Abstract
In vitro drug sensitivity and molecular analyses of Plasmodium falciparum track drug resistance. DNA-binding fluorescent dyes like SYBR Green I may allow field laboratories, proximal to P. falciparum collection sites, to conduct drug assays. In 2007–2008, we assayed 121 P. falciparum field isolates from western Kenya for 50% inhibitory concentrations (IC50) against 6 antimalarial drugs using a SYBR Green I in vitro assay: 91 immediate ex vivo (IEV) and 30 culture-adapted, along with P. falciparum reference clones D6 (chloroquine [CQ] sensitive) and W2 (CQ resistant). We also assessed P. falciparum mdr1 (Pfmdr1) copy number and single nucleotide polymorphisms (SNPs) at four codons. The IC50s for IEV and culture-adapted P. falciparum isolates were similar, and approximated historical IC50s. For Pfmdr1, mean copy number was 1, with SNPs common at codons 86 and 184. The SYBR Green I assay adapted well to our field-based laboratory, for both IEV and culture-adapted P. falciparum, warranting continued use.
Introduction
Since the 1980s, measuring in vitro drug 50% inhibitory concentrations (IC50) against Plasmodium falciparum field isolates, coupled with molecular analysis, has been useful for tracking development of in vivo drug resistance, from Southeast Asia to sub-Saharan Africa.1–5 However, one commonly used assay, 3H-hypoxanthine uptake, is cumbersome, expensive, and often limits assay conduct to urban, well-resourced laboratories.4,5 Moreover, blood samples containing P. falciparum parasites for testing may be stored at collection sites for days, transported long distances to the laboratory, and then culture adapted for assay, all potential sources of clonal selection.
Recently, non-radioisotope microtests using fluorescent DNA dyes such as SYBR Green I, which reliably depict in vitro P. falciparum parasite replication, have gained popularity by reducing some hurdles associated with in vitro P. falciparum drug sensitivity assays.6–9 The SYBR Green I assays are considered convenient, relatively rapid, reproducible, and less costly than radioisotope assays.8,10 These features suggest SYBR Green I drug sensitivity assays could be deployed to field laboratories, proximal to P. falciparum collection sites. The SYBR Green I is increasingly accepted as an alternate to 3H-hypoxanthine uptake assays.11
In Kenya, malaria continues to cause significant morbidity and mortality, and is often a location where drug resistance emerges in Africa. For example, the development of P. falciparum resistance to chloroquine diphosphate (CQ), and later sulfadoxine-pyrimethamine, are well described.12–15 This warrants continued in vitro drug IC50 monitoring of P. falciparum field isolates, which is becoming more practical and safer, as more reliable assays like SYBR Green I become established, combined with simpler field isolate processing such as “immediate ex vivo” (IEV).
Subjects and Methods
Protocol, sites, and subjects.
This study was approved by the Kenya Medical Research Institute (KEMRI) and Walter Reed Army Institute of Research (WRAIR) institutional review boards (protocol numbers: KEMRI 1330, WRAIR 1384). Participating clinical centers, all Kenya Ministry of Health facilities located in West Kenya (Figure 1), including Kisumu, Kisii, and Kericho District Hospitals, and Chulaimbo Sub-District Hospital. Kisumu and Chulaimbo are lowland, malaria holoendemic areas, whereas Kisii and Kericho are highland, malaria hypoendemic areas.16 At each participating facility, training of medical staff, capacity building, and facility upgrades were provided by the Global Emerging Infections Surveillance (GEIS) Program, U.S. Department of Defense.
Figure 1.
Plasmodium falciparum field isolates were collected from four western Kenya sites, denoted by (*).
Subjects attending outpatient clinics in 2007 and 2008, at least 6 months old and suspected of having non-complicated P. falciparum malaria were invited to participate. Written informed consent was obtained from adult subjects (≥ 18 years of age) or legal guardians for subjects < 18 years of age. Persons treated for malaria within the last 2 weeks were excluded.
Sample collection and preparation.
Consented subjects with a positive P. falciparum rapid diagnostic test (RDT; Parascreen [Pan/Pf], Zephyr Biomedicals, Verna Goa, India) provided 2–3 mL of blood for transport to the laboratory. Parascreen detects P. falciparum-specific histidine protein-2 (Pf HRP-2) and pan-specific parasite lactate dehydrogenase (pLDH) in whole blood, the latter is useful for follow-up of antimalarial therapy. Three blood spots of about 100 μL each were placed on FTA filter paper (Whatman Inc., Bound Brook, NJ) for P. falciparum DNA extraction and molecular analysis, and two blood films on glass slides were made for Giemsa staining at the laboratory for microscopic examination, to confirm RDT results and determine parasitemia. For discrepancies between RDT and microscopy, microscopy determined the final result.
Plasmodium falciparum isolates from Kisumu District Hospital and Chulaimbo Health Center, 15-minute drive from the laboratory, were collected in acid citrate dextrose (ACD) vacutainer tubes (Becton-Dickinson, Inc., Franklin Lakes, NJ) and transported within 4 hours to begin IEV drug IC50 testing (described below). Plasmodium falciparum isolates from Kericho and Kisii District Hospitals, 2-hour drive from the laboratory, were placed in storage-transport media, and refrigerated at 4°C until transported to the laboratory, generally within 72 hours, for culture adaptation and drug testing.17
Subjects were treated with oral artemether-lumefantrine (AL; Coartem) administered over three consecutive days, a standard of care for P. falciparum malaria in Kenya. The first dose was observed by the study team and remaining doses were self-administered at home. Subjects living near study centers were asked to return on Day 7, for repeat malaria testing.
In vitro drug sensitivity testing.
A SYBR Green I-based in vitro IC50 drug sensitivity assay, described earlier,6,7,9 was used to test each P. falciparum field isolate against a panel of six conventional antimalarials supplied as chloroquine disphosphate (CQ), mefloquine hydrochloride (MQ), quinine sulfate hydrate (QN), artemisinin (AR), amodiaquine hydrochloride (AQ), and doxycycline hyclate (DX). Because all test drugs, except AR (pure base), were provided as a salt and prepared by weight:volume convention, we used salt formulation weights (except AR) to calculate IC50 values. Test drugs, except DX, were obtained from Walter Reed Army Institute of Research, (Silver Spring, MD). DX was obtained commercially (Sigma-Aldrich, Co., St. Louis, MO; catalog number D9891). Anti-folate drugs were omitted because P. falciparum resistance in Kenya is established.15,18,19
Reference P. falciparum clones assayed periodically for internal control against all six drugs included D6, considered CQ-sensitive and MQ-resistant (CQ-S; MQ-R) and W2, considered CQ-resistant and MQ-sensitive (CQ-R; MQ-S), as well as AR-sensitive.5 Quinine sulfate hydrate generally parallels CQ for D6 and W2 IC50 trends.7 The D6 and W2 clones were obtained from frozen stocks and culture adapted for SYBR Green I IC50 assays.
To prepare test drugs, stock drug solutions at 1 mg/mL were prepared in 5 mL 70% ethanol for MQ and QN or 100% dimethyl sulfoxide for AR, AQ, and DX. For CQ, 1.5 mL deionized water was combined with 3.5 mL of absolute ethanol. Further dilutions were in complete RPMI 1640 media to the desired starting concentration, followed by serial 2-fold dilutions to generate 10 concentrations for IC50 testing.17 The concentration ranges (ng/mL), from highest to lowest, were CQ (1,000 to 1.953), MQ (250 to 0.488), QN (2,000 to 3.906), AR (100 to 0.195), AQ (100 to 0.195), and DX (50,000 to 97.656). The drug solutions were prepared and used immediately, or stored at −80°C for not longer than 1 month before use. Basic and complete RPMI 1640 culture media, the latter with glucose and hypoxanthine enrichment, were prepared as described.7
Plasmodium falciparum field isolates refrigerated in transport media, and the two P. falciparum laboratory reference clones D6 and W2, were culture adapted before IC50 SYBR Green I assay. The parasites were cultured at 6% hematocrit for 7–30 days, to reach 3–8% parasitemia.5 For IC50 drug assays, culture-adapted parasites were adjusted to 2% hematocrit and 1% parasitemia, and antimalarial drug aliquots in complete RPMI 1640 and added to the wells as described below.
Plasmodium falciparum isolates processed IEV were placed into assay within 6 hours of phlebotomy, without culture adaptation. Blood samples with > 1% parasitemia were adjusted to 1% parasitemia at 2% hematocrit, and those with ≤ 1% parasitemia were used unadjusted at 2% hematocrit.
For culture-adapted and IEV assays, transfer of parasite sample and antimalarial drug aliquots in complete RPMI 1640 drug aliquots onto 96-well microculture plates and addition of lysis buffer after 72 hours incubation was done as previously described.7 The plates were then placed at room temperature in the dark, for 5–15 minutes. Parasite replication inhibition was quantified and the IC50 for each drug calculated by an equation generating a sigmoidal concentration-response curve (variable slope), with log transformed drug concentrations on the X axis and relative fluorescent units (RFUs) on the Y axis (Graphpad Prism for Windows, version 4.0; Graphpad Software, Inc., San Diego, CA).6,7
Assessing possible sources of SYBR Green I background signal.
In experiment set 1, to measure the intrinsic effect of each drug alone on SYBR Green I RFU, in comparison with complete assay wells, we plated each drug over the standard 10 dilutions in complete RPMI 1640 media alone (“drug only” wells), or in complete RPMI 1640 media containing 1% P. falciparum parasitemia and 32,000 peripheral blood mononuclear cells (PBMC)/well (“complete assay” wells), respectively. The PBMC were isolated by density gradient from the blood of a healthy donor. Drug only and 1 set of complete assay wells were processed without incubation, and a second set of complete assay wells were incubated for 72 hours at 37°C in a gas mixture, described earlier.7 All plates were then processed for SYBR Green I RFU measurements by Genios Tecan. For complete assay wells, 32,000 PBMC/well approximated the number of white blood cells (WBC) in a typical IEV well, based on an estimate of 8,000 WBC/uL of whole blood in a subject. We also assessed a range of P. falciparum parasitemias, from 1% to 0.112% (2-fold dilutions), to determine the capability of SYBR Green I to discern IC50s in IEV samples with low parasitemias.
In experiment set 2, to assess the influence of PBMC concentration on SYBR Green I assay RFU, we plated a range of PBMC (61 to 250,000 cells/well) in complete RPMI 1640 media with 1% P. falciparum parasitemia, without incubation, and then measured RFUs within 15 minutes by Genios Tecan. All experiments were conducted in triplicate.
Pfmdr1 copy number.
Plasmodium falciparum DNA for all molecular assays was extracted from FTA filter paper blots or whole blood using QIAamp DNA Blood Mini Kit (Qiagen, Inc., Alameda, CA), according to manufacturer instructions.
The 2−ΔΔCt method of relative quantification was used to estimate P. falciparum multiple drug resistance gene 1 (Pfmdr1) copy numbers.20 For this method, there should be at least 1 calibrator with a known number of copies of the gene under study, and a housekeeping gene with constant copy number to normalize the quantitative data.21 For the calibrator, we used genomic DNA from P. falciparum reference clone 3D7, known to have 1 copy of Pfmdr1 gene.21 Plasmodium falciparum tubulin served as a housekeeping gene, and Dd2 P. falciparum clone DNA served as a Pfmdr1 multiple copy control.
The relative quantification of the Pfmdr1 gene was determined as previously described.20 Briefly, ΔΔCt = (Ct, Pfmdr1-Ct, P. falciparum-tubulin)x − (Ct, Pfmdr1-Ct, P. falciparum-tubulin)y, where x is the field isolate and y is 3D7. The results were then calculated as n-fold changes in P. falciparum isolate Pfmdr1 gene copies, normalized to P. falciparum tubulin, relative to the copy number of Pfmdr1 in P. falciparum 3D7 using the equation 2−ΔΔCt, as described.20,21 Each sample was run in triplicate and the Ct value of each well recorded at the end of the reaction. The average of the Ct values for each sample was used for the determination of Pfmdr1 copy number.
Pfmdr1 codon single nucleotide polymorphism (SNP) analysis.
SNP analysis was conducted for Pfmdr1 codons 86 (N86Y), 184 (Y184F), 1034 (S1034C), and 1042 (N1042D) using real-time polymerase chain reaction (PCR).22,23 Probes consisted of VIC for wild-type and 6-FAM (6-carboxyfluoroscein) for the mutation.23 Plasmodium falciparum reference clones 3D7 (MQ-S) and Dd2 (MQ-R), as well as D6 and W2, were also assayed.
Results
Subject enrollment and follow up.
Three hundred fifty-two P. falciparum RDT (+) subjects provided blood samples. For Day 7 post-treatment appointments, 216 subjects tested by Parascreen remained PfHRP-2 (+), but were pLDH (−), the latter indicating absence of P. falciparum. All blood smears were (−) for P. falciparum asexual stages; five had gametocytes.
SYBR Green I IC50 drug assays.
We assayed 121 P. falciparum field isolates, 91 IEV, and 30 culture-adapted, against a panel of six drugs. A “successful” assay, defined as a concentration-response across the 10 drug concentrations for 1 or more drugs per each P. falciparum field isolate, occurred in 68% of all isolates. For IEV, the success rate for all assays was 70%, 78% with parasitemias > 0.5% and 61% for parasitemias ≤ 0.5%. For culture adaptation, 63% of the assays were successful.
Figure 2A–F shows individual and median IC50s for IEV and culture-adapted P. falciparum field isolates, against each of the six drugs, along with D6 (CQ-S, MQ-R) and W2 (CQ-R, MQ-S) P. falciparum reference clones. For CQ, QN, AQ, and DX median IC50 values for IEV versus culture-adapted P. falciparum field isolates were similar (P ≥ 0.05, Mann-Whitney rank sum test). For MQ and AR, median IC50 values of culture-adapted P. falciparum isolates were higher than IEV isolates (P < 0.05, Mann-Whitney rank sum test), at 1.8- and 1.5-fold, respectively. The IC50 ranges for all drugs, except AR, were generally larger for IEV than culture-adapted P. falciparum isolates in the upper half of the IC50 range.
Figure 2.
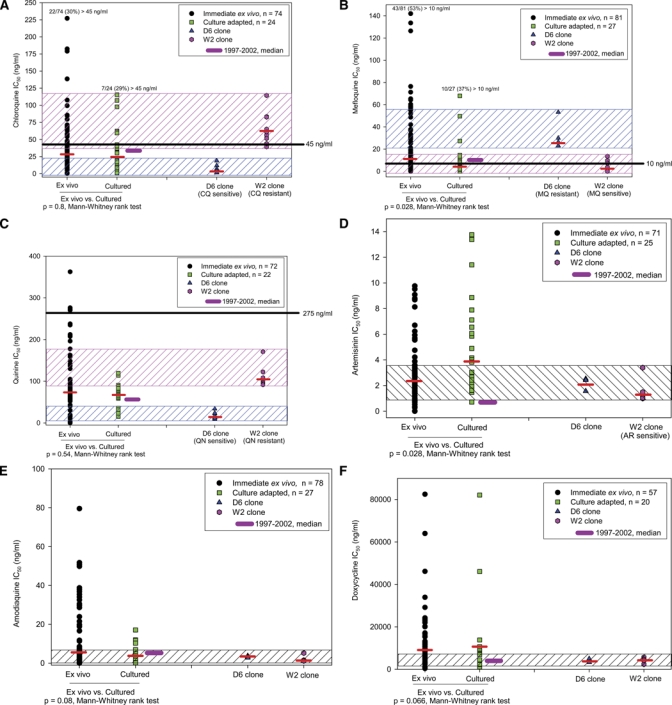
(A–F) SYBR Green I drug IC50s for Plasmodium falciparum field isolates, shown as scatter plot and median (thin black bar). IC50 ranges of P. falciparum reference clones D6 and W2 for chloroquine diphosphate (CQ), mefloquine hydrochloride (MQ), and quinine sulfate hydrate (QN) shown in two hatched boxes; for artemisinin (AR), amodiaquine hydrochloride (AQ), and doxycycline hyclate (DX), in single hatched box. Thick black bars: median IC50 of P. falciparum field isolates obtained 1997–2002 (culture-adapted, radioisotope uptake assay).17 Horizontal solid lines on CQ, MQ, and QN graphs: IC50 threshold values considered discriminative for “resistant” P. falciparum isolates (culture-adapted, radioisotope uptake assay).
For all drugs, except AR, SYBR Green I median IC50s for IEV and culture-adapted P. falciparum isolates were similar to historical median IC50s from 3H-hypoxanthine uptake assays on culture-adapted P. falciparum isolates collected in Kenya from 1997 to 2002 (Figure 2A–F).17 The AR median IC50 values for SYBR Green I IEV and culture-adapted P. falciparum isolates were about 2- to 4-fold higher than the earlier 3H-hypoxanthine assays, respectively.
SYBR Green I background RFU experiments.
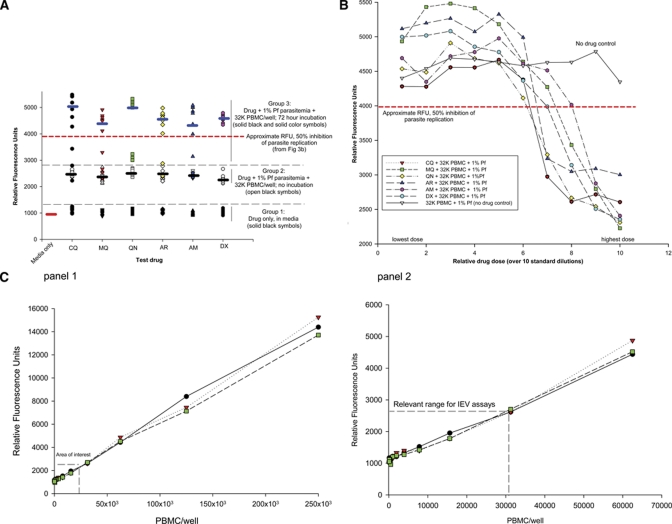
Experiments are summarized in Figure 3A–C. In Figure 3A (Group 1), drug + media versus complete assay wells (drug + 1% parasitemia + 32,000 PBMC/well), both without incubation, showed a difference of about 1,000 RFU for each drug, with little overlap. Media alone values were similar to drug + media. These observations implied that none of the drugs, at any concentration, would obscure IC50 readouts. Complete assay wells with no incubation (Group 2) versus with incubation for 72 hours at 37°C (Group 3; akin to IC50 P. falciparum drug assays), the latter median RFU values were ∼2-fold higher, indicative of parasite replication.
Figure 3.
Experiments assessing SYBR Green I relative fluorescent unit (RFU) background signal of each drug, and peripheral blood mononuclear cells (PBMCs). (A) SYBR Green I individual and median RFUs among three groups, shown as scatter plot and median (short black bars): Group 1: each drug in RPMI 1640 media only. Group 2: Each drug in RPMI 1640 media + 32K PBMC + 1% parasitemia/well; no incubation. Group 3: Same as Group 2, after 72 hours incubation (37°C). Symbols represent RFU value for each drug, across 10 concentrations used in the SYBR Green I assay. For Groups 2 and 3, differences in medians and minimal RFU value overlap implies determinable IC50s after 72 hours incubation. Horizontal dotted line at 4000 RFU approximates 50% inhibition of parasite replication, obtained from Figure 3B. (B) SYBR Green I RFUs in complete assay wells, over 10 drug concentrations, with 32,000 PBMC and 1% parasitemia/well, incubated 72 hours (37°C). Horizontal dotted line approximates 50% inhibition of parasite replication. (C) Panels 1 and 2 show SYBR Green I RFUs for PBMC concentrations ranging from 61 to 250 K cells/well, plated in complete RPMI 1640 media. Each symbol is 1 PBMC spiked replicate, diluted over the range shown. Panel 2: “Area of interest” denotes range for number of white blood cells in a typical immediate ex vivo blood sample, relevant to IEV SYBR Green I IC50 assays. Linear relationships occur over entire range.
In Figure 3B, drug concentrations used in the IC50 assays generated sigmoidal curves, indicating 50% parasite replication inhibition at about 4,000 RFU. Data from Figure 3A and other experiments (not shown) indicated background RFU signal (drug + medium + 32 K PBMC) subtracted from the RFU value of each IC50 replicate was unnecessary, consistent with earlier work.6 Finally, SYBR Green I assay discerned IC50s for complete assay wells with P. falciparum parasitemias ranging from 1% to 0.112%, against all six test drugs (data not shown). These observations, and Figure 3B, are relevant to IEV processing.
In Figure 3C, to determine the RFU signal of PBMC plated across a wide concentration range in complete assays wells (+1% P. falciparum parasitemia), RFU values for wells with ≤ 30,000 PBMC were about 2,500 RFU, a value that would not be expected to obscure the identification of drug IC50 values, at about 4,000 RFU.
Molecular assays of P. falciparum samples.
Plasmodium falciparum field isolates (N = 308) had a mean Pfmdr1 gene copy number of 1, quantified relative to the P. falciparum reference clones 3D7, with a mean of 1 copy, and multiple drug-resistant Dd2 clone, with a mean of 3.5 copies.
The mutation (86Y) rates in Pfmdr1 codons were highest at codon 86 (66%), with fewer at codons 184 (38%) and 1034 (12%), and none at codon 1042, regardless of the collection site. The SNP rates at the four codons were similar among the four locations where P. falciparum isolates were collected, including lowland versus highland sites. As in earlier work, mutant and mixed (mutant + wild) genotypes were combined to depict total mutant genotype populations.17
Associations between the Pfmdr1 codon 86 SNPs and CQ, MQ, and AR IC50 values are shown in Figure 4A–C. Plasmodium falciparum field isolates with Pfmdr1 codon 86 mutation (86Y) (66% of isolates) had a higher median CQ IC50 (lower CQ sensitivity), in comparison with Pfmdr1 codon 86 wild (N86) samples. For MQ and AR, P. falciparum isolates with Pfmdr1 codon 86Y had a lower median IC50, in comparison with Pfmdr1 codon 86 wild, both statistically significant (P < 0.05).
Figure 4.
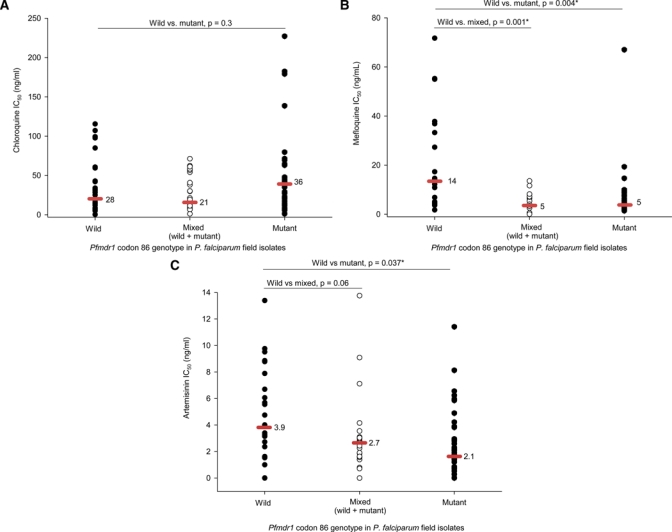
Association between Pfmdr1 codon 86 genotype and chloroquine diphosphate (CQ), mefloquine hydrochloride (MQ), and artemisinin (AR) IC50 values, respectively. P values determined by Mann-Whitney rank test (* denotes P ≤ 0.05). (A) Pfmdr1 codon 86 mutation (86Y), modestly associated with higher CQ IC50. (B) Pfmdr1 codon 86 mutation (86Y), associated with lower MQ IC50. (C) Pfmdr1 codon 86 mutation (86Y), associated with lower AR IC50.
Discussion
This is the first report from Kenya describing a SYBR Green I assay for measuring in vitro drug IC50s against P. falciparum field isolates. Notably, SYBR Green I assay was conducted at our field-based laboratory, proximal to P. falciparum collection sites, using six drugs. Several earlier reports describe SYBR Green I assays in developed countries, assessing P. falciparum clones against a few drugs.7,9 Second, we assayed P. falciparum field isolates IEV and by culture adaptation, obtaining IC50 values in 70% and 63% of assays, respectively. These success rates, higher than historical radioisotope uptake assays in our laboratory, may reflect simpler processing. The overall expedience of SYBR Green I supports continued use.
Median IC50s in SYBR Green I assays for each of six drugs, among IEV and culture-adapted samples, generally paralleled our earlier data using 3H-hypoxanthine uptake in culture-adapted P. falciparum isolates.17 For example, about 30% of CQ IC50s and 50% of MQ IC50s in SYBR Green I assays exceeded previously established in vitro discriminative “resistance” values of 45 ng/mL and 10 ng/mL, respectively.17 The IC50 ranges for two P. falciparum reference clones, D6 and W2, paralleled their expected CQ and MQ sensitivity profiles. These observations underscored adaptability of the SYBR Green I IC50 assay to a field-based laboratory.
We processed P. falciparum field isolates IEV or culture adapted, depending on proximity to the laboratory. For P. falciparum isolates stored for several days at 4°C, we waited until parasitemias reached 3–8% before drug IC50 testing. Here, IEV was feasible because of laboratory proximity to two collection sites, with P. falciparum isolates placed into assay soon after collection. Encouragingly, against each of four drugs (CQ, QN, AQ, DX), median IC50s of IEV and culture-adapted isolates were similar, differing only for MQ and AR, at 1.5- and 1.8-fold, respectively, albeit relatively low magnitudes for IC50 testing. The IEV was simpler than culture adaptation, as others observed.6
To partly address background concerns for SYBR Green I assay, we confirmed each drug alone did not affect RFU values. Second, complete RPMI 1640 media with 1% P. falciparum and 32,000 PBMC/well, approximating WBC in an IEV assay well, appeared unlikely to elevate RFUs to a level obscuring IC50 interpretations. Finally, relevant to IEV, we found that SYBR Green I assay discerns drug IC50s with P. falciparum parasitemias as low as 0.112%, an issue in debate.7,24
Among IEV assays, the upper half of IC50 ranges was notably higher for five drugs, in comparison with culture adaptation. This might reflect greater biological variability in IEV P. falciparum isolates, including better preserved sub-populations of drug-resistant and sensitive parasites.11,25,26 It may be useful to determine multiplicity of infection of each P. falciparum isolate, processed IEV, and culture adapted, along with comparative IEV and culture-adapted SYBR Green I assays.
AR was the lone drug without larger IEV IC50 ranges, in comparison with culture-adapted isolates, perhaps because there are few, if any, AR-resistant clones in Kenya. Alternatively, this may reflect the activity of AR against most P. falciparum blood stages.27 Median IC50 values for AR, among IEV and culture-adapted assays, were 2- to 3-fold higher in comparison with earlier radioisotope assays, and a recent report of SYBR Green I assessing freeze-thawed, culture-adapted P. falciparum samples.7,17 Higher AR IC50s are unlikely to represent resistance, but perhaps methodological variability.
The SYBR Green I IC50 values for six drugs, establishing a modest baseline for western Kenya, support continued work to better define IC50s. For radioisotope assays using culture-adapted P. falciparum isolates, IC50 values discriminative for in vivo “resistance” exist (CQ, MQ, QN, and five others).17 If SYBR Green I is widely implemented for in vitro P. falciparum drug testing, reference IC50 values discriminative for resistance, ideally linked with in vivo responses, could enhance usefulness.
In Kenya, with artemether + lumefantrine (Coartem) implemented as the artemisinin combination therapy (ACT) standard in 2003, and with no clinical evidence of resistance, there is an opportunity with SYBR Green I assay to track A+ L IC50 values. Indeed, in East Africa, Kenya is often a sentinel site for drug resistance.15,28 As such, we have developed SYBR Green I assays for artemether, lumefantrine, artemisinin combinations, as well as atovaquone, halofantrine, primaquine, and tafenoquine.11
Pfmdr1 is a constitutive gene in P. falciparum that confers multiple drug resistance when more than 1 copy is present.2 Parasites with multiple Pfmdr1 copies are most common in Southeast Asia, where multiple drug-resistant P. falciparum malaria, including MQ, exists.2,29,30 As expected, our P. falciparum field isolates from western Kenya contained an average of 1 copy number of Pfmdr1,31 consistent with a wild-type genotype not necessarily associated with multidrug resistance.32 MQ has not been widely used in Kenya, precluding comment on in vitro MQ IC50s and in vivo responsiveness.
SNPs in certain Pfmdr1 codons may confer drug resistance.32–34 Among P. falciparum isolates we collected earlier (1997–2002), Pfmdr1 codon 86 mutation rates (86Y) rates in two western sites (Kisumu, Kericho) were 35% and 68%, respectively (mean, 51%), whereas two eastern sites (Entosopia, Magadi) had rates > 85%.17 This reflected CQ resistance in Kenya.17,35 Here, in P. falciparum isolates from western Kenya, mutation rates in codon 86 were 66%, far less at 184 and 1,034. Mutation rates in codons 86, 184, and 1034 were similar for lowland and highland sites, suggesting Pfmdr1 SNPs are not necessarily associated with severe malaria presentation, often anemia and cerebral malaria, respectively.36
We interpret the similarity in Pfmdr1 codon 86 mutation rates for 1997–2002 (51%) and this study (66%), along with similar CQ IC50s, as sustained CQ resistance in Kenya.14,17,37 Indeed, most P. falciparum isolates contained codon 86 mutations (86Y), modestly associated with higher CQ IC50s. Mutation in pfcrt, well established in Kenya, also confers CQ resistance. In contrast, Pfmdr1 codon 86 mutations (86Y) were associated with lower median IC50s for MQ, and AR. This parallels earlier reports describing Pfmdr1 codon 86Y and drug IC50 relationships, typically direct with CQ, and inverse for MQ.38 For AR, a lower median IC50 in P. falciparum isolates expressing Pfmdr1 codon mutation (86Y) supports the notion this mutation is unlikely associated with emerging AR resistance.39,40
ACKNOWLEDGMENTS
We thank Duke Omariba, Stella Apollo, and all GEIS clinical staff serving at Kisumu, Kisii, and Kericho District Hospitals for their assistance. We also thank Harald Noedl for helpful advice.
Disclaimer: The opinions and assertions contained in this work are the private views of the authors and are not to be construed as official or as reflecting the views of the U.S. Department of the Army or the Department of Defense.
Footnotes
Financial support: This work was supported by the U.S. Department of Defense Global Emerging Infections System, Silver Spring, Maryland.
Disclosure: Presented in part, 2008 Annual Meeting of the American Society of Tropical Medicine and Hygiene, New Orleans, LA.
Authors' addresses: Hoseah M. Akala, Fredrick L. Eyase, Agnes C. Cheruiyot, Angela A. Omondi, Bernhards R. Ogutu, and Jacob D. Johnson, U.S. Army Medical Research Unit-Kenya (USAMRU-K), KEMRI-Walter Reed Project, Kisumu, Kenya, E-mails: hakala@wrp-ksm.org, feyase@wrp-ksm.org, acheruiyot@wrp-ksm.org, aomondi@wrp-ksm, bogutu@wrp-ksm.org, and jjohnson@wrp-ksm.org. Norman C. Waters, U.S. Military Academy, West Point, NY, E-mail: norman.waters@us.army.mil. Mark E. Polhemus, USA ASAALT, Pentagon, Washington, DC, E-mail: mark.polhemus@conus.army.mil. David C. Schnabel, U.S. Army Medical Research Unit-Kenya (USAMRU-K), KEMRI-Walter Reed Project, Kitsumu and Nairobi, Kenya, E-mail: dschnabel@wrp-ksm.org. Douglas S. Walsh, AFRIMS, Bangkok, Thailand, E-mail: douglas.walsh@afrims.org.
References
- 1.Anderson T. Mapping the spread of malaria drug resistance. PLoS Med. 2009;6:1–2. doi: 10.1371/journal.pmed.1000054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wongsrichanalai C, Pickard AL, Wernsdorfer WH, Meshnick SR. Epidemiology of drug-resistant malaria. Lancet Infect Dis. 2002;2:209–218. doi: 10.1016/s1473-3099(02)00239-6. [DOI] [PubMed] [Google Scholar]
- 3.Shretta RO, Rapuoda B, Snow RW. Using evidence to change antimalarial drug policy in Kenya. Trop Med Int Health. 2000;5:753–754. doi: 10.1046/j.1365-3156.2000.00643.x. [DOI] [PubMed] [Google Scholar]
- 4.Webster HK, Boudreau EF, Pavanand K, Yongvanitchit K, Pang LW. Antimalarial drug susceptibility testing of Plasmodium falciparum in Thailand using a microdilution radioisotope method. Am J Trop Med Hyg. 1985;34:228–235. doi: 10.4269/ajtmh.1985.34.228. [DOI] [PubMed] [Google Scholar]
- 5.Desjardins RE, Canfield CJ, Haynes JD, Chulay JD. Quantitative assessment of antimalarial activity in vitro by a semiautomated microdilution technique. Antimicrob Agents Chemother. 1979;16:710–718. doi: 10.1128/aac.16.6.710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bacon DJ, Latour C, Lucas C, Colina O, Ringwald P, Picot S. Comparison of a SYBR green I-based assay with a histidine-rich protein II enzyme-linked immunosorbent assay for in vitro antimalarial drug efficacy testing and application to clinical isolates. Antimicrob Agents Chemother. 2007;51:1172–1178. doi: 10.1128/AAC.01313-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Johnson JD, Dennull RA, Gerena L, Lopez-Sanchez M, Roncal NE, Waters NC. Assessment and continued validation of the malaria SYBR green I-based fluorescence assay for use in malaria drug screening. Antimicrob Agents Chemother. 2007;51:1926–1933. doi: 10.1128/AAC.01607-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rason MA, Randriantsoa T, Andrianantenaina H, Ratsimbasoa A, Menard D. Performance and reliability of the SYBR Green I based assay for the routine monitoring of susceptibility of Plasmodium falciparum clinical isolates. Trans R Soc Trop Med Hyg. 2008;102:346–351. doi: 10.1016/j.trstmh.2008.01.021. [DOI] [PubMed] [Google Scholar]
- 9.Smilkstein M, Sriwilaijaroen N, Kelly JX, Wilairat P, Riscoe M. Simple and inexpensive fluorescence-based technique for high-throughput antimalarial drug screening. Antimicrob Agents Chemother. 2004;48:1803–1806. doi: 10.1128/AAC.48.5.1803-1806.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bennett TN, Paguio M, Gligorijevic B, Seudieu C, Kosar AD, Davidson E, Roepe PD. Novel, rapid, and inexpensive cell-based quantification of antimalarial drug efficacy. Antimicrob Agents Chemother. 2004;48:1807–1810. doi: 10.1128/AAC.48.5.1807-1810.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Co EM, Dennull RA, Reinbold DD, Waters NC, Johnson JD. Assessment of malaria in vitro drug combination screening and mixed-strain infections using the malaria Sybr green I-based fluorescence assay. Antimicrob Agents Chemother. 2009;53:2557–2563. doi: 10.1128/AAC.01370-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Plowe CV, Cortese JF, Djimde A, Nwanyanwu OC, Watkins WM, Winstanley PA, Estrada-Franco JG, Mollinedo RE, Avila JC, Cespedes JL, Carter D, Doumbo OK. Mutations in Plasmodium falciparum dihydrofolate reductase and dihydropteroate synthase and epidemiologic patterns of pyrimethamine-sulfadoxine use and resistance. J Infect Dis. 1997;176:1590–1596. doi: 10.1086/514159. [DOI] [PubMed] [Google Scholar]
- 13.Falaschi F, Ansaloni L. Chloroquine versus pyrimethamine/sulphadoxine in the treatment of uncomplicated P. falciparum malaria in northern Kenya. East Afr Med J. 1997;74:275–277. [PubMed] [Google Scholar]
- 14.Mwai L, Ochong E, Abdirahman A, Kiara SM, Ward S, Kokwaro G, Sasi P, Marsh K, Borrmann S, Mackinnon M, Nzila A. Chloroquine resistance before and after its withdrawal in Kenya. Malar J. 2009;8:106. doi: 10.1186/1475-2875-8-106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Khan B, Omar S, Kanyara JN, Warren-Perry M, Nyalwidhe J, Peterson DS, Wellems T, Kaniaru S, Gitonga J, Mulaa FJ, Koech DK. Antifolate drug resistance and point mutations in Plasmodium falciparum in Kenya. Trans R Soc Trop Med Hyg. 1997;91:456–460. doi: 10.1016/s0035-9203(97)90284-4. [DOI] [PubMed] [Google Scholar]
- 16.Malakooti MA, Biomndo K, Shanks GD. Reemergence of epidemic malaria in the highlands of western Kenya. Emerg Infect Dis. 1998;4:671–676. doi: 10.3201/eid0404.980422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mbaisi A, Liyala P, Eyase F, Achilla R, Akala H, Wangui J, Mwangi J, Osuna F, Alam U, Smoak BL, Davis JM, Kyle DE, Coldren RL, Mason C, Waters NC. Drug susceptibility and genetic evaluation of Plasmodium falciparum isolates obtained in four distinct geographical regions of Kenya. Antimicrob Agents Chemother. 2004;48:3598–3601. doi: 10.1128/AAC.48.9.3598-3601.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sibley CH, Hyde JE, Sims PF, Plowe CV, Kublin JG, Mberu EK, Cowman AF, Winstanley PA, Watkins WM, Nzila AM. Pyrimethamine-sulfadoxine resistance in Plasmodium falciparum: what next? Trends Parasitol. 2001;17:582–588. doi: 10.1016/s1471-4922(01)02085-2. [DOI] [PubMed] [Google Scholar]
- 19.Certain LK, Briceno M, Kiara SM, Nzila AM, Watkins WM, Sibley CH. Characteristics of Plasmodium falciparum dhfr haplotypes that confer pyrimethamine resistance, Kilifi, Kenya, 1987–2006. J Infect Dis. 2008;197:1743–1751. doi: 10.1086/588198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-delta delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 21.Ferreira ID, Rosario VE, Cravo PV. Real-time quantitative PCR with SYBR Green I detection for estimating copy numbers of nine drug resistance candidate genes in Plasmodium falciparum. Malar J. 2006;5:1. doi: 10.1186/1475-2875-5-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nelson AL, Purfield A, McDaniel P, Uthaimongkol N, Buathong N, Sriwichai S, Miller RS, Wongsrichanalai C, Meshnick SR. pfmdr1 genotyping and in vivo mefloquine resistance on the Thai-Myanmar border. Am J Trop Med Hyg. 2005;72:586–592. [PubMed] [Google Scholar]
- 23.Purfield A, Nelson A, Laoboonchai A, Congpuong K, McDaniel P, Miller RS, Welch K, Wongsrichanalai C, Meshnick SR. A new method for detection of pfmdr1 mutations in Plasmodium falciparum DNA using real-time PCR. Malar J. 2004;3:9. doi: 10.1186/1475-2875-3-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vossen MG, Pferschy S, Chiba P, Noedl H. The SYBR Green I malaria drug sensitivity assay: performance in low parasitemia samples. Am J Trop Med Hyg. 2009;82:398–401. doi: 10.4269/ajtmh.2010.09-0417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jafari S, Le Bras J, Bouchaud O, Durand R. Plasmodium falciparum clonal population dynamics during malaria treatment. J Infect Dis. 2004;189:195–203. doi: 10.1086/380910. [DOI] [PubMed] [Google Scholar]
- 26.Mookherjee S, Howard V, Nzila-Mouanda A, Watkins W, Sibley CH. Identification and analysis of dihydrofolate reductase alleles from Plasmodium falciparum present at low frequency in polyclonal patient samples. Am J Trop Med Hyg. 1999;61:131–140. doi: 10.4269/ajtmh.1999.61.131. [DOI] [PubMed] [Google Scholar]
- 27.Wein S, Maynadier M, Tran Van Ba C, Cerdan R, Peyrottes S, Fraisse L, Vial H. Reliability of antimalarial sensitivity tests depends on drug mechanisms of action. J Clin Microbiol. 2010;48:1651–1660. doi: 10.1128/JCM.02250-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Omar SA, Adagu IS, Gump DW, Ndaru NP, Warhurst DC. Plasmodium falciparum in Kenya: high prevalence of drug-resistance-associated polymorphisms in hospital admissions with severe malaria in an epidemic area. Ann Trop Med Parasitol. 2001;95:661–669. doi: 10.1080/00034980120103234. [DOI] [PubMed] [Google Scholar]
- 29.Wongsrichanalai C, Sirichaisinthop J, Karwacki JJ, Congpuong K, Miller RS, Pang L, Thimasarn K. Drug resistant malaria on the Thai-Myanmar and Thai-Cambodian borders. Southeast Asian J Trop Med Public Health. 2001;32:41–49. [PubMed] [Google Scholar]
- 30.Price RN, Uhlemann AC, Brockman A, McGready R, Ashley E, Phaipun L, Patel R, Laing K, Looareesuwan S, White NJ, Nosten F, Krishna S. Mefloquine resistance in Plasmodium falciparum and increased pfmdr1 gene copy number. Lancet. 2004;364:438–447. doi: 10.1016/S0140-6736(04)16767-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Holmgren G, Bjorkman A, Gil JP. Amodiaquine resistance is not related to rare findings of pfmdr1 gene amplifications in Kenya. Trop Med Int Health. 2006;11:1808–1812. doi: 10.1111/j.1365-3156.2006.01742.x. [DOI] [PubMed] [Google Scholar]
- 32.Nkhoma S, Nair S, Mukaka M, Molyneux ME, Ward SA, Anderson TJ. Parasites bearing a single copy of the multi-drug resistance gene (pfmdr-1) with wild-type SNPs predominate amongst Plasmodium falciparum isolates from Malawi. Acta Trop. 2009;111:78–81. doi: 10.1016/j.actatropica.2009.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.de Monbrison F, Raynaud D, Latour-Fondanaiche C, Staal A, Favre S, Kaiser K, Peyron F, Picot S. Real-time PCR for chloroquine sensitivity assay and for pfmdr1-pfcrt single nucleotide polymorphisms in Plasmodium falciparum. J Microbiol Methods. 2003;54:391–401. doi: 10.1016/s0167-7012(03)00086-1. [DOI] [PubMed] [Google Scholar]
- 34.Uhlemann AC, Krishna S. Antimalarial multi-drug resistance in Asia: mechanisms and assessment. Curr Top Microbiol Immunol. 2005;295:39–53. doi: 10.1007/3-540-29088-5_2. [DOI] [PubMed] [Google Scholar]
- 35.Duraisingh MT, Jones P, Sambou I, von Seidlein L, Pinder M, Warhurst DC. The tyrosine-86 allele of the pfmdr1 gene of Plasmodium falciparum is associated with increased sensitivity to the anti-malarials mefloquine and artemisinin. Mol Biochem Parasitol. 2000;108:13–23. doi: 10.1016/s0166-6851(00)00201-2. [DOI] [PubMed] [Google Scholar]
- 36.Zhong D, Afrane Y, Githeko A, Cui L, Menge DM, Yan G. Molecular epidemiology of drug-resistant malaria in western Kenya highlands. BMC Infect Dis. 2008;8:105. doi: 10.1186/1471-2334-8-105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Siringi S. Over-the-counter sale of antimalaria drugs stalls Kenyan disease strategy. Lancet. 2001;357:1862. doi: 10.1016/S0140-6736(00)05025-X. [DOI] [PubMed] [Google Scholar]
- 38.Mwai L, Kiara SM, Abdirahman A, Pole L, Rippert A, Diriye A, Bull P, Marsh K, Borrmann S, Nzila A. In vitro activities of piperaquine, lumefantrine, and dihydroartemisinin in Kenyan Plasmodium falciparum isolates and polymorphisms in pfcrt and pfmdr1. Antimicrob Agents Chemother. 2009;53:5069–5073. doi: 10.1128/AAC.00638-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chaijaroenkul W, Wisedpanichkij R, Na-Bangchang K. Monitoring of in vitro susceptibilities and molecular markers of resistance of Plasmodium falciparum isolates from Thai-Myanmar border to chloroquine, quinine, mefloquine and artesunate. Acta Trop. 2010;113:190–194. doi: 10.1016/j.actatropica.2009.10.016. [DOI] [PubMed] [Google Scholar]
- 40.Imwong M, Dondorp AM, Nosten F, Yi P, Mungthin M, Hanchana S, Das D, Phyo AP, Lwin KM, Pukrittayakamee S, Lee SJ, Saisung S, Koecharoen K, Nguon C, Day NP, Socheat D, White NJ. Exploring the contribution of candidate genes to artemisinin resistance in Plasmodium falciparum. Antimicrob Agents Chemother. 2010;54:2886–2892. doi: 10.1128/AAC.00032-10. [DOI] [PMC free article] [PubMed] [Google Scholar]