yes | Yes | 39.6 | ||
| C24 × RRS10 (P129) | Yes | Yes | 61.6 | ||
| Tsu-1 × RRS10 (P9) | Yes | Yes | Yes | 64.3 | |
| Ts-1 × Tsu-1 (P169) | Yes | Yes | 51.9 | ||
| Van-0 × Bor-4 (P6) | Yes | Yes | 20.75 | ||
| NFA-8 ×x Van-0 (P7) | Yes | Yes | 28.7 | ||
| Bor-4 × NFA-8 (P20) | Yes | Yes | 21.1 | ||
| Total count | 12 | 8 | 11 | 8 | |
| Variance mean | 14.8% | 12.1% | 18.8% | 13% | 39.2 |
| Variance range | 3.4–35.7 | 5.9–18.5 | 4–53.5 | 3.2–32.5 | 9.7–64.3 |
Measured effects and prediction of parental flowering times:
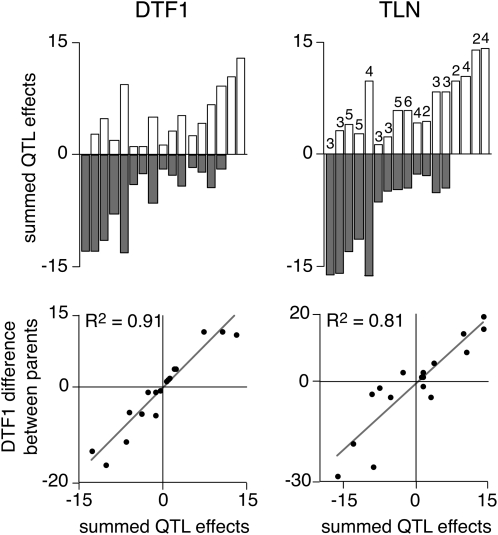
We extracted individual effects associated with parental alleles at each QTL and arranged the 17 F2 populations on the basis of the sum of effects (Figure 4, A and B). Some populations displayed comparable positive and negative effects, which canceled each other out to give mean population values close to zero. Other populations were dominated by effects in a single direction. For example, the effects in Lov-5 × Sha (P2), Bur-0 × Bay-0 (P3), and Bur-0 × Cvi-0 (P10) were mostly negative, while those in Bay-0 × Lov-5 (P19), C24 × RRS10 (P129), and Ts-1 × Tsu-1 (P169) were largely positive (Figure 4, A and B). Although the detected QTL peaks accounted, on average, for 39% of the observed variation in flowering time, the measured cumulated effects could predict well the differences in flowering time between parental accessions (Figure 4, A and B).
Figure 4.—
Congruence between QTL effects and flowering-time difference of parents. Top, sum of negative (gray) and positive (white) effects; numbers indicate count of QTL. Bottom, correlation between summed QTL effects and difference in flowering time of parents.
Comparative power of QTL detection in F2 and RIL populations:
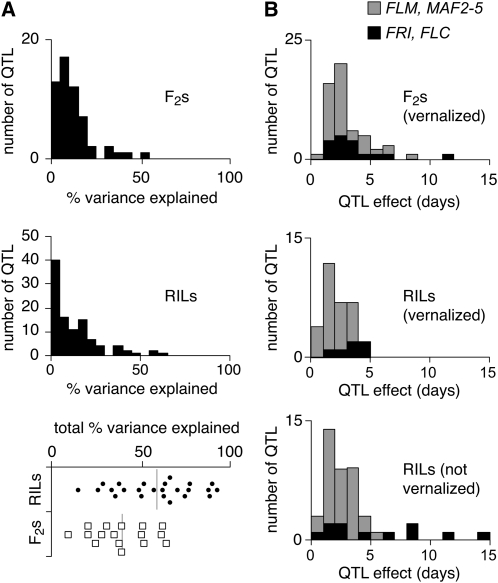
We compared the power of QTL detection reported in RILs (Alonso-Blanco et al. 1998; Loudet et al. 2002; Weinig et al. 2002; El-Lithy et al. 2004, 2006; Werner et al. 2005; O’neill et al. 2008; Simon et al. 2008) with our F2 populations. Only small-effect QTL, explaining <3% of variance, were more frequently reported in RIL studies. Since much of the variation is due to large-effect QTL, the total variance explained in our F2 populations is not greatly different than that in RIL populations (Figure 5A). Effects measured in our study for specific QTL such as FRI and FLC are comparable to those reported in RIL studies (Figure 5B; and see below).
Figure 5.—
Comparison of the power for QTL discovery in F2 populations and RILs. (A) Variance explained by individual or all QTL in our F2 populations and in RILs (see Figure 6). Because of our significance threshold for QTL, individual QTL explain at least 3% of the total variance, while RIL QTL may explain less. Gray line indicates mean for all populations. (B) Relative contribution of QTL at either FRI and FLC or FLM and MAF2-5.
Common flowering-time QTL and the FLC clade:
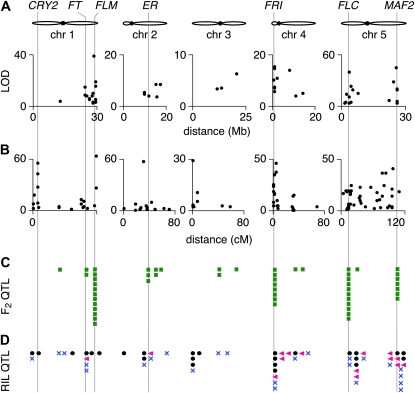
Since we measured flowering time in 17 large populations under the same conditions, we were in a good position to assess the contribution of genomic regions identified by QTL mapping to the global control of flowering time in the species. Simple interval mapping (IM) and composite interval mapping (CIM) identified the same QTL peaks (Figure S6). As many as 39 of the 55 detected QTL overlapped across populations, suggesting that distinct alleles segregate among accessions (Figure 3B, Table 2, and Table S2). Flowering-time QTL were overrepresented in four genomic regions, on chromosomes 1, 4, and 5. A joint analysis across all 17 F2 populations identified in addition to the same four regions only the middle of chromosome 2 as making a small but significant contribution to flowering-time variation (Figure 3C), thus confirming the importance of these four regions in shaping the genetic architecture responsible for the measured variation in flowering time.
Major contributors to flowering-time variation in our populations include QTL that map to three genomic regions containing members of the FLC/MAF clade of transcription factors. Without vernalization, FLC and its activator FRI can explain over 70% of the total flowering-time variation between wild accessions (Lempe et al. 2005; Shindo et al. 2005). QTL associated with the genomic regions containing members of the FLC clade (FLM, on the bottom of chromosome 1; FLC, on the top of chromosome 5; MAF2-5 on the bottom of chromosome 5) were found in up to 12 F2 populations (Table 2). It is worth noting that our 6-week vernalization treatment did not eliminate the effects of FRI and FLC, the expression of which is strongly vernalization dependent (Michaels and Amasino 1999; Sheldon et al. 1999). However, it has been shown before that accessions with an active FLC allele differ in their vernalization requirement, with much of this variation mapping to FLC itself (Shindo et al. 2006).
QTL mapping to the FLC and FRI genomic regions were found in 11 and 8 of the 17 F2 populations, respectively, and contribute about 19 and 12%, respectively, of the variance (Figure 3 and Table 2). The MAF2–MAF5 cluster, which is very polymorphic between accessions and has been recently implicated in flowering-time variation (Caicedo et al. 2009; Rosloski et al. 2010), is a candidate for 8 QTL on the bottom of chromosome 5. These QTL can explain 15% of measured variance. Finally, the region containing the FLC ortholog FLM, which is deleted in the Nd-0 accession (Werner et al. 2005), is included in 12 QTL explaining 15% of variance.
Overall, the 39 QTL associated with the genomic regions of FRI and the FLC clade contribute over 85% of explained variation. The remaining 15 QTL likely reflect accession-specific variation (Table 2). Two QTL studies have identified a QTL at FT (Shindo et al. 2006; Schwartz et al. 2009). One showed that Est-1 carries an allele that is less active than the reference allele (Schwartz et al. 2009), and we could detect FT QTL in the Est-1 × RRS7 (P8) and Est-1 × Br-0 (P12) populations, which share Est-1 as one of the grandparents (Figures 3 and 6). Several modest QTL peaks were detected near ERECTA, and the EARLY-FLOWERING 3 (ELF3) gene is a candidate causal locus (Hicks et al. 2001).
Figure 6.—
Comparison of flowering-time QTL mapping in F2 populations (this study) and published RIL populations. (A) Fifty-five QTL from simple interval mapping of F2 populations (this study). (B) QTL from the analysis of published RIL populations. (C) QTL locations in F2 populations (this study, green squares). (D) QTL locations in RIL studies: solid circles (Loudet et al. 2002; El-Lithy et al. 2004, 2006; Werner et al. 2005); pink triangles (O’neill et al. 2008); blue X’s (Simon et al. 2008). In all cases, detected QTL are for days to flower (DTF1).
A broader comparison of our results from simple interval mapping with previously published QTL revealed that the FRI, FLC, FLM, and the MAF2–MAF5 regions (Figure 6, Table 2, and Table 3) were probably shared with other studies (Alonso-Blanco et al. 1998; Werner et al. 2005; El-Lithy et al. 2006; O’neill et al. 2008). It is difficult to determine how well QTL detected in early studies overlap with our candidate genomic regions, as reported positions were not reported relative to the physical map (Alonso-Blanco et al. 1998; Loudet et al. 2002; Weinig et al. 2002; El-Lithy et al. 2004). More recent studies have, however, taken advantage of the Arabidopsis genome sequence information to generate a consensus physical map onto which QTL were mapped (El-Lithy et al. 2006; O’neill et al. 2008; Simon et al. 2008; Brachi et al. 2010). Many of the QTL identified with RILs over the past decade mapped to the same genomic regions and overlapped with the locations of FLC (13 instances), FRI (11), MAF2-5 (11), and, to a lesser extent, with FLM (six times; Table 3). Mean explained variance in RILs contributed by FRI and the FLC clade reached 37.4%, very similar to the variance of 39.2% we observed to be associated with the same genomic regions (Tables 2 and 3). Additional QTL seen in RILs but not in our F2 populations could explain another 20% of the standing variation, but these are likely to reflect single-gene variants specific for a parental accession. One example is the well-known loss-of-function allele at the receptor kinase gene ERECTA (ER) found in the accession Ler (a founding accession for 6 RILs; Table S5). We did not detect a significant QTL for CRY2, known to be functionally divergent in Cvi-0, the parent for two of our populations (Bur-0 × Cvi-0 [P10] and Cvi-0 × RRS7 [P17]), reflecting the short-day-dependent nature of the early flowering phenotype conferred by the CRY2Cvi-0 allele (Figure 6C).
TABLE 3.
QTL candidates in other studies
| Reference | FLM | FRI | FLC/CO | MAF2–MAF5 | Total variance (%) |
| Alonso-Blanco et al. (1998) | Yes | Yes | 28.3 | ||
| Weinig et al. (2002) | Yes | Yes | 34 | ||
| Loudet et al. (2002) | Yes | Yes | 52 | ||
| El-Lithy et al. (2004) | Yes | Yes | Yes | Yes | 53.5 |
| Werner et al. (2005) | Yes | 26.6 | |||
| El-Lithy et al. (2006) | Yes | Yes | 29.6 | ||
| El-Lithy et al. (2006) | Yes | Yes | Yes | Yes | 52.5 |
| El-Lithy et al. (2006) | Yes | Yes | Yes | 52.2 | |
| Simon et al. (2008) | Yes | Yes | 21 | ||
| Simon et al. (2008) | Yes | Yes | Yes | Yes | 18 |
| Simon et al. (2008) | Yes | Yes | 28 | ||
| Simon et al. (2008) | Yes | Yes | 12 | ||
| Simon et al. (2008) | Yes | Yes | 32 | ||
| O’neill et al. (2008) | Yes | Yes | Yes | 55.3 | |
| O’neill et al. (2008) | Yes | Yes | 29 | ||
| O’neill et al. (2008) | Yes | 38 | |||
| O’neill et al. (2008) | Yes | Yes | Yes | 64 | |
| O’neill et al. (2008) | Yes | 37 | |||
| O’neill et al. (2008) | — | — | — | — | |
| Total count | 6 | 11 | 13 | 11 (12) | |
| Variance mean | 11.5% | 21.5% | 12.7% | 14.2 (15.2)% | 37.4 |
| Variance range | 3–26.6 | 5–46.6 | 2–48 | 2–37 | 12–64 |
Evidence for allelic series within the FLC clade:
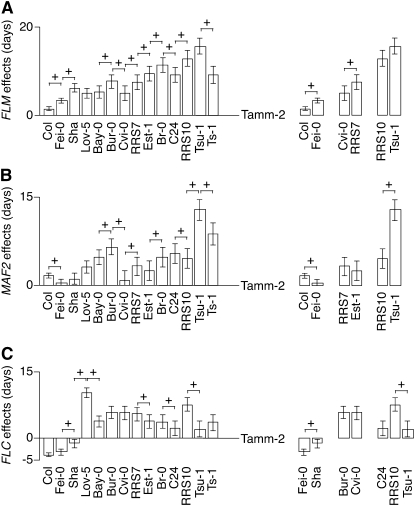
The round-robin design (Figure S1) allowed us to draw a logic chain linking 14 of our founding accessions, and thus predict effects between accessions not directly connected in a cross. A clear gradient in the strength of FLM, MAF2-5, and FLC alleles was apparent (Figure 7), indicating that not only FLC (Shindo et al. 2006), but also other members of the FLC clade, contributed quantitatively to the observed variation in flowering time through allelic series.
Figure 7.—
Allelic series among QTL at members of the FLC clade. Effects relative to the Tamm-2 allele were plotted along the logic chain of our simple round-robin crossing design, and then added up. + indicates detected QTL. (A) FLM effects, (B) MAF2 effects, and (C) FLC effects.
The proposed allelic series generally agreed with the presence or absence of a QTL between two consecutive accessions. For example, large differences in effects separated the FLM locus of Col-0 and Fei-0 (grandparents of P66), as well as Fei-0 and Sha (grandparents of P145), but not those of Sha and Lov-5 (grandparents of P2). In agreement, a significant QTL was detected in the FLM region in P66 and P145, but not in P2 (Figure 3C). There were, however, some limitations of our analysis: the FLM QTL of Tsu-1 appeared to confer slightly later flowering than that of RRS10, but this difference in effect did not result in a QTL in the FLM region in the P9 population derived from these two parents (Figures 3 and 7A). The behavior of MAF2 QTL also generally followed the predicted results from our QTL discovery (Figure 7B), although a very-late-flowering FLC QTL, on the top of chromosome 5, appeared to mask MAF2 QTL effects on the bottom of chromosome 5. Populations lacking a QTL near MAF2 but showing strong differences in effect for the parental alleles of this region share the Lov-5 accession, which has a very-late-flowering FLC QTL. Only following composite interval mapping was a QTL detected in the MAF2 region, and only in the Bay-0 × Lov-5 population (P19). This does not reflect a missed epistatic interaction, as we could not detect any epistasis between the upper and lower arm of chromosome 5 in Lov-5 × Sha (P2) or P19. It is worth noting that FLCLov-5 confers the strongest effects among our populations, especially in the P2 population with a measured effect of 11.5 days for the trait DTF1.
The allelic series of the QTL at FLC was dominated by the very strong effects associated with FLCLov-5 and FLCRRS10 (Figure 7C). FLCC24 is probably not inactive, as was previously indicated by crosses to plants with known functional or inactive alleles of FLC (Sanda and Amasino 1996). When C24 is crossed to flc-3, an FLC loss-of-function allele in Col-0, a fraction of F2 plants exhibited a late-flowering phenotype that cosegregates with FLCC24 (not shown). In addition, a flowering-time QTL was detected around the FRI region in a Col-0 × C24 RIL set, while no QTL was found around the FLC region in the same population, indicating that the Col-0 and C24 alleles of FLC are similar (S. Balasubramanian, T. Altmann and D. Weigel, unpublished results). C24 might therefore carry a functional FLC copy whose effect is canceled by an extragenic modifier.
The observed gradient in flowering time caused by the parental alleles at the FLM, MAF2, and FLC loci suggested that the late-flowering accessions might not share the same allele but instead each carry a rare allele. We attempted to test this hypothesis by querying existing sequence data sets. Clark and Colleagues (2007) determined polymorphisms in all founding accessions. The oligonucleotide-based resequencing technology, however, revealed only about half of all coding SNPs and a considerably smaller portion of noncoding SNPs. It was therefore not surprising that the presence of a Clark SNP between two accessions (regardless of their position: promoter, coding sequence, or within introns) was not correlated with the existence of a QTL for any of our candidates (Table S6, Table S7, Table S8). Available sequence information at the MAF2–MAF5 gene cluster is unfortunately of limited use in our case, as only two of our accessions are represented in the 168 accessions characterized by Caicedo and Colleagues (2009). The current resolution in known common polymorphisms therefore suggests an allelic series contributed by rare alleles for our candidate genes rather than a single SNP segregating in our F2 populations with a QTL at a given flowering-time candidate locus.
DISCUSSION
The control of flowering time in A. thaliana has been the focus of much study over the past decade. Yet, despite the wealth of resources at our disposal, a clear picture of the species-wide genetic architecture of flowering time has not yet emerged, since the simultaneous analysis of populations representing several parents has been the exception (Simon et al. 2008; Kover et al. 2009; Brachi et al. 2010).
Most of the previous work mapping flowering-time QTL has used RILs. Because RILs represent immortalized, largely fixed recombinant genotypes that can be phenotyped many times, genotyping costs could be amortized over many phenotyping trials. In the past few years, expenses associated with genotyping have dropped considerably, and adoption of next-generation sequencing platforms promises to further lower costs while increasing the resolution of genotyping (e.g., Baird et al. 2008; Huang et al. 2009; Xie et al. 2010). Apart from marker analysis, polymorphism discovery used to be a major bottleneck, before the advent of ultra-high-resolution microarrays and new sequencing methods (Clark et al. 2007; Ossowski et al. 2008). We have investigated the potential of F2 populations as an alternative to immortal RILs, by making full use of our knowledge of hundreds of thousands of polymorphisms described for 20 accessions (Clark et al. 2007).
The major QTL that we detected could explain on average about 40% of the overall variation, indicating that the remaining 60% of flowering-time variation must be associated with modest-effect QTL that lie below our significance threshold. That the unexplained variance does not hinder us from predicting the parental flowering times suggests that the remaining effects must be (1) very small, and therefore remain undetectable in our populations, and (2) equally distributed between negative and positive effects, thus canceling each other out. We also observed extensive variation in the onset of flowering in all F2 populations, even when the parental accessions flowered at very similar times. Because hybridization of A. thaliana accessions occurs regularly in the wild (Abbott and Gomes 1989; Bergelson et al. 1998; Nordborg et al. 2005; Picó et al. 2008), our results have important implications for the initial stages of adaptation via flowering time.
We also compared our results to a recently published species-wide study of flowering-time QTL in maize. In our populations, 54 of 55 QTL alleles altered flowering time by at least 1 day, while this was true for only 7 of 333 QTL in maize (Figure S4, Buckler et al. 2009). In A. thaliana, an average of 3–4 QTL per F2 population explained 3.1–22.7 days difference in flowering (mean 10.1 days), while the combined effects of 13–14 maize QTL per population in maize ranged from 1.5 to 13.0 (mean 6.2).
Also in contrast to maize, a small number of regions was overrepresented for flowering-time QTL. Two of them include FRI and FLC, although our F2 populations were all exposed to prolonged cold in an attempt to identify vernalization-independent loci. Variation at the FRI region strongly contributed to flowering-time variation in RILs, reaching values as high as 46%, and averaging 21.5% across all RILs. The relative importance of the FRI genomic region in our 17 F2 populations was not quite as strong, averaging only 12.1% of total variance, and was never higher than 19%, indicating that 6 weeks of vernalization was effective in limiting the contribution of FRI to flowering time. QTL mapping to the FLC genomic region explained between 4 and 53.5% of the standing variation in our populations (Table 2), and between 2 and 37% in RILs (Table 3), confirming FLC as a major gene for flowering time. Lov-5 carries a strong, vernalization-insensitive FLC allele (Shindo et al. 2006), which may skew the mean and range associated with FLC: after removal of FLCLov-5 from our list, mean variance dropped to 13.3 (range of 4–41.5) and was then more in line with results obtained with RILs. Two additional regions where QTL clustered overlapped with the locations of the remaining members of the FLC clade, FLM, and MAF2-5, in both our F2 and RIL populations. Mean variance and range were comparable in both sets of populations, suggesting that the observed allelic series at FLM and MAF2-5 between 14 of our 18 founding accessions might also apply to the RIL parental accessions as well. We detected QTL overlapping with the FLM genomic regions twice as often as in RIL studies, possibly reflecting the partial bias in RIL parental accessions. Indeed, the common laboratory accessions Col and Ler were crossed, either to each other or to other accessions, to create 12 of the 19 RIL populations characterized for flowering-time QTL (Table S5). In field experiments such as Brachi and Colleagues (2010), the decision to flower results from the integration of daily temperature cycles and gradual photoperiod changes. Only under these conditions—where daily temperatures often did not raise above 10°—were QTL in genes associated with the circadian clock detected, indicating that low temperatures may define a sensitized condition for variation in clock function in the specific context of flowering time. In all other studies, including this study, growth conditions included a constant temperature >16° and long and nonchanging photoperiods sufficient to saturate the photoperiodic pathway, thus allowing the emergence of effects caused by general mediators of flowering time and providing an explanation for the absence of clock-associated loci in our list of QTL.
Although we vernalized seedlings for 6 weeks before release at 16°, we still detected QTL mapping to the FRI, FLC, and FLM/MAF2-5 genomic regions. Our growth chambers maintain very good control of temperature, light intensity, and air humidity, which likely greatly limited phenotypic variation due to microenvironmental noise and therefore enhanced our ability to detect QTL. In addition, the relatively low temperature of 16° generally delays flowering in long days compared to 23° (Lempe et al. 2005). Responses to ambient temperature involve SVP, as demonstrated by a similar flowering time at 16° and 23° for svp mutants (Lee et al. 2007). SVP function is dependent on FLM, as an svp loss of function can suppress the late flowering caused by FLM overexpression (Scortecci et al. 2003). It is thus conceivable that flm mutants might be similarly insensitive to changes in ambient temperature and that growing plants at 16° allowed us to measure differences in the strength of FLM alleles that had escaped detection in several previous studies.
Although two of our major QTL clusters overlap with the locations of FLM and MAF2-5, initial genome-wide association studies failed to identify significant SNPs at either FLM or MAF2–MAF5 (Atwell et al. 2010; Brachi et al. 2010). Genome-wide association studies fail when they include too few accessions with functionally variant alleles, or if too many of the functionally variant alleles are distinct from each other. The evidence for allelic series at all our QTL is in support of the latter hypothesis. Only after an increase in sample size from 96 to 473 unique accessions did MAF2 emerge as a possible flowering-time QTL candidate following association mapping (Li et al. 2010). In all cases described in Arabidopsis, one constant feature remains: QTL for flowering time are few, but are associated with large effects.
The chromosomal location of most strong-effect QTL is in itself quite striking: aside from ER, which is close to the centromere of chromosome 2, all other flowering-time QTL candidate genes (FLC, FLM, MAF2-5, and FRI) are located at the ends of their respective chromosomes. Following hybridization, parental genomes recombine and segregate to form novel combinations of alleles in the progeny. The low frequency of crossovers each generation means that large, intact fragments of parental chromosome will be transmitted to the progeny. The large-effect QTL that we detected in our populations would thus generate distinct pools of alleles in the F2 and subsequent generations, which could have adaptive significance due to variation in flowering time. On the other hand, growth-related traits tend to display more complex genetic architectures than flowering time, with many small-effect QTL, and are often ripe with epistatic interactions (Vlad et al. 2010). This delicate balance of alleles will be severely disrupted after hybridization and formation of pools of early and late-flowering plants; however, positioning flowering-time QTL to ends of chromosomes will limit the extent of genetic drag imposed on the rest of the chromosome.
In conclusion, we have identified a small number of genomic regions with strong effects on flowering time. Some of the same regions, and indeed candidate genes, are now coming to the forefront through genome-wide association mapping studies. That FLM has yet to be described as being associated with flowering-time variation in association studies might mean only that the number of accessions remains too small, if many rare alleles contribute. The complete sequencing of hundreds, and soon thousands, of genomes from A. thaliana accessions (Weigel and Mott 2009) is a prerequisite for the genome-wide annotation of potential functional polymorphisms; apart from the direct analysis of QTL candidates, this will also improve the power of genome-wide association studies, since alleles that are the consequence of convergent changes in activity can be combined.
Acknowledgments
We thank Richard Clark and Suresh Balasubramanian for discussions during the design stages of the project. We also are indebted to Josip Perkovič, Hannah Helms, Waldemar Hauf, and Marcella Amorim for help with phenotyping and seed collection. K.B. and D.W. conceived and designed the experiments, P.A.S., K.B., R.A.E.L., and L.Y. performed the experiments, P.A.S. and R.M. analyzed the data, and P.A.S. and D.W. wrote the paper. This research was supported by postdoctoral fellowships from the European Molecular Biology Organization (P.A.S.), National Institutes of Health (K.B.), Human Frontiers Science Program (R.A.E.L.), grant FP6 IP AGRON-OMICS (contract LSHG-CT-2006-037704), from a Gottfried Wilhelm Leibniz Award of the Deutsche Forschungsgemeinschaft, and the Max Planck Society (D.W.).
LITERATURE CITED
- Abbott R. J., Gomes M. F., 1989. Population genetic structure and outcrossing rate of Arabidopsis thaliana (L.) Heynh. Heredity 62: 411–418 [Google Scholar]
- Alonso-Blanco C., El-Assal S. E., Coupland G., Koornneef M., 1998. Analysis of natural allelic variation at flowering time loci in the Landsberg erecta and Cape Verde Islands ecotypes of Arabidopsis thaliana. Genetics 149: 749–764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amasino R., 2010. Seasonal and developmental timing of flowering. Plant J. 61: 1001–1013 [DOI] [PubMed] [Google Scholar]
- Atwell S., Huang Y. S., Vilhjalmsson B. J., Willems G., Horton M., et al. , 2010. Genome-wide association study of 107 phenotypes in Arabidopsis thaliana inbred lines. Nature 465: 627–631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baird N. A., Etter P. D., Atwood T. S., Currey M. C., Shiver A. L., et al. , 2008. Rapid SNP discovery and genetic mapping using sequenced RAD markers. PLoS One 3: e3376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balasubramanian S., Sureshkumar S., Agrawal M., Michael T. P., Wessinger C., et al. , 2006. The PHYTOCHROME C photoreceptor gene mediates natural variation in flowering and growth responses of Arabidopsis thaliana. Nat. Genet. 38: 711–715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bäurle I., Dean C., 2006. The timing of developmental transitions in plants. Cell 125: 655–664 [DOI] [PubMed] [Google Scholar]
- Bergelson J., Stahl E., Dudek S., Kreitman M., 1998. Genetic variation within and among populations of Arabidopsis thaliana. Genetics 148: 1311–1323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brachi B., Faure N., Horton M., Flahauw E., Vazquez A., et al. , 2010. Linkage and association mapping of Arabidopsis thaliana flowering time in nature. PLoS Genet. 6: e1000940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broman K. W., Wu H., Sen S., Churchill G. A., 2003. R/qtl: QTL mapping in experimental crosses. Bioinformatics 19: 889–890 [DOI] [PubMed] [Google Scholar]
- Buckler E. S., Holland J. B., Bradbury P. J., Acharya C. B., Brown P. J., et al. , 2009. The genetic architecture of maize flowering time. Science 325: 714–718 [DOI] [PubMed] [Google Scholar]
- Caicedo A. L., Richards C., Ehrenreich I. M., Purugganan M. D., 2009. Complex rearrangements lead to novel chimeric gene fusion polymorphisms at the Arabidopsis thaliana MAF2–5 flowering time gene cluster. Mol. Biol. Evol. 26: 699–711 [DOI] [PubMed] [Google Scholar]
- Clark R. M., Schweikert G., Toomajian C., Ossowski S., Zeller G., et al. , 2007. Common sequence polymorphisms shaping genetic diversity in Arabidopsis thaliana. Science 317: 338–342 [DOI] [PubMed] [Google Scholar]
- El-Din El-Assal S., Alonso-Blanco C., Peeters A. J., Raz V., Koornneef M., 2001. A QTL for flowering time in Arabidopsis reveals a novel allele of CRY2. Nat. Genet. 29: 435–440 [DOI] [PubMed] [Google Scholar]
- El-Lithy M. E., Clerkx E. J., Ruys G. J., Koornneef M., Vreugdenhil D., 2004. Quantitative trait locus analysis of growth-related traits in a new Arabidopsis recombinant inbred population. Plant Physiol. 135: 444–458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Lithy M. E., Bentsink L., Hanhart C. J., Ruys G. J., Rovito D., et al. , 2006. New Arabidopsis recombinant inbred line populations genotyped using SNPWave and their use for mapping flowering-time quantitative trait loci. Genetics 172: 1867–1876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filiault D. L., Wessinger C. A., Dinneny J. R., Lutes J., Borevitz J. O., et al. , 2008. Amino acid polymorphisms in Arabidopsis phytochrome B cause differential responses to light. Proc. Natl. Acad. Sci. USA 105: 3157–3162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenup A., Peacock W. J., Dennis E. S., Trevaskis B., 2009. The molecular biology of seasonal flowering-responses in Arabidopsis and the cereals. Ann. Bot. 103: 1165–1172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hicks K. A., Albertson T. M., Wagner D. R., 2001. EARLY FLOWERING3 encodes a novel protein that regulates circadian clock function and flowering in Arabidopsis. Plant Cell 13: 1281–1292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X., Feng Q., Qian Q., Zhao Q., Wang L., et al. , 2009. High-throughput genotyping by whole-genome resequencing. Genome Res. 19: 1068–1076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johanson U., West J., Lister C., Michaels S., Amasino R., et al. , 2000. Molecular analysis of FRIGIDA, a major determinant of natural variation in Arabidopsis flowering time. Science 290: 344–347 [DOI] [PubMed] [Google Scholar]
- Jurinke C., van den Boom D., Cantor C. R., Koster H., 2001. Automated genotyping using the DNA MassArray technology. Methods Mol. Biol. 170: 103–116 [DOI] [PubMed] [Google Scholar]
- Kobayashi Y., Weigel D., 2007. Move on up, it’s time for change: mobile signals controlling photoperiod-dependent flowering. Genes Dev. 21: 2371–2384 [DOI] [PubMed] [Google Scholar]
- Kover P. X., Valdar W., Trakalo J., Scarcelli N., Ehrenreich I. M., et al. , 2009. A multiparent advanced generation inter-cross to fine-map quantitative traits in Arabidopsis thaliana. PLoS Genet. 5: e1000551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J. H., Yoo S. J., Park S. H., Hwang I., Lee J. S., et al. , 2007. Role of SVP in the control of flowering time by ambient temperature in Arabidopsis. Genes Dev. 21: 397–402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lempe J., Balasubramanian S., Sureshkumar S., Singh A., Schmid M., et al. , 2005. Diversity of flowering responses in wild Arabidopsis thaliana strains. PLoS Genet. 1: e6–0109–0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Huang Y., Bergelson J., Nordborg M., Borevitz J. O., 2010. Association mapping of local climate-sensitive quantitative trait loci in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 107: 21199–21204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loudet O., Chaillou S., Camilleri C., Bouchez D., Daniel-Vedele F., 2002. Bay-0 × Shahdara recombinant inbred line population: a powerful tool for the genetic dissection of complex traits in Arabidopsis. Theor. Appl. Genet. 104: 1173–1184 [DOI] [PubMed] [Google Scholar]
- Michaels S. D., Amasino R. M., 1999. FLOWERING LOCUS C encodes a novel MADS domain protein that acts as a repressor of flowering. Plant Cell 11: 949–956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nordborg M., Hu T. T., Ishino Y., Jhaveri J., Toomajian C., et al. , 2005. The pattern of polymorphism in Arabidopsis thaliana. PLoS Biol. 3: e196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Neill C. M., Morgan C., Kirby J., Tschoep H., Deng P. X., et al. , 2008. Six new recombinant inbred populations for the study of quantitative traits in Arabidopsis thaliana. Theor. Appl. Genet. 116: 623–634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ossowski S., Schneeberger K., Clark R. M., Lanz C., Warthmann N., et al. , 2008. Sequencing of natural strains of Arabidopsis thaliana with short reads. Genome Res. 18: 2024–2033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picó F. X., Mendez-Vigo B., Martinez-Zapater J. M., Alonso-Blanco C., 2008. Natural genetic variation of Arabidopsis thaliana is geographically structured in the Iberian peninsula. Genetics 180: 1009–1021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratcliffe O. J., Nadzan G. C., Reuber T. L., Riechmann J. L., 2001. Regulation of flowering in Arabidopsis by an FLC homologue. Plant Physiol. 126: 122–132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratcliffe O. J., Kumimoto R. W., Wong B. J., Riechmann J. L., 2003. Analysis of the Arabidopsis MADS AFFECTING FLOWERING gene family: MAF2 prevents vernalization by short periods of cold. Plant Cell 15: 1159–1169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosloski S. M., Jali S. S., Balasubramanian S., Weigel D., Grbic V., 2010. Natural diversity in flowering responses of Arabidopsis thaliana caused by variation in a tandem gene array. Genetics 186: 263–276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanda S. L., Amasino R., 1996. Genetic and physiological analysis of flowering time in the C24 line of Arabidopsis thaliana. Weeds World 2: 2–8 [Google Scholar]
- Schwartz C., Balasubramanian S., Warthmann N., Michael T. P., Lempe J., et al. , 2009. Cis-regulatory changes at FLOWERING LOCUS T mediate natural variation in flowering responses of Arabidopsis thaliana. Genetics 183: 723–732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scortecci K., Michaels S. D., Amasino R. M., 2003. Genetic interactions between FLM and other flowering-time genes in Arabidopsis thaliana. Plant Mol. Biol. 52: 915–922 [DOI] [PubMed] [Google Scholar]
- Sheldon C. C., Burn J. E., Perez P. P., Metzger J., Edwards J. A., et al. , 1999. The FLF MADS box gene: a repressor of flowering in Arabidopsis regulated by vernalization and methylation. Plant Cell 11: 445–458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shindo C., Aranzana M. J., Lister C., Baxter C., Nicholls C., et al. , 2005. Role of FRIGIDA and FLOWERING LOCUS C in determining variation in flowering time of Arabidopsis. Plant Physiol. 138: 1163–1173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shindo C., Lister C., Crevillen P., Nordborg M., Dean C., 2006. Variation in the epigenetic silencing of FLC contributes to natural variation in Arabidopsis vernalization response. Genes Dev. 20: 3079–3083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon M., Loudet O., Durand S., Berard A., Brunel D., et al. , 2008. Quantitative trait loci mapping in five new large recombinant inbred line populations of Arabidopsis thaliana genotyped with consensus single-nucleotide polymorphism markers. Genetics 178: 2253–2264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turck F., Fornara F., Coupland G., 2008. Regulation and identity of florigen: FLOWERING LOCUS T moves center stage. Annu. Rev. Plant Biol. 59: 573–594 [DOI] [PubMed] [Google Scholar]
- Vlad D., Rappaport F., Simon M., Loudet O., 2010. Gene transposition causing natural variation for growth in Arabidopsis thaliana. PLoS Genet. 6: e1000945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q., Sajja U., Rosloski S., Humphrey T., Kim M. C., et al. , 2007. HUA2 caused natural variation in shoot morphology of A. thaliana. Curr. Biol. 17: 1513–1519 [DOI] [PubMed] [Google Scholar]
- Weigel D., Mott R., 2009. The 1001 Genomes Project for Arabidopsis thaliana. Genome Biol. 10: 107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinig C., Ungerer M. C., Dorn L. A., Kane N. C., Toyonaga Y., et al. , 2002. Novel loci control variation in reproductive timing in Arabidopsis thaliana in natural environments. Genetics 162: 1875–1884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner J. D., Borevitz J. O., Warthmann N., Trainer G. T., Ecker J. R., et al. , 2005. Quantitative trait locus mapping and DNA array hybridization identify an FLM deletion as a cause for natural flowering-time variation. Proc. Natl. Acad. Sci. USA 102: 2460–2465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie W., Feng Q., Yu H., Huang X., Zhao Q., et al. , 2010. Parent-independent genotyping for constructing an ultrahigh-density linkage map based on population sequencing. Proc. Natl. Acad. Sci. USA 107: 10578–10583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yandell B. S., Mehta T., Banerjee S., Shriner D., Venkataraman R., et al. , 2007. R/qtlbim: QTL with Bayesian Interval Mapping in experimental crosses. Bioinformatics 23: 641–643 [DOI] [PMC free article] [PubMed] [Google Scholar]