Abstract
Proper regulation of metabolism is essential to maximizing fitness of organisms in their chosen environmental niche. Nitrogen metabolite repression is an example of a regulatory mechanism in fungi that enables preferential utilization of easily assimilated nitrogen sources, such as ammonium, to conserve resources. Here we provide genetic, transcriptional, and phenotypic evidence of nitrogen metabolite repression in the human pathogen Cryptococcus neoformans. In addition to loss of transcriptional activation of catabolic enzyme-encoding genes of the uric acid and proline assimilation pathways in the presence of ammonium, nitrogen metabolite repression also regulates the production of the virulence determinants capsule and melanin. Since GATA transcription factors are known to play a key role in nitrogen metabolite repression, bioinformatic analyses of the C. neoformans genome were undertaken and seven predicted GATA-type genes were identified. A screen of these deletion mutants revealed GAT1, encoding the only global transcription factor essential for utilization of a wide range of nitrogen sources, including uric acid, urea, and creatinine—three predominant nitrogen constituents found in the C. neoformans ecological niche. In addition to its evolutionarily conserved role in mediating nitrogen metabolite repression and controlling the expression of catabolic enzyme and permease-encoding genes, Gat1 also negatively regulates virulence traits, including infectious basidiospore production, melanin formation, and growth at high body temperature (39°–40°). Conversely, Gat1 positively regulates capsule production. A murine inhalation model of cryptococcosis revealed that the gat1Δ mutant is slightly more virulent than wild type, indicating that Gat1 plays a complex regulatory role during infection.
NITROGEN is a major component of macromolecules ranging from proteins to nucleic acids that are essential to all living organisms (Carroll and Salt 2004). Yet while almost 80% of the earth’s atmosphere is made up of diatomic nitrogen gas, very few species are able to break the strong triple bond that exists between the two atoms to reduce gaseous nitrogen into a state that is biologically available (Postgate 1998). In organisms unable to utilize nitrogen directly from the atmosphere, the ability to scavenge reduced forms of nitrogen from alternative sources is crucial and elaborate control mechanisms are needed to ensure its constant supply.
As major decomposers of organic material in the biosphere, fungi are well known for their ability to utilize numerous compounds as nitrogen sources by expressing catabolic enzymes and permeases in a range of pathways. Readily assimilated compounds such as ammonium and glutamine are usually the preferred nitrogen sources of fungi, although in the absence of these sources, less easily assimilated nitrogen sources such as amines, amides, purines, and pyrimidines may also be utilized (Marzluf 1997). In members of the phylum Ascomycota, the selective utilization of these secondary nitrogen sources is typically tightly controlled by transcriptionally regulating the synthesis of the enzymes and permeases required for their scavenging and degradation (Mitchell and Magasanik 1984; Marzluf 1997; Fraser et al. 2001; Magasanik and Kaiser 2002). Achieved through a global regulatory circuit known as either nitrogen metabolite repression or nitrogen catabolite repression, this process ensures secondary nitrogen source degrading pathways are not expressed when more easily assimilated nitrogen sources are available, thereby maximizing the fitness of the organism in its ever-changing local environment (Wiame et al. 1985; Marzluf 1997; Fraser et al. 2001). By combining nitrogen metabolite repression with pathway-specific induction mediated by an array of dedicated transcription factors, resources are even more tightly conserved until preferred nitrogen sources are depleted (Davis et al. 1993; Feng and Marzluf 1998; Berger et al. 2006, 2008). In this way, the fungus activates the transcription of catabolic genes only when their substrates are immediately available.
In ascomycete species where it has been observed, nitrogen metabolite repression is mediated by transcription factors belonging to the GATA family. These proteins are distinguished by their highly conserved DNA-binding domain consisting of a Cys2/Cys2-type zinc finger motif followed by an adjacent basic region and are named after the DNA sequence 5′-GATA-3′ that they recognize (Ko and Engel 1993; Merika and Orkin 1993; Ravagnani et al. 1997). GATA transcription factors are widely found in eukaryotes, including metazoans and plants, and have diverse biological functions. For example, in the nematode worm Caenorhabditis elegans, the GATA factor ELT-2 is required for immunity to bacterial and fungal pathogens such as Salmonella enterica and C. neoformans (Kerry et al. 2006). In the plant Arabidopsis thaliana, the GATA factor HAN regulates the development of flower and shoot apical meristems (Zhao et al. 2004).
The first GATA factor mutant, however, was identified in Neurospora crassa on the basis of its central role in nitrate metabolism (Fincham 1950). Subsequent analyses showed that these positively acting nitrogen regulatory GATA factors often act synergistically with pathway-specific transcription factors to activate the expression of catabolic enzyme and permease-encoding genes required to assimilate secondary nitrogen sources when preferred sources are lacking (Davis et al. 1993; Feng and Marzluf 1998; Berger et al. 2006, 2008). As a consequence, loss of such global trans-acting GATA regulatory genes including areA in Aspergillus nidulans and nit-2 in N. crassa render these species incapable of utilizing nitrogen sources other than ammonium or glutamine (Arst and Cove 1973; Stewart and Vollmer 1986; Fu and Marzluf 1987; Kudla et al. 1990; Caddick 1992; Marzluf 1997). These GATA factors have also been shown to affect virulence in a number of pathogenic fungi. Aspergillus fumigatus areA∆, clinical Saccharomyces cerevisiae gln3Δ, and Candida albicans gat1Δ or gln3Δ mutants all exhibit reduced virulence compared to wild type in murine virulence assays (Hensel et al. 1998; Limjindaporn et al. 2003; Kingsbury et al. 2006; Liao et al. 2008).
The importance of nitrogen metabolism in the study of C. neoformans, an opportunistic pathogen of the phylum Basidiomycota that causes life-threatening meningoencephalitis predominantly in immunocompromised individuals, is also evident (Casadevall and Perfect 1998). The primary ecological niche of C. neoformans is nutrient-rich pigeon guano; 70% of the nitrogen present is in the form of uric acid with the rest consisting primarily of xanthine, urea, and creatinine (Staib et al. 1976; Staib et al. 1978; Casadevall and Perfect 1998). In contrast, nitrogen availability becomes scarce upon infection of humans. Beyond these obvious differences in nitrogen availability that C. neoformans often encounters as part of its infection cycle, there are more subtle indications that nitrogen metabolism may also play a role in the regulation of virulence. The antiphagocytic polysaccharide capsule is a well-known virulence factor that is highly induced in the presence of uric acid (Staib et al. 1976, 1978). Assimilation of uric acid as a nitrogen source requires a number of catabolic enzymes including the virulence factor urease, a nitrogen-scavenging enzyme shown to play a role in central nervous system invasion (Cox et al. 2000; Olszewski et al. 2004; Shi et al. 2010). While evidence that nitrogen availability may play a role in pathogenicity of C. neoformans is mounting, little is known about this process.
In this study we provide the first proof of canonical ascomycete-like nitrogen metabolite repression in the basidiomycete C. neoformans. We also reveal that beyond the traditional role in nitrogen scavenging, this regulatory mechanism controls the coordinated production of the virulence factors capsule and melanin. Study of deletion mutants of every predicted GATA-type gene in the C. neoformans genome revealed the existence of only one positively acting nitrogen regulatory GATA factor: Gat1. Gat1 mediates nitrogen metabolite repression and is required for utilization of most nitrogen sources including the preferred ammonium; this represents an anomaly from archetypical nitrogen metabolite repression. Gat1 also functions to negatively regulate mating, melanin production at human body temperature (37°), and growth at high body temperature (39°–40°) and is required for capsule synthesis. Importantly, this complex series of changes in the regulation of C. neoformans virulence composite combines to create an unexpected phenotype during murine infection: the gat1Δ mutant is slightly more virulent than wild type. Together, these studies indicate that in addition to controlling the regulon of genes for nitrogen acquisition, Gat1 is a key coordinator of multiple virulence-associated phenotypes.
MATERIALS AND METHODS
Strains and media:
Cryptococcus strains (supporting information, Table S1) were grown in YPD (1% yeast extract, 2% Bacto-peptone, 2% glucose) or YNB (0.45% yeast nitrogen base w/o amino acids and ammonium sulfate, 2% glucose, 10 mm nitrogen source) unless specified otherwise. Biolistic transformants were selected on YPD medium supplemented with 100 µl/ml G418 (Sigma) and/or nourseothricin (Werner BioAgents). Pigeon guano medium was prepared as described previously while the derivative pigeon guano (unfiltered) medium was prepared with the exclusion of the filtration step to ensure that insoluble nitrogen sources such as uric acid would not be removed (Nielsen et al. 2007). l-3,4-Dihydroxyphenylalanine (l-DOPA) medium was prepared as described previously with the original nitrogen source (asparagine) replaced with 10 mm of the specified nitrogen source (D’souza et al. 2001). V8 (5% V8 juice, 3 mm KH2PO4, 0.1% myo-inositol, 4% Bacto-agar) and Murashige & Skoog (MS) mating media (PhytoTechnology Laboratories) at pH 5.0 were prepared as described previously (Kwon-Chung et al. 1982; Xue et al. 2007). Christensen’s urea agar was prepared as described previously (Cox et al. 2000). Nematode growth medium (NGM) was prepared as described previously (Brenner 1974). Escherichia coli Mach-1 cells served as the host strain for transformation and propagation of all plasmids, which were carried out according to methods of Sambrook et al. (1989). C. elegans strain N2 was maintained at 15° and propagated on its normal laboratory food source E. coli OP50 cells as described previously (Brenner 1974; Honda et al. 1993; Garsin et al. 2001).
Bioinformatic analyses:
The genome sequence of C. neoformans var. grubii reference strain, H99 (http://www.broadinstitute.org/annotation/genome/cryptococcus_neoformans/MultiHome.html), was used in our study to gain insights into the regulation of nitrogen metabolism and virulence in C. neoformans. Sequence analyses were performed using BLAST and MacVector 9.5 (MacVector Inc, Cary, NC) (Altschul et al. 1990). Sequence alignments were created using ClustalW v. 1.4 within MacVector (Thompson et al. 1994). Sequence traces generated at the Australian Genome Research Facility (Brisbane, Queensland, Australia) were analyzed using Sequencher 4.7 (Gene Codes Corporation, Ann Arbor, MI).
Construction and complementation of mutant strains:
GATA-type gene deletion mutants were created using overlap PCR and biolistic transformation as described previously (Davidson et al. 2000, 2002). All primers used for the generation of deletion mutants are listed in Table S2. For example, to construct the gat1∆ mutant, the 4239-bp GAT1 (CNAG00193.2) coding sequence was replaced with the neomycin phosphotransferase II-encoding selectable marker NEO using a construct created by overlap PCR combining an ∼1-kb fragment upstream from the GAT1 start codon, the NEO marker, and an ∼1-kb fragment downstream from the GAT1 stop codon. Strain H99 genomic DNA and the plasmid pJAF1 were used as PCR templates (Fraser et al. 2003). The construct was transformed into the wild-type strain H99 via particle bombardment. Transformants were selected on YPD plates supplemented with 100 µl/ml G418. Deletion of GAT1 was confirmed by Southern blot, creating strain RL1 (Southern 2006). To complement the gat1∆ mutant, the wild-type GAT1 gene was amplified from genomic DNA via PCR, the product cloned into pCR2.1-TOPO (Invitrogen) to give pIRL1, and sequenced. The 5.95-kb AscI/SacI GAT1 fragment of pIRL1 was subcloned into the AscI/SacI sites of the Cryptococcus nourseothricin resistance vector pPZP-NATcc, creating the complementation construct pIRL3 (Walton et al. 2005). pIRL3 was subsequently linearized with AscI and XmaI, then biolistically transformed into the gat1∆ mutant. Stable transformants were selected on YPD plates supplemented with 100 µl/ml nourseothricin and transformants containing a single copy of the wild-type GAT1 gene were identified by Southern blot.
Quantitative real-time PCR:
Strains were grown in YNB supplemented with 10 mm of the specified nitrogen source or YPD and shaken at 30° for 16 hr. Overnight cultures were harvested, cell pellets frozen and lyophilised, total RNA isolated using TRIzol reagent (Invitrogen), and cDNA generated using the SuperscriptIII first-strand synthesis system (Invitrogen). Primers for genes URO1 (CNAG04305.2), DAL1 (CNAG00934.2), URE1 (CNAG05540.2), GDH1 (CNAG01577.2), GLN1 (CNAG00457.2), GLT1 (CNAG04862.2), GDH2 (CNAG00879.2), AMT1 (CNAG00235.2), AMT2 (CNAG04758.2), PUT1 (CNAG02049.2), PUT5 (CNAG02048.2), and PUT2 (CNAG05602.2) (Table S2) were designed to span exon–exon boundaries and tested to verify that they bind specifically to their respective cDNA genes but not H99 genomic DNA (data not shown). Quantitative real-time PCR (qRT-PCR) was performed using SYBR green supermix (Applied Biosystems) and an Applied Biosystems 7900HT fast real time PCR System with the following cycling conditions: denaturation at 95° for 10 min, followed by amplification and quantification in 45 cycles at 95° for 15 sec and 60° for 1 min, with melting curve profiling at 95° for 2 min, 60° for 15 sec, and 95° for 15 sec. Dissociation analysis confirmed the amplification of a single PCR product for each primer pair and an absence of primer dimer formation. Relative gene expression was quantified using SDS software 1.3.1 (Applied Biosystems) on the basis of the 2-∆∆CT method (Livak and Schmittgen 2001). Several housekeeping genes including GPD1 (glyceraldehyde-3-phosphate dehydrogenase), HHT1 (histone H3), and TUB2 (β-tubulin) were tested, but the ACT1 (actin) gene was eventually used as a control for normalization since its gene expression demonstrated the highest consistency across all growth conditions tested. Statistical analysis of variance (one-way analysis of variance) was performed using the unpaired, two-tailed t-test in GraphPad Prism Version 5.0c. P values <0.05 were considered statistically significant.
Capsule assays:
Strains were inoculated onto YNB plates supplemented with 10 mm specified nitrogen source or into RPMI/DMEM supplemented with 10% fetal calf serum (Gibco), and incubated at 37° for 2 days. To visualize capsule, cells were stained with India ink (Becton Dickinson) and observed using a ZEISS Axioplan 2 epifluorescent/light microscope. Pictures were taken with an Axiocam grayscale digital camera using the AxioVision AC imaging software. Quantitative analysis of capsule diameter was performed as described previously (Zaragoza et al. 2003).
Growth, melanization, and urease assays:
Starter cultures were prepared by growth in YPD at 30° overnight with shaking, diluted to OD595 nm = 0.05 in water, and then further diluted 10-fold in series. For nitrogen utilization, melanization, and urease assays as well as growth test on pigeon guano and at human body temperature, each diluted cell suspension was spotted onto YNB or l-DOPA supplemented with 10 mm specified nitrogen source, Christensen’s urea agar, 25% (wt/vol) pigeon guano medium, or YPD medium, respectively. Results were imaged after 2 days’ incubation at 30° (nitrogen utilization, growth test on pigeon guano, and urease assays), 30° and 37° (melanisation assays), or 37°–40° (growth test at human body temperature).
Mating assays:
Mating assays were conducted as described previously (Nielsen et al. 2003). Briefly, strains were pregrown on YPD plates for 2 days, after which a small amount of cells was removed using a flat-end toothpick and patched onto V8 and MS mating media (pH 5.0) either alone or mixed in equal proportions with a strain of the opposite mating-type (MAT) (Kwon-Chung et al. 1982; Xue et al. 2007). Plates were then incubated at room temperature in the dark for 1 week and assessed by light microscopy for formation of filaments and basidia.
C. elegans killing assays:
Starter cultures of C. neoformans strains were prepared by growth in YPD at 30° overnight with shaking. Overnight cultures, 10 µl, were spread onto both brain–heart infusion (BHI) (Becton Dickinson) and 2.5% pigeon guano agar plates (35 mm), and incubated at 25° overnight. Approximately 50 young adult C. elegans worms were then transferred from a lawn of E. coli OP50 on NGM to the lawn of BHI and pigeon guano media-grown C. neoformans (Mylonakis et al. 2002). Plates were incubated at 25° and worms examined for viability at 24-hr intervals using a dissecting microscope, with worms that did not respond to a touch with a platinum wire pick considered dead. Each experimental condition was performed in triplicate. Survival was plotted against time, and P values were calculated by plotting a Kaplan–Meier survival curve and performing a log-rank (Mantel–Cox) test using Graphpad Prism v. 5.0c. P values <0.05 were considered statistically significant.
Murine virulence assays:
For murine killing assays, C. neoformans strains were used to infect 7-week-old female BALB/c mice by nasal inhalation (Cox et al. 2000). Ten mice were inoculated each with a 50-µl drop containing 1 × 105 cells of each strain. Mice were weighed before infection and daily thereafter; animals were sacrificed by cervical dislocation once their body weight had decreased by 20% of the preinfection body weight. Survival was plotted against time, and P values were calculated by plotting a Kaplan–Meier survival curve and performing a log-rank (Mantel–Cox) test using Graphpad Prism v. 5.0c. P values <0.05 were considered statistically significant.
Ethics statement:
This study was carried out in strict accordance with the recommendations in the Australian Code of Practice for the Care and Use of Animals for Scientific Purposes by the National Health and Medical Research Council. The protocol was approved by the Molecular Biosciences Animal Ethics Committee of The University of Queensland (AEC approval no. SCMB/473/09/UQ/NHMRC). Infection and sacrifice were performed under methoxyflurane anesthesia, and all efforts were made to minimize suffering through adherence to the Guidelines to Promote the Wellbeing of Animals Used for Scientific Purposes as put forward by the National Health and Medical Research Council.
RESULTS
Cryptococcus assimilates a limited subset of nitrogen sources:
Work on Cryptococcus has demonstrated the ability of this organism to assimilate a number of different nitrogen sources (Chaskes and Tyndall 1975; Staib et al. 1976; Polacheck and Kwon-Chung 1980; Fiskin et al. 1990; Kmetzsch et al. 2010). However, these studies have not taken into consideration the significant advances that have been made in understanding the population structure of the species: the former variety Cryptococcus gattii has now been classified as a separate species, and eight distinct Cryptococcus haploid molecular types have been identified, each of which displays distinct physiological properties and virulence features (Kwon-Chung and Varma 2006; Litvintseva et al. 2006). To begin to better understand nitrogen metabolism in this pathogen, we sought to extensively identify the panel of usable nitrogen sources in the context of this complex population structure.
We assessed the ability of an array of 16 strains that represent all eight molecular types of the Cryptococcus species complex to assimilate a diverse range of 42 different compounds as sole nitrogen sources on YNB defined medium, including the common l-amino acids as well as a range of purines, pyrimidines, amides, and amines (Table S3). As commercial formulations of agar have been shown to contain substantial concentrations of nitrogen as impurities (up to 4 mm when 20 g/liter is present), we performed all nitrogen assimilation assays using agarose (SeaKem LE) as the gelling agent (Scholten and Pierik 1998). Unlike other well-known fungal genera such as Aspergillus and Neurospora, which can assimilate a remarkably large collection of nitrogen sources, the nitrogen assimilation profile of Cryptococcus more closely resembled that of Saccharomyces; both species are largely restricted to utilization of ammonium and more complex macromolecules such as amino acids and purines (Kinsky 1961; Hynes 1970). Only a subset of strains was able to utilize xanthine, lysine, and nicotinamide as the sole nitrogen source. None of the strains were able to utilize cadaverine, nitrite, or nitrate as sole nitrogen sources. Importantly, while interstrain growth variability on different nitrogen sources was observed, this did not correlate with molecular type.
The explanation for some of these significant differences between Cryptococcus and Aspergillus or Neurospora is evident at the genomic level. As previously reported, analysis of the C. neoformans genome confirmed an absence of genes required for molybdenum cofactor biosynthesis, an essential component in all previously characterized fungal nitrate metabolism pathways (Heck et al. 2002; Cultrone et al. 2005). Genes encoding homologs of key catabolic enzymes for cadaverine, nitrate, and nitrite utilization were also not found in the genome of VNI strain H99 via BLASTp analysis, explaining the lack of growth on these nitrogen sources. We are therefore unable to explain the Cryptococcus growth phenotype associated with nitrate utilization that was observed by Jiang et al.; consistent with our bioinformatic analyses, none of the 16 Cryptococcus strains tested were able to utilize nitrate or nitrite as the sole nitrogen source (Jiang et al. 2009).
Nitrogen metabolite repression of transcription exists in C. neoformans:
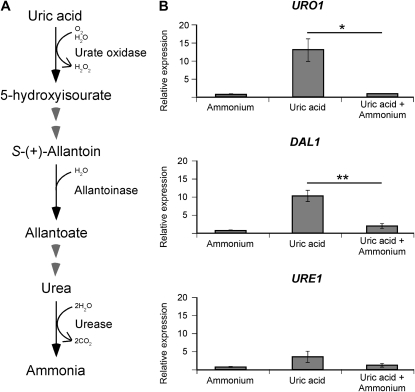
In many members of the phylum Ascomycota, nitrogen metabolite repression ensures that of the options available, favored nitrogen sources such as ammonium are assimilated before all others. To address if the same regulatory mechanism exists in the most clinically prevalent form of C. neoformans (VNI), we bioinformatically identified several predicted catabolic genes belonging to the uric acid degradation pathway in the unpublished H99 genome and employed qRT-PCR to analyze their transcriptional regulation: URO1 (encoding urate oxidase), DAL1 (encoding allantoinase), and URE1 (encoding the virulence factor urease) (Figure 1A). When compared to an ammonium-grown control, the level of expression of URO1 and DAL1 was significantly upregulated (>10-fold) during growth in uric acid as the sole nitrogen source, while that of URE1 was only slightly upregulated (∼3-fold) (Figure 1B). When both uric acid and ammonium were present, this upregulation was abolished, the hallmark of traditional nitrogen metabolite repression (URO1, P = 0.0224; DAL1, P = 0.0058; URE1, P = 0.1448) (Figure 1B). It is worth noting that nitrogen metabolite regulation of URE1 was not statistically significant, indicating that nitrogen metabolite repression may not affect every nitrogen catabolism-associated gene in C. neoformans. However, it is possible that differential expression of URE1 may be observed under other untested nitrogen conditions such as nitrogen starvation.
Figure 1.—
The predicted catabolic enzyme-encoding genes of uric acid, URO1 and DAL1, are sensitive to nitrogen metabolite repression. (A) Scheme representing the predicted (partial) uric acid degradation pathway of C. neoformans. (B) cDNA from wild-type H99 grown in YNB supplemented with ammonium, uric acid, or uric acid plus ammonium (10 mm each nitrogen source) were amplified via qRT-PCR using primers against URO1 (urate oxidase), DAL1 (allantoinase), URE1 (urease), and the control gene ACT1 (actin). In the presence of uric acid as the sole nitrogen source, the expression of URO1 and DAL1 was significantly increased while that of URE1 was slightly increased, but this upregulation was abolished when ammonium was also present. This nitrogen metabolite repression sensitivity of URO1 (* denotes P < 0.05) and DAL1 (** denotes P < 0.01) was statistically significant. Error bars represent standard errors across three biological replicates.
Nitrogen metabolite repression regulates well-established virulence factors:
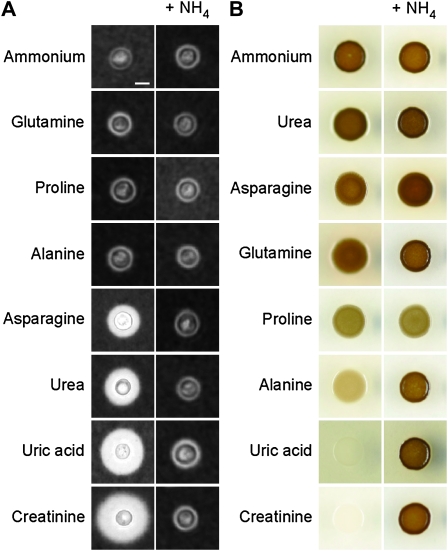
Expanding our analysis, we sought to determine whether nitrogen metabolite repression plays a role in regulating the expression of other key virulence factors. Early studies have shown that the virulence factors capsule and melanin are expressed differently during growth on different nitrogen sources (Chaskes and Tyndall 1975; Staib et al. 1976, 1978). To expand on these classic analyses, we grew C. neoformans on a wider selection of easily assimilated (e.g., ammonium) and less easily assimilated (e.g., uric acid) nitrogen sources and found that varying capsule size was indeed observed in the presence of different nitrogen-yielding substrates (Figure 2A and Figure S1). The size of capsule was uniformly small when C. neoformans was grown on ammonium, glutamine, proline, or alanine as the sole nitrogen source. In contrast, capsule production was much more prolific when cells were grown on asparagine, compounds from the uric acid degradation pathway (urea, uric acid), or creatinine. Significantly, simultaneous addition of ammonium to each of these nitrogen sources repressed capsule production to a level equivalent to that seen on ammonium alone, providing the first evidence that nitrogen metabolite repression directly influences the formation of the virulence determinant capsule (Figure 2A and Figure S1).
Figure 2.—
Nitrogen metabolite repression influences capsule and melanin formation. (A) India ink cell staining under light microscopy showed that wild-type H99 produces capsules that vary in size when grown on YNB supplemented with different nitrogen sources (10 mm each). Capsule size increased in the following order: (ammonium, glutamine, proline, alanine), asparagine, urea, uric acid, and creatinine. Upon coculture of each of these nitrogen sources with ammonium, capsule size was restored to that of the ammonium control. Scale bar, 10 µm. (B) Wild-type H99 produces varying amounts of melanin when grown on l-DOPA supplemented with different nitrogen sources (10 mm each). Melanization increased in the following order: (creatinine, uric acid), alanine, proline and (glutamine, asparagine, urea, ammonium). Upon coculture of each of these nitrogen sources with ammonium, melanin production was restored to that of the ammonium control, with the exception of the proline-grown cells, which melanized to the same extent with or without ammonium.
We next investigated C. neoformans formation of the dark antioxidant pigment melanin on l-DOPA medium supplemented with the same panel of nitrogen sources. Again, varying virulence factor production was observed in response to the available nitrogen source (Figure 2B). Creatinine, uric acid, and alanine-grown cells synthesized the least melanin, proline synthesized an intermediate amount, and glutamine, asparagine, urea, and ammonium were the most prolific producers of melanin. It was interesting to note that the effect of creatinine and uric acid on capsule production was inversely proportional to melanin production: while uric acid and creatinine induce capsule formation to the largest extent, melanization on these nitrogen sources was the poorest. Similarly, while ammonium induces melanization to the largest extent, capsule formation was extremely poor on this nitrogen source. Consistent with conservation of the nitrogen metabolite repression regulatory paradigm, simultaneous addition of ammonium restored melanin production to the same level as that of the ammonium control (Figure 2B). The only unusual exception occurred with proline-grown cells, which melanized to the same extent with or without ammonium, suggesting that proline catabolism in C. neoformans may be insensitive to the repressing effects of ammonium. This observation was not seen with the capsule assays previously, as both proline and ammonium were equally poor inducers of capsule synthesis. Overall, we observed that the general effect of nitrogen metabolite repression on melanin was opposite to that observed for capsule—rather than blocking virulence factor production, the presence of ammonium induces the highest level of melanin formation.
The C. neoformans genome encodes up to seven GATA factors:
Our phenotypic analyses showed that beyond controlling nitrogen catabolism, nitrogen metabolite repression plays a role in regulating at least two important virulence factors in C. neoformans: capsule and melanin. The production of these factors varied on different nitrogen sources, with the addition of ammonium dominating over other nitrogen signals (apart from proline) to either abolish (capsule) or highly activate (melanin) virulence factor production. In the phylum Ascomycota, this dominance of ammonium in controlling a physiological response is mediated by transcription factors belonging to the GATA zinc finger family. We therefore chose to pursue potential GATA factors of C. neoformans as likely candidates for the underlying mechanism controlling these responses to nitrogen availability.
GATA transcription factors are distinguished by their highly conserved DNA-binding domain consisting of a Cys2/Cys2-type zinc finger motif followed by an adjacent basic region (Fu and Marzluf 1990; Marzluf 1997). Within the consensus “Cys–X2–Cys–Xvariable–Cys–X2–Cys,” there are always two residues separating cysteines 1 and 2, and always two residues separating cysteines 3 and 4 of the zinc finger motif. In contrast, the number of residues comprising the central loop separating cysteines 2 and 3 can vary. For example, the zinc finger motif of the A. nidulans nitrogen regulatory protein AreA contains a 17-amino-acid central loop while that of N. crassa blue light response factor WC1 contains 18 residues (Kudla et al. 1990; Ballario et al. 1996). We sought to establish a more precise description of this consensus domain in fungi to assist in accurately predicting all of the bona fide GATA factor-encoding genes in the genome of C. neoformans strain H99.
To more accurately define the sequence constraints associated with fungal GATA factors, we aligned the DNA-binding motif sequences of all 38 known characterized GATA factors, originating from 17 fungal species (Figure S2). By comparing the number of amino acid residues at the central loop of the zinc finger motif and the distribution of basic residues within 25 amino acids adjacent to the zinc finger motif, we were able to define a more stringent GATA motif consensus for fungi: all reported examples matched the motif “Cys–X2–Cys–X17–20–Cys–X2–Cys + 5–12 basic residues within the 25 beyond the most C-terminal cysteine.”
Parallel searches were subsequently taken to identify potential GATA factors in the C. neoformans genome. In the first approach, the protein sequences of all 38 known characterized fungal GATA factors were searched against the Broad Institute genome sequence database of strain H99 for potential homologs using BLASTp, leading to the identification of five potential GATA factor-encoding genes (Table S4). However, this approach was heavily biased in requiring the GATA factors detectable in the C. neoformans genome to have first been identified in other fungi. In the second approach, we searched the C. neoformans genome for the consensus “Cys–X2–Cys–X17–20–Cys–X2–Cys” using a Perl script-based approach, and hits analyzed for a match with the “5–12 basic residues within the 25 beyond the most C-terminal cysteine” consensus. This broader analysis identified the same five GATA factor-encoding genes as the BLASTp analysis, plus two additional candidates. In the third approach, we BLASTed all seven hits back against the H99 genome, finding no additional homologs. Through these combined bioinformatic analyses, we therefore identified a total of seven possible GATA factor-encoding genes. Of the seven predicted GATA ORFs identified, all four previously characterized C. neoformans members of this family were present: Bwc2, Cir1, Gat201, and Gat1—thus validating the combination approach taken (Idnurm and Heitman 2005; Jung et al. 2006; Liu et al. 2008; Kmetzsch et al. 2010). Furthermore, sequence searches of these candidates in Pfam confirmed the presence of a predicted GATA zinc finger, supporting the hypothesis that all seven are bona fide GATA factors. Consistent with the observation of Wong et al., all C. neoformans GATA factors lack putative leucine zippers that are present in negatively acting nitrogen regulatory GATA factors such as Dal80 and AreB of S. cerevisiae and A. nidulans, respectively (Wong et al. 2008).
Only one positively acting GATA factor controls global nitrogen source utilization in C. neoformans:
Despite the fact that four GATA factors have been previously characterized in C. neoformans, to our knowledge any potential role in nitrogen metabolite repression has not been explored in these transcription factors. It is possible that a GATA factor may play a regulatory role in more than one global response or that more than one GATA transcriptional activator may play a role in nitrogen regulation as in systems of S. cerevisiae and C. albicans (Stanbrough et al. 1995; Limjindaporn et al. 2003; Liao et al. 2008). All seven ORFs containing our strict consensus for a fungal GATA DNA-binding domain were therefore deleted via homologous recombination.
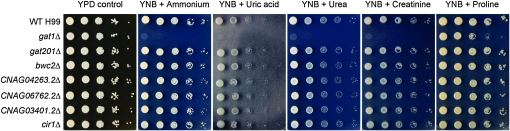
The seven GATA mutants were viable and had similar growth rate as the wild-type strain on YPD medium at 30° (Figure 3). However, all but one of the C. neoformans GATA mutant strains exhibited growth equivalent to wild-type H99 on YNB defined medium supplemented with a wide panel of 38 usable nitrogen sources (Figure 3 and Table S5). Consistent with previous studies conducted by Kmetzsch et al. (2010), deletion of ORF CNAG00193.2 (GAT1) rendered C. neoformans incapable of utilizing a wide repertoire of nitrogen sources. The gat1Δ mutant, however, could still utilize a limited selection of amino acids, notably proline, strengthening our hypothesis that proline utilization is largely independent of nitrogen metabolite repression. Importantly, we found that the gat1Δ mutant was also unable to utilize uric acid and creatinine, two predominant nitrogen constituents found in the ecological niche (pigeon guano) of C. neoformans (Staib et al. 1976, 1978; Casadevall and Perfect 1998).
Figure 3.—
The gat1∆ mutant is unable to utilize a wide variety of nitrogen sources. Tenfold spot dilution assays of wild-type H99 and GATA-type deletion mutants for nitrogen utilization showed that the gat1∆ mutant is unable to grow on YNB supplemented with 10 mm ammonium, uric acid, urea, or creatinine but exhibits only a slight growth defect compared to wild type on 10 mm proline. Complementation of the gat1∆ mutant with the GAT1 gene restored wild-type nitrogen utilization phenotype (Figure S3).
Gat1 mediates nitrogen metabolite repression:
The fact that the gat1∆ mutant does not grow on a wide variety of nitrogen sources does not ipso facto prove that Gat1 mediates nitrogen metabolite repression. To investigate the regulatory mechanism of Gat1, we performed qRT-PCR to analyze how the deletion of GAT1 affects transcriptional regulation of genes associated with nitrogen metabolism. The unusual observation that loss of GAT1 merely reduces growth on the less easily assimilated nitrogen source proline rather than completely abolish it provided an ideal condition under which to study the transcriptional effects of the loss of this regulatory gene. Growth in proline enabled the isolation of RNA from defined medium without concern for starvation-related artifacts of the gat1∆ mutant associated with growth in nonutilizable nitrogen sources such as uric acid. Hence, by choosing proline as the sole source of nitrogen for culturing of both the wild-type and mutant strains, we were able to isolate RNA from a steady-state, actively growing culture with a defined nitrogen source.
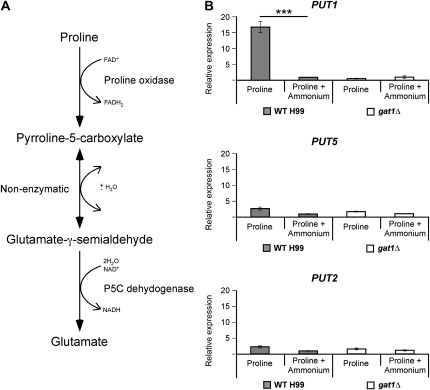
Bioinformatic analyses of the H99 genome identified several genes predicted to encode enzymes required for degradation of proline: PUT1 and PUT5, two paralogs that each encode proline oxidase, and PUT2, which encodes pyrroline-5-carboxylate dehydrogenase (Figure 4A). qRT-PCR using RNA isolated from wild-type cells revealed that only one of these three proline catabolic genes was regulated by nitrogen metabolite repression, with higher levels of transcription (>15-fold) on proline than when both proline and ammonium were present (PUT1, P < 0.0001) (Figure 4B). In the gat1∆ mutant, transcription of PUT1 was not highly activated but instead showed the same level of transcription irrespective of the presence of ammonium, confirming the role of Gat1 as the mediator of nitrogen metabolite repression (Figure 4B). Unlike PUT1, both PUT5 and PUT2 were unaffected by nitrogen metabolite repression, explaining why the gat1∆ mutant can utilize proline as the sole nitrogen source, albeit less effectively than wild type. Although the expression of PUT1 was drastically reduced in the gat1∆ mutant, PUT5 was still expressed at wild-type levels sufficient to produce proline oxidase, as was PUT2 (encoding pyrroline-5-carboxylate dehydrogenase), enabling Gat1-independent growth on proline as the sole nitrogen source.
Figure 4.—
Gat1 regulates nitrogen metabolite repression. (A) Scheme representing the predicted proline degradation pathway of C. neoformans. (B) cDNA from wild-type H99 and gat1∆ mutant grown in YNB supplemented with proline or proline plus ammonium (10 mm each nitrogen source) were amplified via qRT-PCR using primers against PUT1 (proline oxidase), PUT5 (proline oxidase), PUT2 (pyrroline-5-carboxylate dehydrogenase), and the control gene ACT1 (actin). One of the predicted catabolic enzyme-encoding genes of proline, PUT1, was sensitive to nitrogen metabolite repression in wild type (*** denotes P < 0.0001) but not in the gat1∆ mutant. The remaining two catabolic genes, PUT5 and PUT2, were nitrogen metabolite repression insensitive in both strains. Error bars represent standard errors across three biological replicates.
Gat1 positively regulates the expression of key ammonium assimilation and permease genes:
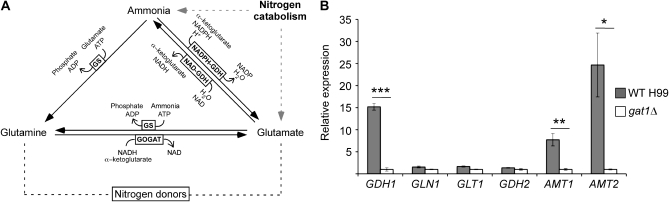
While certain phenotypic similarities to ascomycete nitrogen regulatory GATA factor mutants were clear, this basidiomycete ortholog mutant exhibited an ammonium phenotype much more extreme than that seen in previously described GATA factor mutants. The gat1Δ mutant is unable to utilize the most easily assimilated nitrogen source in both ammonium replete and sufficient conditions (1–100 mm) (data not shown), paradoxically suggesting that the nitrogen metabolite repression mechanism that occurs in the presence of ammonium blocks the ability to utilize ammonium itself. While a slight reduction in growth on ammonium has been observed for other fungal nitrogen regulatory GATA mutants, the complete loss of growth is unprecedented and counterintuitive (Christensen et al. 1998; Hensel et al. 1998; Liao et al. 2008). To investigate this difference in phenotype, we bioinformatically identified the predicted central nitrogen metabolism genes in the H99 genome: GDH2 [encoding NAD+-dependent glutamate dehydrogenase (NAD-GDH)] as well as the ammonium assimilation enzyme-encoding genes GDH1 [encoding NADP+-dependent glutamate dehydrogenase (NADPH-GDH)], GLN1 [encoding glutamine synthetase (GS)], and GLT1 [encoding NAD+-dependent glutamate synthase (GOGAT)] (Figure 5A).
Figure 5.—
Gat1 regulates the expression of the ammonium assimilation enzyme and permease-encoding genes. (A) Scheme representing the predicted pathway of central nitrogen metabolism in C. neoformans. (B) cDNA from wild-type H99 and gat1∆ mutant grown in YPD were amplified via qRT-PCR using primers against GDH1 (NADPH-GDH), GLN1 (GS), GLT1 (GOGAT), GDH2 (NAD-GDH), AMT1 (ammonium transporter 1), AMT2 (ammonium transporter 2), and the control gene ACT1 (actin). The expression of GDH1, AMT1, and AMT2 was significantly lower in the gat1∆ mutant compared to wild type, while that of GLN1, GLT1, and GDH2 was slightly lower. This difference in level of expression for GDH1 (*** denotes P < 0.0001), AMT1 (** denotes P < 0.01), and AMT2 (* denotes P < 0.05) was statistically significant. Error bars represent standard errors across three biological replicates.
qRT-PCR was again employed, this time to analyze the transcriptional regulation of these central nitrogen metabolism genes as well as the two known ammonium permease-encoding genes AMT1 and AMT2 in both the wild-type and gat1Δ mutant strains (Figure 5B) (Rutherford et al. 2008). When grown in rich undefined YPD medium that contains a wide array of nitrogen sources including the generally preferred ammonium, glutamine, and glutamate, the level of expression of the major ammonium assimilation enzyme-encoding gene GDH1 as well as the permease-encoding genes AMT1 and AMT2 was significantly lower (∼10–20-fold) in the gat1Δ mutant compared to wild type (GDH1, P < 0.0001; AMT1, P = 0.0086; AMT2, P = 0.0308). The level of expression of the other central nitrogen metabolism genes was also consistently lower (∼1.5-fold) in the gat1Δ mutant than in wild type, although these differences for the remaining two ammonium assimilation enzyme-encoding genes GLN1 and GLT1 were not quite statistically significant (GLN1, P = 0.0666; GLT1, P = 0.0523; GDH2, P = 0.1308). This result indicates that Gat1 controls the expression of the ammonium assimilation enzyme-encoding gene GDH1 and permease-encoding genes AMT1 and AMT2 in C. neoformans.
Gat1 is not required for the growth of C. neoformans on pigeon guano:
The gat1∆ mutant is unable to utilize uric acid, urea, or creatinine, the most abundant nitrogen sources found in the natural habitat of C. neoformans. This raises the question of whether the GAT1 gene is essential for optimum growth on pigeon guano, a growth medium previously validated for both C. neoformans and C. gattii (Nielsen et al. 2007). We therefore investigated whether the loss of GAT1 affected growth on this substrate that mimics the environmental niche. Our growth assays indicate that the wild-type and gat1∆ mutant strains displayed equivalent growth on both filtered and unfiltered 25% pigeon guano agar media, revealing that pigeon guano contains at least one nitrogen source that supports robust growth and whose utilization is Gat1 independent (Figure S4).
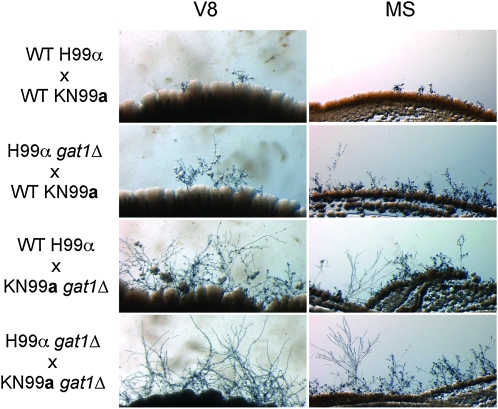
Gat1 negatively regulates mating in both mating-type α and a strains:
During nitrogen limiting or starvation conditions, C. neoformans can undergo sexual reproduction that leads to the production of potentially infectious air-borne basidiospores (Kwon-Chung 1976; Lengeler et al. 2000; Giles et al. 2009). To address whether Gat1 plays a role in this sexual development, we created a gat1Δ derivative of MATa C. neoformans strain KN99a (VNI) by replacing the GAT1 ORF with the nourseothricin acetyl-transferase (NAT) selectable marker, to serve as a mating partner for our existing gat1::NEO of MATα strain H99. Wild type (wild type × wild type), unilateral (gat1 × wild type), and bilateral (gat1 × gat1) crosses were then performed by mixing strains of opposite mating type on V8 and MS media. The ability to mate was examined by microscopically inspecting the production of filaments and basidia at the edges of the mating patches. Unexpectedly, filamentation and production of basidia were enhanced in both unilateral and bilateral gat1Δ crosses in comparison to wild-type crosses (Figure 6). Furthermore, deletion of GAT1 in both the MATα and MATa strains led to the most robust filament formation and production of basidia, indicating that the effect of this gene deletion is additive. This result indicates that Gat1 plays a critical role in regulating morphological differentiation in both mating types of C. neoformans.
Figure 6.—
Gat1 inhibits filament formation during mating. Filamentation assays on V8 and MS mating media (pH 5.0) showed that filament formation was enhanced in unilateral gat1Δ crosses in comparison to the wild-type crosses. Filamentation was enhanced even further in a bilateral gat1Δ cross. Complementation of the H99α gat1Δ and KN99a gat1Δ mutants with the GAT1 gene restored wild-type mating in all tested crosses (data not shown).
Gat1 positively regulates capsule synthesis, but negatively regulates melanin formation and growth at high body temperature:
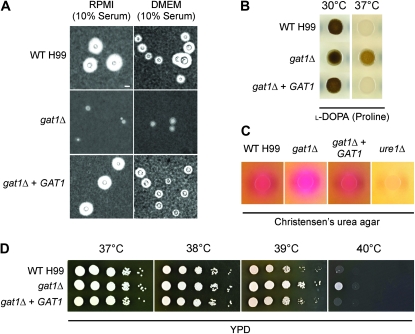
Given that nitrogen metabolite repression plays a role in regulation of capsule synthesis, we next chose to examine capsule production by the gat1Δ mutant. Strains were cultured in amino-acid-rich RPMI and DMEM media supplemented with 10% fetal calf serum; both media supported growth of all strains including the gat1∆ mutant (Zaragoza et al. 2003). India ink staining revealed that while the wild-type strain produced characteristic halos around its cells representing enlarged capsule, the gat1Δ mutant produced reduced amounts of capsule under these serum-induced conditions (Figure 7A and Figure S5). This result indicates that Gat1 plays a positive role in the regulation of capsule synthesis.
Figure 7.—
Gat1 is required for capsule synthesis, but negatively regulates melanization and growth at 39° or 40°. (A) India ink cell staining under light microscopy revealed that the wild-type H99 and gat1Δ + GAT1 strains produce enlarged capsules while the gat1∆ mutant produces residual amount of capsule when strains were cultured under serum-induced growth conditions. Scale bar, 10 µm. (B) The gat1∆ mutant produces more melanin compared to both the wild-type and gat1∆ + GAT1 strains when grown on l-DOPA medium supplemented with 10 mm proline at 37°. In contrast, all three strains melanized to the same extent when grown at 30°. (C) Unlike the negative control ure1∆ mutant, the wild-type, gat1∆ mutant and gat1∆ + GAT1 strains all had the ability to produce urease when grown on Christensen’s urea agar as reflected by the bright pink clearing surrounding the aliquot of spotted cells. (D) Tenfold spot dilution assays on YPD medium at human body temperature showed that the gat1∆ mutant exhibits enhanced growth compared to both the wild-type and gat1∆ + GAT1 strains at 39° and 40°.
Like capsule, melanin synthesis is also under the influence of nitrogen metabolite repression control. The role of Gat1 was therefore evaluated for melanin production at both 30° and 37° on l-DOPA medium supplemented with proline as the sole nitrogen source, since proline supports the most robust growth in the gat1∆ mutant on defined medium. Interestingly, the gat1∆ mutant melanized to the same extent as wild type at 30° but produced more melanin than wild type at 37° (Figure 7B). This result indicates that Gat1 plays a negative role in regulating melanin production at human body temperature.
We also evaluated the role of Gat1 in regulating the activity of the nitrogen-scavenging enzyme urease on Christensen’s urea agar. Although both the gat1∆ and negative control ure1∆ mutants are unable to utilize urea as the sole nitrogen source, the mutants could still grow on Christensen’s medium as impurities found in agar likely provide alternative sources of consumable nitrogen. In support of our previous qRT-PCR data indicating that urease is not significantly regulated by nitrogen metabolite repression, the gat1Δ mutant still produces urease even though the mutant is unable to utilize the ammonia resulting from urea catabolism (Figure 7C).
To complete a thorough phenotypic analysis of the gat1Δ mutant, we evaluated the role of Gat1 for its ability to grow at high temperature (37°–40°), a common stress that C. neoformans encounters both in a mammalian host and in its ecological niche. While growth up to 38° was equivalent in the wild-type and gat1∆ mutant strains, the mutant exhibited better growth in comparison to wild type at 39°, the febrile temperature commonly experienced by most patients upon the onset of cryptococcal meningoencephalitis (Figure 7D). Furthermore, at 40°, growth of the wild-type strain was completely abolished while the gat1∆ mutant was still able to proliferate, albeit at a slow rate. This unanticipated finding indicates that Gat1 plays a negative role in regulating growth at high temperature.
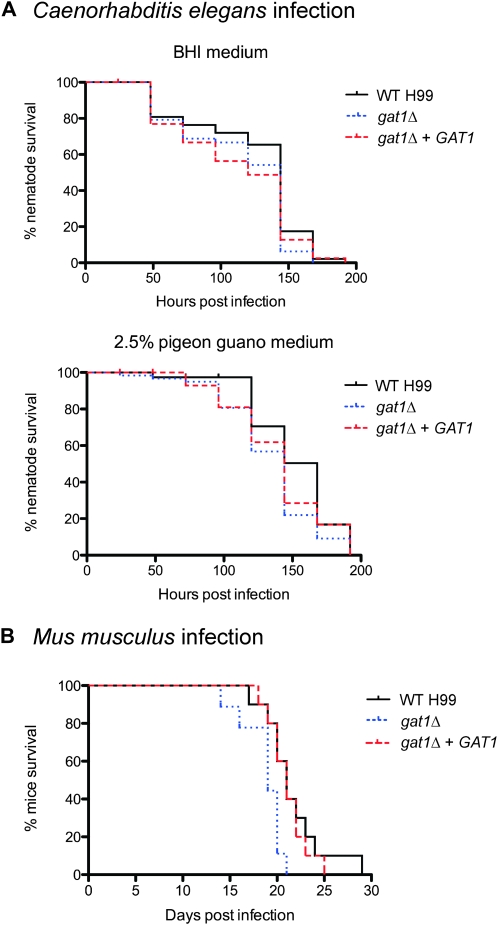
Gat1 is not required for the killing of C. elegans but modestly represses virulence in a murine inhalation model of cryptococcosis:
Our ultimate interest in C. neoformans lies in better understanding its pathogenicity. In this context our data thus far were confusing, as Gat1 arguably contributes to production of some virulence attributes but repression of others. For example, the gat1Δ mutant’s reduction in capsule synthesis and abated ability to utilize a variety of nitrogen sources suggest that this strain should exhibit a decrease in virulence, just like that seen in equivalent ascomycete GATA factor mutants. In contrast, the increase in melanin production and enhanced growth at high temperature would support an increase in virulence. To determine the effect of Gat1 on virulence, we performed C. elegans killing assays and a murine inhalation model of cryptococcosis.
C. neoformans presumably interacts with C. elegans in the environment and killing of C. elegans by C. neoformans in vitro has previously been validated as a model for studying pathogenesis (Mylonakis et al. 2002). We performed C. elegans virulence assays using two different media: the standard BHI medium for nematode killing experiments, as well as 2.5% pigeon guano medium to mimic C. neoformans ecological niche. Under both growth conditions, killing of C. elegans by the gat1Δ mutant [LT50 (time for half of the worms to die) = 6 days for both BHI and pigeon guano media] was not significantly different to that observed for wild type (LT50 = 6 days and 7 days, for BHI and pigeon guano media, respectively) (Figure 8A). Gat1 is therefore not required for C. neoformans-mediated killing of the invertebrate C. elegans.
Figure 8.—
The gat1Δ mutant kills C. elegans as efficiently as wild-type H99, but exhibits modestly enhanced virulence in a murine host. (A) C. elegans infection: ∼50 nematode worms were transferred to a lawn of wild-type, gat1Δ, or gat1Δ + GAT1 cells as the sole food source on both BHI and 2.5% pigeon guano media, and survival was monitored at 24-hr intervals. There was no observable difference in C. elegans killing by all three strains on both BHI (wild type vs. gat1Δ, P = 0.1066; gat1Δ vs. gat1Δ + GAT1, P = 0.9230) and pigeon guano media (wild type vs. gat1Δ, P = 0.0250; gat1Δ vs.gat1Δ + GAT1, P = 0.3614). (B) Mus musculus infection: 10 mice were infected intranasally with either 1 × 105 cells of wild type, gat1Δ, or gat1Δ + GAT1 strains, and survival was monitored daily. Mice infected with the wild-type and gat1Δ + GAT1 strains progress to morbidity at the same rate (wild type vs. gat1Δ + GAT1, P = 0.6691), whereas mice infected with the gat1Δ strain progress to morbidity more rapidly (wild type vs. gat1Δ, P = 0.0151).
On the other hand, the murine inhalation model of cryptococcosis more closely mimics human infection by C. neoformans—inhaled cryptococcal cells first infect the lungs before disseminating to the brain to cause meningoencephalitis. Interestingly, mice infected with the gat1Δ mutant succumbed to infection slightly faster (between 14 and 21 days postinfection, median survival of 19 days) than mice infected with the wild-type strain (between 17 and 28 days postinfection, median survival of 23 days) (wild type vs. gat1Δ, P = 0.0151) (Figure 8B). This result suggests that the gat1Δ mutant is modestly more virulent than wild type during murine infection. Thus, in addition to regulating nitrogen metabolism and virulence factor expression in vitro, Gat1 also represses virulence in a vertebrate host.
DISCUSSION
The ability to acquire and catabolize nutrients from the environment is imperative for survival of an organism. While fungi are well known for their ability to utilize a broad range of nitrogen sources, the study of molecular mechanisms controlling nitrogen acquisition and catabolism in model fungi have until now been primarily limited to members of the phylum Ascomycota (Wong et al. 2008). Our studies of transcriptional regulation and phenotypic responses to the presence of ammonium have shown that utilization of nitrogen sources by C. neoformans is controlled in a similar fashion. The results we report here represent the first definitive characterization of the regulatory phenomenon of nitrogen metabolite repression in a second phylum of the kingdom fungi, the Basidiomycota. In addition to functional conservation between these two phyla, the repertoire of physiological responses controlled by nitrogen metabolite repression in C. neoformans has expanded to encompass regulation of virulence factor expression. We have also shown that the GATA factor Gat1 is responsible for mediating nitrogen metabolite repression in C. neoformans.
Bioinformatically, the predicted GATA factor of C. neoformans Gat1 shows little similarity to nitrogen regulatory GATA factors of the Ascomycota for the majority of the protein, except within the GATA motif itself (Table S6). With 19% overall protein identity, it showed poor but still highest similarity to the various nitrogen regulatory GATA factors of the Ascomycota. This similarity, however, was largely restricted to the predicted GATA zinc finger where identity to A. nidulans AreA and N. crassa Nit2 was 88 and 90%, respectively.
The dependence of C. neoformans nitrogen metabolism on a single GATA factor is in stark contrast to S. cerevisiae or C. albicans, where multiple nitrogen regulatory GATA factors are required (Stanbrough et al. 1995; Limjindaporn et al. 2003; Liao et al. 2008). It is important to note that the existence of the two positively acting GATA factors, Gln3 and Gat1, in the hemiascomycete S. cerevisiae is not the result of the whole-genome duplication event that occurred around 100 million years ago, since these two orthologs are also present in C. albicans, a species that did not undergo this event (Wong et al. 2008; Gordon et al. 2009). C. neoformans global nitrogen regulatory circuit instead more closely resembles the single positively acting GATA factor systems in A. nidulans and N. crassa (Stewart and Vollmer 1986; Kudla et al. 1990). Given that N. crassa nit-2 was named after the mutant’s inability to utilize nitrate, which C. neoformans is unable to do, and the apparent existence of only one GATA factor responsible for nitrogen utilization, we therefore recommend an alternative and more informative name to this gene: ARE1.
The similarities in the global nitrogen regulatory circuit between C. neoformans and A. nidulans or N. crassa continue when NmrA and Nmr1 are considered. The Nmr proteins are inhibitors of the functions of A. nidulans AreA and N. crassa Nit2, and a potential homolog of this protein named Tar1 has recently been characterized in C. neoformans, suggesting that the regulation of GATA factor activity may operate in a similar fashion to these filamentous ascomycetes (Pan et al. 1997; Andrianopoulos et al. 1998; Jiang et al. 2009). However, this model is confounded by conflicting observations between the studies of Jiang et al. and our own. Most notably, we have bioinformatics and phenotypic evidence indicating that C. neoformans is unable to utilize nitrate as a nitrogen source. Confusingly, Jiang et al. observed growth changes on this same nitrogen source as evidence that Tar1 plays a role in nitrogen metabolism, an observation that we are unable to explain. Furthermore, C. neoformans Gat1/Are1 lacks the highly conserved C-terminus sequence of AreA and Nit2, which is involved in Nmr recognition (Platt et al. 1996; Pan et al. 1997; Andrianopoulos et al. 1998). Nevertheless, Nmr proteins also interact with the DNA-binding domain of AreA and Nit2, and further study will be required to determine if Tar1 is a true functional ortholog of NmrA/Nmr1.
Unlike regulation of AreA and Nit2 by Nmr, the S. cerevisiae GATA transcriptional activators Gln3 and Gat1 are negatively regulated by the structurally unrelated prion-forming glutathione-S-transferase Ure2, predicted to have been horizontally transferred from the bacterial species Burkholderia vietnamiensis (Masison and Wickner 1995; Xu et al. 1995; Hall and Dietrich 2007; Wong et al. 2008). Our analyses did not identify a Ure2-like candidate encoded in the C. neoformans genome. Together, these data support the model that C. neoformans and the filamentous ascomycetes share a nitrogen regulatory mechanism that more closely resembles the ancestral nitrogen metabolism regulatory pathway than that seen in S. cerevisiae. We therefore propose that the last common ancestor of the Ascomycota and Basidiomycota likely had one positively acting GATA factor (the AreA/Nit2/Gat1/Are1 ortholog) and corepressor protein Nmr; after the phyla separated, a second GATA factor (Gln3) and Ure2 coevolved in S. cerevisiae, and the nmr gene was lost.
While the effect of the loss of GAT1/ARE1 on nitrogen metabolism largely met our predicted phenotype, a key aspect of it did not. One of the most confusing, and seemingly contradictory, phenotypes of the gat1/are1∆ mutant is its inability to grow on ammonium as the sole nitrogen source. We subsequently gained insights into this paradox by proving that along with its established role as an activator of secondary catabolic gene expression in the absence of ammonium, Gat1/Are1 is also functional in the presence of ammonium. Like A. nidulans AreA, C. neoformans Gat1/Are1 also regulates the expression of the major ammonium assimilation enzyme-encoding gene GDH1 as well as the permease-encoding genes AMT1 and AMT2 (Christensen et al. 1998; Monahan et al. 2002, 2006; Rutherford et al. 2008). Since the alternative pathway of ammonium assimilation (via GLN1 and GLT1) is not significantly impaired in the gat1/are1Δ mutant, we speculate that the basal levels of transcription of AMT1 and AMT2 may be insufficient for ammonium uptake. It is worth noting that our nitrogen metabolite repression study followed the traditional use of ammonium as the “repressing” nitrogen source. In reality, it is likely that the true signal affecting Gat1/Are1 activity may be intracellular concentrations of glutamine and/or glutamate since these metabolites reflect the nitrogen status of the cell in other species. Further work using glutamine, glutamate, and perhaps even nitrogen starvation will be required to dissect this aspect of the nitrogen-sensing mechanism.
The opposing regulatory effect of Gat1/Are1 on melanin and capsule production has been observed in previously characterized GATA factors of C. neoformans: Cir1 and Gat201 (Jung et al. 2006; Liu et al. 2008). Notably, although LAC1 is the major contributor to melanin biosynthesis, we were unable to find putative Gat1/Are1 HGATAR binding sites within a 1-kb region upstream from the start codon of LAC1 (Pukkila-Worley et al. 2005). In contrast, numerous HGATAR sites were found in the promoter regions of CAP60 and CAP64, two key genes that are regulated to control capsule biosynthesis (Chang et al. 1996; Chang and Kwon-Chung 1998). Together, these data suggest that Gat1/Are1 may activate the transcription of capsule biosynthesis genes directly, but indirectly regulate melanin production, perhaps by regulating the expression of a repressor of LAC1 transcription.
To gain insights into the role of Gat1/Are1 in the environment we employed a novel approach, combining an established virulence model based on a known predator (C. elegans) with a medium designed to emulate the environmental niche (pigeon guano). This study enabled us to make an important observation: Gat1/Are1 does not appear to have a significant effect on virulence in the environmental niche; instead it may play a role in nitrogen scavenging. However, in a murine host, this gene has likely been co-opted into regulating various aspects of the virulence composite.
It is nevertheless worth noting that the C. elegans killing assays were conducted at 25° instead of mammalian body temperature. Whereas the enhanced growth and melanization abilities of the gat1/are1Δ mutant may be exhibited during the infection process in mice, these virulence attributes could not be displayed in C. elegans. While the slightly quicker progression to morbidity in mice infected with the gat1/are1Δ mutant would not be considered great enough to classify this strain as “hypervirulent,” it certainly highlights the complexity of the role of Gat1/Are1 in gene regulation of C. neoformans during infection. We note that Kmetzsch et al. (2010) recently reported that the gat1/are1Δ mutant exhibited equivalent virulence to wild type when high doses (1 × 107 cells per strain) were used, which resulted in mice succumbing to infection as early as the third day postinfection. In contrast, we employed the more traditional inoculum of 1 × 105 cells to ensure the gradual progression of disease. We believe this increased sensitivity helped identify the subtle difference in virulence between the wild-type and gat1/are1Δ mutant strains that had previously been missed.
Notably, all other known GATA factors of C. neoformans (Bwc2, Cir1, and Gat201) play a role in virulence factor expression in vitro and affect virulence during murine infection (Idnurm and Heitman 2005; Jung et al. 2006; Liu et al. 2008). Bwc2, Cir1, Gat201, and Gat1/Are1 together with the three other uncharacterized GATA factors (CNAG03401.2, CNAG04263.2, CNAG06762.2) we have identified are predicted to bind to 5′-GATA-3′ sites in the genome (Ko and Engel 1993; Merika and Orkin 1993; Ravagnani et al. 1997). Whether these GATA factors antagonize and compete for the same 5′-GATA-3′ binding sites in the promoter regions of genes, act in synergy to activate transcription of genes, or act interchangably remains to be determined. Notwithstanding, it is becoming increasingly apparent that Bwc2, Cir1, Gat201, and Gat1/Are1 can act in parallel to regulate multiple virulence pathways in C. neoformans.
In summary, Gat1/Are1 not only regulates nitrogen metabolite repression and the expression of catabolic enzyme and permease-encoding genes required for nitrogen assimilation, it is also a key regulator of essential virulence traits in this important human pathogen. While the significance of Gat1/Are1 is now clearly demonstrated, full appreciation of its role awaits further analysis of the gene targets and processes regulated by this global transcription factor as well as its potential interactions with other GATA factors. Certainly, a more complete understanding of the complex regulatory circuit governing nitrogen metabolism and virulence mechanisms in C. neoformans will require further study.
Acknowledgments
We thank Bob Simpson for assistance with qRT-PCR, Jason Stajich for assistance with Perl script-based bioinformatic analyses, Jim Kronstad for the provision of the cir1Δ strains, and Gary Newell of the Queensland Racing Pigeon Federation, Inc., for the provision of pigeon guano.
LITERATURE CITED
- Altschul S. F., Gish W., Miller W., Myers E. W., Lipman D. J., 1990. Basic local alignment search tool. J. Mol. Biol. 215: 403–410 [DOI] [PubMed] [Google Scholar]
- Andrianopoulos A., Kourambas S., Sharp J. A., Davis M. A., Hynes M. J., 1998. Characterization of the Aspergillus nidulans nmrA gene involved in nitrogen metabolite repression. J. Bacteriol. 180: 1973–1977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arst H. N., Jr, Cove D. J., 1973. Nitrogen metabolite repression in Aspergillus nidulans. Mol. Gen. Genet. 126: 111–141 [DOI] [PubMed] [Google Scholar]
- Ballario P., Vittorioso P., Magrelli A., Talora C., Cabibbo A., et al. , 1996. White collar-1, a central regulator of blue light responses in Neurospora, is a zinc finger protein. EMBO J. 15: 1650–1657 [PMC free article] [PubMed] [Google Scholar]
- Berger H., Pachlinger R., Morozov I., Goller S., Narendja F., et al. , 2006. The GATA factor AreA regulates localization and in vivo binding site occupancy of the nitrate activator NirA. Mol. Microbiol. 59: 433–446 [DOI] [PubMed] [Google Scholar]
- Berger H., Basheer A., Bock S., Reyes-Dominguez Y., Dalik T., et al. , 2008. Dissecting individual steps of nitrogen transcription factor cooperation in the Aspergillus nidulans nitrate cluster. Mol. Microbiol. 69: 1385–1398 [DOI] [PubMed] [Google Scholar]
- Brenner S., 1974. The genetics of Caenorhabditis elegans. Genetics 77: 71–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caddick M. X., 1992. Characterization of a major Aspergillus regulatory gene, areA, pp. 141–152 Molecular Biology of Filamentous Fungi, edited by Stal U., Tudzynski P. VCH Press, Weinheim, Germany [Google Scholar]
- Carroll S. B., Salt S. D., 2004. Ecology for Gardeners. Timber Press, Portland, OR [Google Scholar]
- Casadevall A., Perfect J. R., 1998. Cryptococcus neoformans. ASM Press, Washington, DC [Google Scholar]
- Chang Y. C., Kwon-Chung K. J., 1998. Isolation of the third capsule-associated gene, CAP60, required for virulence in Cryptococcus neoformans. Infect. Immun. 66: 2230–2236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang Y. C., Penoyer L. A., Kwon-Chung K. J., 1996. The second capsule gene of Cryptococcus neoformans, CAP64, is essential for virulence. Infect. Immun. 64: 1977–1983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaskes S., Tyndall R. L., 1975. Pigment production by Cryptococcus neoformans from para- and ortho-Diphenols: effect of the nitrogen source. J. Clin. Microbiol. 1: 509–514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen T., Hynes M. J., Davis M. A., 1998. Role of the regulatory gene areA of Aspergillus oryzae in nitrogen metabolism. Appl. Environ. Microbiol. 64: 3232–3237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox G. M., Mukherjee J., Cole G. T., Casadevall A., Perfect J. R., 2000. Urease as a virulence factor in experimental cryptococcosis. Infect. Immun. 68: 443–448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cultrone A., Scazzocchio C., Rochet M., Montero-Moran G., Drevet C., et al. , 2005. Convergent evolution of hydroxylation mechanisms in the fungal kingdom: molybdenum cofactor-independent hydroxylation of xanthine via alpha-ketoglutarate-dependent dioxygenases. Mol. Microbiol. 57: 276–290 [DOI] [PubMed] [Google Scholar]
- Davidson R. C., Cruz M. C., Sia R. A., Allen B., Alspaugh J. A., et al. , 2000. Gene disruption by biolistic transformation in serotype D strains of Cryptococcus neoformans. Fungal Genet. Biol. 29: 38–48 [DOI] [PubMed] [Google Scholar]
- Davidson R. C., Blankenship J. R., Kraus P. R., de Jesus Berrios M., Hull C. M., et al. , 2002. A PCR-based strategy to generate integrative targeting alleles with large regions of homology. Microbiology 148: 2607–2615 [DOI] [PubMed] [Google Scholar]
- Davis M. A., Kelly J. M., Hynes M. J., 1993. Fungal catabolic gene regulation: molecular genetic analysis of the amdS gene of Aspergillus nidulans. Genetica 90: 133–145 [DOI] [PubMed] [Google Scholar]
- D'Souza C. A., Alspaugh J. A., Yue C., Harashima T., Cox G. M., et al. , 2001. Cyclic AMP-dependent protein kinase controls virulence of the fungal pathogen Cryptococcus neoformans. Mol. Cell Biol. 21: 3179–3191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng B., Marzluf G. A., 1998. Interaction between major nitrogen regulatory protein NIT2 and pathway-specific regulatory factor NIT4 is required for their synergistic activation of gene expression in Neurospora crassa. Mol. Cell Biol. 18: 3983–3990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fincham J. R. S., 1950. Mutant strains of Neurospora deficient in aminating ability. J. Biol. Chem. 182: 61–74 [Google Scholar]
- Fiskin A. M., Zalles M. C., Garrison R. G., 1990. Electron cytochemical studies of Cryptococcus neoformans grown on uric acid and related sources of nitrogen. J. Med. Vet. Mycol. 28: 197–207 [PubMed] [Google Scholar]
- Fraser J. A., Davis M. A., Hynes M. J., 2001. The formamidase gene of Aspergillus nidulans: regulation by nitrogen metabolite repression and transcriptional interference by an overlapping upstream gene. Genetics 157: 119–131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser J. A., Subaran R. L., Nichols C. B., Heitman J., 2003. Recapitulation of the sexual cycle of the primary fungal pathogen Cryptococcus neoformans var. gattii: implications for an outbreak on Vancouver Island, Canada. Eukaryot. Cell 2: 1036–1045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y. H., Marzluf G. A., 1987. Characterization of nit-2, the major nitrogen regulatory gene of Neurospora crassa. Mol. Cell Biol. 7: 1691–1696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y. H., Marzluf G. A., 1990. nit-2, the major positive-acting nitrogen regulatory gene of Neurospora crassa, encodes a sequence-specific DNA-binding protein. Proc. Natl. Acad. Sci. USA 87: 5331–5335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garsin D. A., Sifri C. D., Mylonakis E., Qin X., Singh K. V., et al. , 2001. A simple model host for identifying Gram-positive virulence factors. Proc. Natl. Acad. Sci. USA 98: 10892–10897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giles S. S., Dagenais T. R., Botts M. R., Keller N. P., Hull C. M., 2009. Elucidating the pathogenesis of spores from the human fungal pathogen Cryptococcus neoformans. Infect. Immun. 77: 3491–3500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon J. L., Byrne K. P., Wolfe K. H., 2009. Additions, losses, and rearrangements on the evolutionary route from a reconstructed ancestor to the modern Saccharomyces cerevisiae genome. PLoS Genet. 5: e1000485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall C., Dietrich F. S., 2007. The reacquisition of biotin prototrophy in Saccharomyces cerevisiae involved horizontal gene transfer, gene duplication and gene clustering. Genetics 177: 2293–2307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heck I. S., Schrag J. D., Sloan J., Millar L. J., Kanan G., et al. , 2002. Mutational analysis of the gephyrin-related molybdenum cofactor biosynthetic gene cnxE from the lower eukaryote Aspergillus nidulans. Genetics 161: 623–632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hensel M., Arst H. N., Jr, Aufauvre-Brown A., Holden D. W., 1998. The role of the Aspergillus fumigatus areA gene in invasive pulmonary aspergillosis. Mol. Gen. Genet. 258: 553–557 [DOI] [PubMed] [Google Scholar]
- Honda S., Ishii N., Suzuki K., Matsuo M., 1993. Oxygen-dependent perturbation of life span and aging rate in the nematode. J. Gerontol. 48: B57–B61 [DOI] [PubMed] [Google Scholar]
- Hynes M. J., 1970. Induction and repression of amidase enzymes in Aspergillus nidulans. J. Bacteriol. 103: 482–487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Idnurm A., Heitman J., 2005. Light controls growth and development via a conserved pathway in the fungal kingdom. PLoS Biol. 3: e95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang N., Xiao D., Zhang D., Sun N., Yan B., et al. , 2009. Negative roles of a novel nitrogen metabolite repression-related gene, TAR1, in laccase production and nitrate utilization by the basidiomycete Cryptococcus neoformans. Appl. Environ. Microbiol. 75: 6777–6782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung W. H., Sham A., White R., Kronstad J. W., 2006. Iron regulation of the major virulence factors in the AIDS-associated pathogen Cryptococcus neoformans. PLoS Biol. 4: e410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerry S., TeKippe M., Gaddis N. C., Aballay A., 2006. GATA transcription factor required for immunity to bacterial and fungal pathogens. PLoS One 1: e77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kingsbury J. M., Goldstein A. L., McCusker J. H., 2006. Role of nitrogen and carbon transport, regulation, and metabolism genes for Saccharomyces cerevisiae survival in vivo. Eukaryot. Cell 5: 816–824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinsky S. C., 1961. Induction and repression of nitrate reductase in Neurospora crassa. J. Bacteriol. 82: 898–904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kmetzsch L., Staats C. C., Simon E., Fonseca F. L., Oliveira D. L., et al. , 2010. The GATA-type transcriptional activator Gat1 regulates nitrogen uptake and metabolism in the human pathogen Cryptococcus neoformans. Fungal Genet. Biol. [DOI] [PubMed] [Google Scholar]
- Ko L. J., Engel J. D., 1993. DNA-binding specificities of the GATA transcription factor family. Mol. Cell Biol. 13: 4011–4022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kudla B., Caddick M. X., Langdon T., Martinez-Rossi N. M., Bennett C. F., et al. , 1990. The regulatory gene areA mediating nitrogen metabolite repression in Aspergillus nidulans. Mutations affecting specificity of gene activation alter a loop residue of a putative zinc finger. EMBO J. 9: 1355–1364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon-Chung K. J., 1976. Morphogenesis of Filobasidiella neoformans, the sexual state of Cryptococcus neoformans. Mycologia 68: 821–833 [PubMed] [Google Scholar]
- Kwon-Chung K. J., Varma A., 2006. Do major species concepts support one, two or more species within Cryptococcus neoformans? FEMS Yeast Res. 6: 574–587 [DOI] [PubMed] [Google Scholar]
- Kwon-Chung K. J., Bennett J. E., Rhodes J. C., 1982. Taxonomic studies on Filobasidiella species and their anamorphs. Antonie van Leeuwenhoek 48: 25–38 [DOI] [PubMed] [Google Scholar]
- Lengeler K. B., Wang P., Cox G. M., Perfect J. R., Heitman J., 2000. Identification of the MATa mating-type locus of Cryptococcus neoformans reveals a serotype A MATa strain thought to have been extinct. Antonie Van Leeuwenhoek 97: 14455–14460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao W. L., Ramon A. M., Fonzi W. A., 2008. GLN3 encodes a global regulator of nitrogen metabolism and virulence of C. albicans. Fungal Genet. Biol. 45: 514–526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Limjindaporn T., Khalaf R. A., Fonzi W. A., 2003. Nitrogen metabolism and virulence of Candida albicans require the GATA-type transcriptional activator encoded by GAT1. Mol. Microbiol. 50: 993–1004 [DOI] [PubMed] [Google Scholar]
- Litvintseva A. P., Thakur R., Vilgalys R., Mitchell T. G., 2006. Multilocus sequence typing reveals three genetic subpopulations of Cryptococcus neoformans var. grubii (serotype A), including a unique population in Botswana. Genetics 172: 2223–2238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu O. W., Chun C. D., Chow E. D., Chen C., Madhani H. D., et al. , 2008. Systematic genetic analysis of virulence in the human fungal pathogen Cryptococcus neoformans. Cell 135: 174–188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak K. J., Schmittgen T. D., 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)). Method. Methods 25: 402–408 [DOI] [PubMed] [Google Scholar]
- Magasanik B., Kaiser C. A., 2002. Nitrogen regulation in Saccharomyces cerevisiae. Gene 290: 1–18 [DOI] [PubMed] [Google Scholar]
- Marzluf G. A., 1997. Genetic regulation of nitrogen metabolism in the fungi. Microbiol. Mol. Biol. Rev. 61: 17–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masison D. C., Wickner R. B., 1995. Prion-inducing domain of yeast Ure2p and protease resistance of Ure2p in prion-containing cells. Science 270: 93–95 [DOI] [PubMed] [Google Scholar]
- Merika M., Orkin S. H., 1993. DNA-binding specificity of GATA family transcription factors. Mol. Cell Biol. 13: 3999–4010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell A. P., Magasanik B., 1984. Regulation of glutamine-repressible gene products by the GLN3 function in Saccharomyces cerevisiae. Mol. Cell Biol. 4: 2758–2766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monahan B. J., Fraser J. A., Hynes M. J., Davis M. A., 2002. Isolation and characterization of two ammonium permease genes, meaA and mepA, from Aspergillus nidulans. Eukaryot. Cell 1: 85–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monahan B. J., Askin M. C., Hynes M. J., Davis M. A., 2006. Differential expression of Aspergillus nidulans ammonium permease genes is regulated by GATA transcription factor AreA. Eukaryot. Cell 5: 226–237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mylonakis E., Ausubel F. M., Perfect J. R., Heitman J., Calderwood S. B., 2002. Killing of Caenorhabditis elegans by Cryptococcus neoformans as a model of yeast pathogenesis. Proc. Natl. Acad. Sci. USA 99: 15675–15680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen K., Cox G. M., Wang P., Toffaletti D. L., Perfect J. R., et al. , 2003. Sexual cycle of Cryptococcus neoformans var. grubii and virulence of congenic a and alpha isolates. Infect. Immun. 71: 4831–4841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen K., De Obaldia A. L., Heitman J., 2007. Cryptococcus neoformans mates on pigeon guano: implications for the realized ecological niche and globalization. Eukaryot. Cell 6: 949–959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olszewski M. A., Noverr M. C., Chen G. H., Toews G. B., Cox G. M., et al. , 2004. Urease expression by Cryptococcus neoformans promotes microvascular sequestration, thereby enhancing central nervous system invasion. Am. J. Pathol. 164: 1761–1771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan H., Feng B., Marzluf G. A., 1997. Two distinct protein-protein interactions between the NIT2 and NMR regulatory proteins are required to establish nitrogen metabolite repression in Neurospora crassa. Mol. Microbiol. 26: 721–729 [DOI] [PubMed] [Google Scholar]
- Platt A., Langdon T., Arst H. N., Jr, Kirk D., Tollervey D., et al. , 1996. Nitrogen metabolite signalling involves the C-terminus and the GATA domain of the Aspergillus transcription factor AREA and the 3′ untranslated region of its mRNA. EMBO J. 15: 2791–2801 [PMC free article] [PubMed] [Google Scholar]
- Polacheck I., Kwon-Chung K. J., 1980. Creatinine metabolism in Cryptococcus neoformans and Cryptococcus bacillisporus. J. Bacteriol. 142: 15–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Postgate J., 1998. Nitrogen Fixation. Cambridge University Press, Cambridge, UK [Google Scholar]
- Pukkila-Worley R., Gerrald Q. D., Kraus P. R., Boily M. J., Davis M. J., et al. , 2005. Transcriptional network of multiple capsule and melanin genes governed by the Cryptococcus neoformans cyclic AMP cascade. Eukaryot. Cell 4: 190–201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravagnani A., Gorfinkiel L., Langdon T., Diallinas G., Adjadj E., et al. , 1997. Subtle hydrophobic interactions between the seventh residue of the zinc finger loop and the first base of an HGATAR sequence determine promoter-specific recognition by the Aspergillus nidulans GATA factor AreA. EMBO J. 16: 3974–3986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutherford J. C., Lin X., Nielsen K., Heitman J., 2008. Amt2 permease is required to induce ammonium-responsive invasive growth and mating in Cryptococcus neoformans. Eukaryot. Cell 7: 237–246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J., Fritsch E. F., Maniatis T., 1989. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- Scholten H. J., Pierik R. L. M., 1998. Agar as a gelling agent: chemical and physical analysis. Plant Cell Rep. 17: 230–235 [DOI] [PubMed] [Google Scholar]
- Shi M., Li S. S., Zheng C., Jones G. J., Kim K. S., et al. , 2010. Real-time imaging of trapping and urease-dependent transmigration of Cryptococcus neoformans in mouse brain. J. Clin. Invest. 120: 1683–1693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Southern E., 2006. Southern blotting. Nat. Protoc. 1: 518–525 [DOI] [PubMed] [Google Scholar]
- Staib F., Mishra S. K., Able T., Blisse A., 1976. Growth of Cryptococcus neoformans on uric acid agar. Zentralbl Bakteriol. 236: 374–385 [Orig A] [PubMed] [Google Scholar]
- Staib F., Grave B., Altmann L., Mishra S. K., Abel T., et al. , 1978. Epidemiology of Cryptococcus neoformans. Mycopathologia 65: 73–76 [DOI] [PubMed] [Google Scholar]
- Stanbrough M., Rowen D. W., Magasanik B., 1995. Role of the GATA factors Gln3p and Nil1p of Saccharomyces cerevisiae in the expression of nitrogen-regulated genes. Proc. Natl. Acad. Sci. USA 92: 9450–9454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart V., Vollmer S. J., 1986. Molecular cloning of nit-2, a regulatory gene required for nitrogen metabolite repression in Neurospora crassa. Gene 46: 291–295 [DOI] [PubMed] [Google Scholar]
- Thompson J. D., Higgins D. G., Gibson T. J., 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22: 4673–4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walton F. J., Idnurm A., Heitman J., 2005. Novel gene functions required for melanization of the human pathogen Cryptococcus neoformans. Mol. Microbiol. 57: 1381–1396 [DOI] [PubMed] [Google Scholar]
- Wiame J. M., Grenson M., Arst H. N., Jr, 1985. Nitrogen catabolite repression in yeasts and filamentous fungi. Adv. Microb. Physiol. 26: 1–88 [DOI] [PubMed] [Google Scholar]
- Wong K. H., Hynes M. J., Davis M. A., 2008. Recent advances in nitrogen regulation: a comparison between Saccharomyces cerevisiae and filamentous fungi. Eukaryot. Cell 7: 917–925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu S., Falvey D. A., Brandriss M. C., 1995. Roles of URE2 and GLN3 in the proline utilization pathway in Saccharomyces cerevisiae. Mol. Cell Biol. 15: 2321–2330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue C., Tada Y., Dong X., Heitman J., 2007. The human fungal pathogen Cryptococcus can complete its sexual cycle during a pathogenic association with plants. Cell. Host Microbe 1: 263–273 [DOI] [PubMed] [Google Scholar]
- Zaragoza O., Fries B. C., Casadevall A., 2003. Induction of capsule growth in Cryptococcus neoformans by mammalian serum and CO(2). Infect. Immun. 71: 6155–6164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y., Medrano L., Ohashi K., Fletcher J. C., Yu H., et al. , 2004. HANABA TARANU is a GATA transcription factor that regulates shoot apical meristem and flower development in Arabidopsis. Plant Cell 16: 2586–2600 [DOI] [PMC free article] [PubMed] [Google Scholar]