Abstract
In a previous study, a new flavivirus genome sequence was identified in Culex tarsalis mosquitoes obtained in Alberta, Canada and was shown to be genetically related to but distinct from members of the insect-specific flaviviruses. Nonstructural protein 5–encoding sequences amplified from Cx. tarsalis pools from western Canada have shown a high similarity to genome sequences of novel flaviviruses isolated from mosquitoes in California and Colorado. Despite wide distribution of this virus, designated Calbertado virus, strains demonstrate a high degree of nonstructural protein 5 nucleotide (> 90%) and amino acid (> 97%) identity. The ecology and geographic range of Calbertado virus warrants further study because it may potentially influence transmission of mosquito-borne flaviviruses, including important human pathogens such as West Nile and Saint Louis encephalitis viruses.
Introduction
The family Flaviviridae genus Flavivirus comprises more than 70 positive-sense single-stranded RNA viruses, including a number of important human pathogens that are transmitted by arthropods such as mosquitoes and ticks.1,2 In addition, the genus includes vertebrate flaviviruses with no known vector and a newly recognized complex of insect-specific viruses related to cell fusing agent virus (CFAV) and Kamiti River virus (KRV).3,4 Cell fusing agent virus was first isolated from a cultured line of Aedes aegypti mosquito cells by Stollar and Thomas in 1975.5 Subsequently, it was isolated from field-collected Aedes and Culex mosquitoes in Puerto Rico.6 In 2003, KRV was identified in Ae. macintoshi larvae and pupae from Kenya, and isolates were shown to be related to CFAV.7,8
Recently, a number of additional flaviviruses related to CFAV and KRV have been identified in a variety of mosquito species. These newly identified agents include the Culex flavivirus (CxFV),9 Quang Binh virus,10 and Aedes flavivirus.11 The insect-specific Culex flavivirus, first isolated in 2003 and 2004 in Japan from Culex pipiens and other Culex species, displays a wide geographic distribution, with strains of the virus now described in Culex mosquitoes from Guatemala,12 Mexico,13 Trinidad,14 Texas,14 Iowa,15 and Uganda.16 Quang Binh virus was isolated from Cx. tritaeniorhynchus mosquitoes obtained in Vietnam in 2002 and is more closely related to CxFV than to other insect-specific flaviviruses.10 Aedes flavivirus was isolated from Ae. albopictus mosquitoes in Japan and has a close genetic relationship with CFAV.11,17 These viruses replicate in arthropod cells and do not appear to infect vertebrate cells. Phylogenetic analyses indicate that these insect-specific viruses occupy the most ancestral lineage of flaviviruses and may have been the progenitors of arboviruses transmitted to vertebrate hosts.3
We have identified a novel member of the insect-specific flaviviruses in Cx. tarsalis mosquitoes in Alberta.18 Preliminary genetic characterization of the Alberta virus based on nonstructural protein 5 (NS5) gene sequences indicates that although related to other insect-specific flaviviruses, it is distinct. We have designated it Calbertado virus (CLBOV; based on its identification in California, Alberta, and Colorado). The purpose of our study was to further determine the relatedness among virus genomes detected over a wide North American geographic range. Amplification and sequencing of CLBOV-specific NS5 gene segments from California, Colorado, and western Canada indicate a wide distribution and a high degree of genetic relatedness. Also, these viruses are associated almost exclusively with Cx. tarsalis mosquitoes, indicating that this species is most likely the specific host for this agent.
Materials and Methods
Mosquito collections and homogenate preparation.
Culex spp. mosquitoes from northcentral Colorado (Larimer County, Figure 1) were obtained and sorted by species during mosquito seasons in 2006 and 2007.19 California mosquito samples were obtained by the Kern Country Mosquito Control District at various sites in Kern County, California (Figure 1) in 2007 as part of routine surveillance testing for West Nile virus (WNV). The Kern County mosquito pools were processed at the Center for Vector-borne Diseases at the University of California, Davis.
Figure 1.
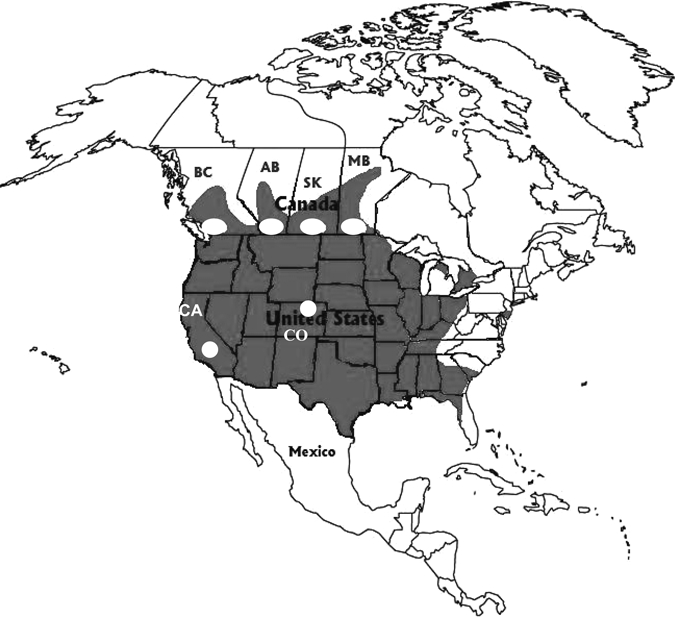
Geographic distribution of Culex tarsalis in North America (shaded) with sites (white dots) indicating where Cx. tarsalis pools were collected in western Canada, California (CA, Kern County), and Colorado (CO, Larimer County). For British Columbia (BC), Alberta (AB), Saskatchewan (SK), and Manitoba (MB), mosquitoes were trapped near the cities of Vancouver, Calgary, Saskatoon, and Winnipeg, respectively. Map modified from Darsie and Ward.27
Culex tarsalis mosquitoes from Canada were obtained at a variety of sites throughout the southern portions of the provinces of Manitoba, Saskatchewan, Alberta, and British Columbia (Figure 1). Mosquito collections in Alberta took place during the summer months (June–September) of 2003 and 2005.18 The collections and sorting of Cx. tarsalis in Manitoba, Saskatchewan, and British Columbia were conducted in the summer of 2007 as part of a WNV surveillance program.
Trapping procedures were similar for all three studies. Standard CO2-baited Centers for Disease Control Miniature Light Traps (John W. Hock Company, Gainesville, FL) were used for mosquito collections. Female mosquitoes were identified to species by trained staff and placed in pools averaging 50 mosquitoes (range 1–80). Mosquito pools were triturated in 1–1.5 mL of diluent and homogenized in minimal essential medium supplemented with 10% fetal bovine serum by using a copper-coated steel shot and agitation by vortexing or a QIAgen Mixer Mill (QIAGEN, Valencia, CA). The homogenates were centrifuged and supernatants were stored at –80°C until nucleic acids were extracted. Minimal infection rates were calculated by using the standard formula: (number of CLBOV-genome positive pools/total mosquitoes tested) × 1,000. (Table 1).
Table 1.
Frequency of Calbertado flavivirus RNA-positive samples among Culex tarsalis pools obtained at various sites in western Canada
| Location | Prevalence (no. positive pools/total pools tested) | Minimum infection rates |
|---|---|---|
| British Columbia | 6.3% (6/95) | 2.7 |
| Alberta | 47.9% (67/140) | 16.5 |
| Saskatchewan | 12.6% (31/246) | 6.0 |
| Manitoba | 8.8% (16/182) | 3.7 |
RNA extraction and reverse transcription–polymerase chain reaction amplification of flavivirus genome.
RNA was extracted from 50 μL of supernatant from mosquito homogenates by using the RNeasy Mini Kit (QIAGEN) according to the manufacturer's instructions. RNA was eluted in 50 μL of sterile distilled water and used for conventional and real-time reverse transcription–polymerase chain reaction (RT-PCR). No evidence for genomic integration of CLBOV sequences was found when PCR was used without an RT step.18
Initial screening of mosquito homogenates for flavivirus genomes was carried out by using universal flavivirus primers CFD2 and MAMD and other primer sets specific for the NS5 coding region of the RNA.18–20 The RT-PCRs were performed by using the QIAGEN One-Step RT-PCR Kit in a total volume of 50 μL. Reaction mixtures followed kit instructions and contained 10 μL of 5× RT-PCR buffer, 2 μL of 10 mM dNTP, 5 μL of template RNA, 1.5 μL (20 mM) of each forward and reverse universal primers, 0.125 μL (40 units/μL of RNaseOUT (Invitrogen, Carlsbad, CA), 2 μL of One-Step RT-PCR Enzyme Mixture, and 28 μL of RNase-free water. Reaction conditions consisted of a 30-minute RT step at 50°C; a 15-minute denaturation step at 95°C; 35 cycles at 94°C, 50°C, and 72°C for 45 seconds each; and a 10-minute extension at 72°C.
On the basis of primary NS5 gene sequence of the CLBOV genome in positive pools of Cx. tarsalis obtained in Alberta, a Taqman real-time RT-PCR assay was designed by using primers CFlaviF (5′-TTGACTCCAACGCCTC-3′), CFlaviR (5′-ACCTTGAGTTCGAAGCG-3′), and an FAM-labeled probe (5′-AAGTTCTCTCGGGAAACCCAATGGTCC-3′) with a TAMRA quencher. Real-time RT-PCR screening of mosquito pools was performed by using RNA extracted from mosquito homogenates and amplified by using an ABI 7900 Sequence Detection System and an RT-PCR Kit for Taqman PCR as per the manufacturer's instructions (Applied Biosystems, Foster City, CA). Real-time RT-PCR mixtures were pre-incubated at 50°C for 30 minutes, followed by denaturation at 95°C for 10 minutes, 40 cycles at 95°C for 15 seconds, and 60°C for 1 minute for annealing and extension. Data were analyzed by using ABI software supplied with the 7900 sequence detection platform. A cycle threshold value < 38 was used to identify and determine Taqman-positive samples.
To generate a larger NS5 cDNA segment for sequencing and phylogenetic analysis, CLBOV RNA-positive extracts underwent RT-PCR amplification with MAMD20 and Flav2 primers,21 which produced a fragment of approximately 1,000 nucleotides. The RT-PCR products were visualized after electrophoresis on a 1% agarose gel stained with ethidium bromide. Negative (no template) and positive controls were included in each RT-PCR.
Detection, sequencing, and phylogenetic analysis of NS5 gene amplicons.
Conventional RT-PCR products were visualized by agarose gel electrophoresis and staining with ethidium bromide. Amplicons were purified by using the QIAquick Gel Extraction Kit (QIAGEN) according to manufacturer's instructions. Amplified cDNA was sequenced in both directions by using the ABI PRISM® BigDye® Terminator v3.1 Cycle Sequencing Kit with the ABI 3730XL DNA Analyzer.
Sequences were aligned in SeqManII (DNAStar) and Mega v4.0 by using the ClustalW method.22 Phylogenetic and molecular evolutionary analyses were conducted by using the neighbor-joining method with Kimura 2-parameter distance corrections and MEGA software.23 Bootstrap analyses were performed by using neighbor-joining and the Kimura 2-parameter (DNA) or Poisson correction (protein) models with 500 replicates.23
Results
Detection of CLBOV RNA in mosquito pools from Canada.
More than 700 pools of Cx. tarsalis obtained during 2003–2007 from various sites in four western Canadian provinces (British Columbia, Alberta, Saskatchewan, and Manitoba; Figure 1) were screened for CLBOV RNA. Collection sites included Vancouver and associated areas of British Columbia, the outskirts of Calgary in southern Alberta, the Swift Current and Saskatoon town sites of Saskatchewan, and the rural regions near Winnipeg in Manitoba (Figure 1). Viral RNA was detected in Cx. tarsalis in all provinces, and the proportion of pools containing NS5-specific sequence ranged from 6.3% in British Columbia to more than 40% in Alberta (Table 1). Minimal infection rates ranged from 2.7 to 16.5 (Table 1). Seasonal differences in CLBOV prevalence were not determined because of limited numbers of pools available for this preliminary study.
Sequence analysis and phylogenetic characterization of representative CLBOV genotypes from Canada, California, and Colorado.
Comparison of amino acid sequences inferred from approximately 1-kb portions of the NS5 gene (270 amino acid fragment) from representative CLBOVs identified in Alberta, Kern County, California, and Larimer County, Colorado with other flaviviruses reaffirmed that this agent is most closely related to insect-specific flaviviruses (Figure 2). The NS5 amino acid sequence homology was more than 97% among CLBOVs identified in these distinct geographic areas. The highest degree of NS5 amino acid similarity with other insect-specific flaviviruses was with CxFV and Quang Binh virus (65–67%), which are also associated primarily with Culex spp. mosquitoes. The CLBOV flavivirus is related to other insect-specific flaviviruses associated with Aedes spp. mosquitoes, but only displayed an average of 60% amino acid identity with these insect-specific flaviviruses. The most closely related viruses based on amino acid sequence homologies for the NS5 region (CxFV, CFAV, and KRV) exhibit less similarity to CLBOV when assessed at the nucleotide level.19 These results indicate that although CLBOVs are related to other insect-specific flaviviruses, they may represent a distinct clade based on amino acid comparisons (Figure 2) and nucleotide comparisons, respectively.
Figure 2.
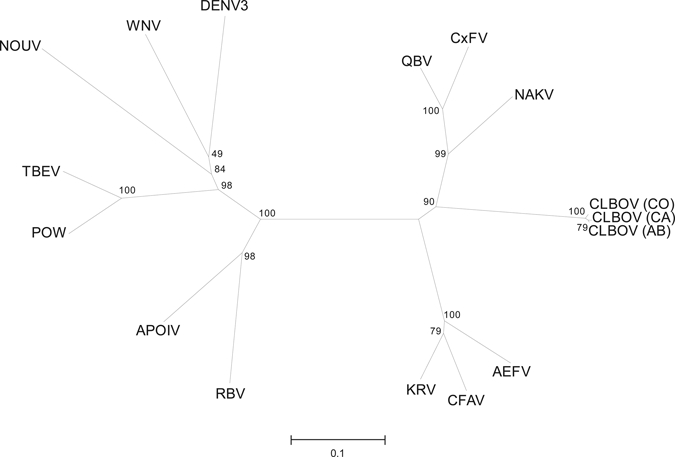
Phylogenetic relationship between Calbertado virus (CLBOV), other members of the cell fusing agent virus–related insect-specific viruses, and other representative flaviviruses. Comparison is based on the amino acid sequence of a segment of the nonstructural protein 5 gene comprising 270 amino acid residues derived from CLBOV. Included in the comparison were AEFV (Aedes flavivirus NC_012932), APOIV (Apoi virus NC_003676), CLBOV (Calbertado virus EU569288 (AB), GU952268 (CA), GU952254 (CO), CFAV (Cell fusing agent virus NC_001564), CxFV (Culex flavivirus NC_008604), DENV3 (Dengue virus type 3 NC_001475), KRV (Kamiti River virus NC_005064), NAKV (Nakiwogo virus GQ165809), NOUV (Nounane virus EU159426), POW (Powassan virus NC_003687), QBV (Quang Binh virus NC_012671), RBV (Rio Bravo virus NC_003675), TBEV (Tick-borne encephalitis virus NC_001672), and WNV (West Nile virus NC_009942). Comparison was performed as described in the Materials and Methods, and the resulting bootstrap values are indicated. Scale bar indicates nucleotide substitutions per site.
Additional CLBOV genotypes from various sites in western Canada, Kern County California, and Larimer County, Colorado were compared with each another by using phylogenetic analysis of nucleic acid sequences (Figure 3). Comparison of NS5 fragments showed three primary subpopulations. Clade 1 is composed of all NS5 nucleotide sequences from British Columbia and California mosquito pools and three sequences from Alberta (27, 1024, 1055), two from Saskatchewan (42 and 43) and one from Colorado (5890). Clade 2 is composed of a Manitoba virus sequence (52–48), a Saskatchewan sequence (90), and two sequences from Alberta (10 and 95). Clade 3 contains the sequences from a second Manitoba pool (44–13) and a Calbertado isolate from Colorado (5901).19 Alberta sequences 1024, 1055, and 11 grouped outside the three clades and exhibited characteristics common to Clades 1 and 2 because of the mixture of genotypes identified (Figure 4).
Figure 3.
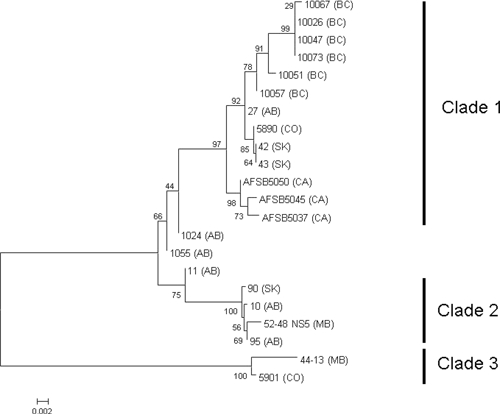
Phylogenetic relatedness among Canadian, California, and Colorado Calbertado virus nonstructural protein 5 (NS5) gene sequences. Nucleotide sequences of the NS5 gene segment obtained from mosquito pools from Manitoba (MB), Colorado (CO), Alberta (AB), British Columbia (BC), and California (CA) were aligned by using ClustalW, and the resulting dendogram was generated by using MEGA version 4.0. GenBank accession numbers of Calbertado virus NS5 sequences are 52–48_(MB) GU952252, 44–13_(MB) GU952253, 5901_(CO) GU952254, 5890_(CO) GU952255, 10_(AB) GU952256, 95_(AB) GU952257, 90_(SK) GU952258, 11_(AB) GU952259, 1055_(AB) GU952260, 27_(AB) GU952261, 10073_(BC) GU952262, 10067_(BC) GU952263, 10057_(BC) GU952264, 10051_(BC) GU952265, 10047_(BC) GU952266, 10026_(BC) GU952267, AFSB5045(CA) GU952268, AFSB5050(CA) GU952269, AFSB5037(CA) GU952270, 42_(SK) GU952271, and 43_(SK) GU952272. Scale bar indicates nucleotide substitutions per site.
Figure 4.
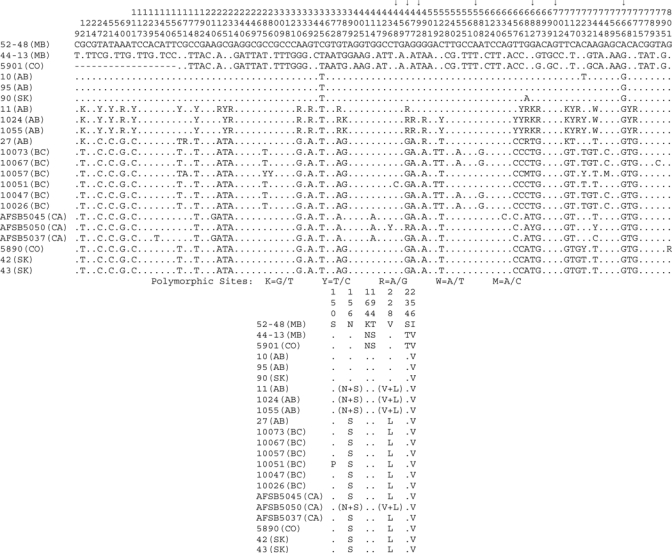
Nucleotide and amino acid variations among Calbertado virus nonstructural protein 5 (NS5) gene sequences. Nucleotide and protein alignments were generated for NS5 sequences, and resulting alignments were condensed to depict only the positions found to be variable between the pools. Alignment coordinates are indicated vertically above each position. Alignment summaries are relative to the 52-48 (MB) sequence, positions found to be identical are represented with a dot, and gaps are indicated with a dash. Variations are indicated with the substitution identified at that location. Standard International Union of Biochemistry coding was used in the nucleotide alignment for positions found to be polymorphic (K = G/T, Y = T/C, R = A/G, W = A/T, M = A/C). Even with the extensive nucleotide variations noted in this region, which codes for 270 amino acids of NS5, only 7 amino acid positions were found to differ between various sequences (nucleotide substitutions responsible for these changes are indicated by arrows). A summary of these amino acid substitutions and positions is included below the nucleotide alignment. Several of the reverse transcription–polymerase chain reaction products sequenced were found to contain polymorphic sites, indicating that the mosquito pool contained a mixture of viruses. Two of these polymorphic sites occurred in codon-altering positions as noted in the amino acid summary.
The amino acid sequence similarity within the three clades was 99.6–99.9% for Clade 1, 98.2–100% for Clade 2, and 99.1% for Clade 3. Clade 3 was the most divergent from the other clades; approximately 10% variation was observed. The degree of divergence within a given clade is influenced by the limited numbers of samples in each. Also, three of the Calbertado virus RT-PCR amplicons generated from Alberta mosquito pools (1024, 1055, and 11) indicated the presence of polymorphic nucleotide and amino acid sites, which suggested a mixture of genotypes with features observed for Clades 1 and 2 (Figures 3 and 4). Two of the polymorphic sites occurred in codon-altering positions resulting in amino acid variations of N and S or V and L (Figure 4).
Despite a number of differences at the nucleotide sequence level, most of the CLBOV NS5 changes were synonymous, and there was little variation in the amino acid sequences. The nucleotide sequence assessed encodes 270 amino acids, yet only 7 positions were found to be variable (Figure 4). Three of these distinguished Clade 3 from the others (K164N, T194S, and S234T), two differentiated Clade 1 from Clade 2 (N156S and V228L) and one each were unique to 52–48 MB (V256I) and 10051 BC (S150P).
Discussion
Our results indicate a wide distribution of a novel insect-specific flavivirus. Calbertado virus was identified in Cx. tarsalis pools obtained throughout western Canada and from Cx. tarsalis and Cx. pipiens obtained in California and Colorado.19 Despite its extensive geographic range, this flavivirus has highly conserved NS5 nucleotide and amino acid sequences.
Phylogenetic analysis of a 270-amino acid sequence in the NS5 protein separated the viruses in this study into 3 main clades. Although the greatest divergence was between the Clade 3 viruses and all other Calbertado genotypes, Clade 3 nucleotide sequence similarity with other viruses in the complex was still almost 90%, and the amino acid sequence divergence was approximately 3%.
The NS5 nucleotide sequences in California viruses harbored a few single nucleotide polymorphisms that made them distinct from the others in Clade 1, but overall they were more similar to this group than they were to the members of Clades 2 or 3 (Figure 4). It is interesting to note that although California genotype AFSB 5050 clearly clustered with Clade 1 on the basis of nucleotide identity, it had the same polymorphic amino acid sites as Alberta sequences 11, 1024 and 1055, which placed it in Clades 1 and 2.
In general, the sequences appeared to cluster on the basis of geographic boundaries. However, there were a number of notable exceptions. One Manitoba and one Colorado CLBOV NS5 sequence (44-13 MB, 5901 CO) exhibited distinct nucleotide and amino acid polymorphisms and were the only representative genotypes in Clade 3, and other sequences from these locations were situated within Clades 1 and 2. Sequence similarity between the three CLBOV sequences from Saskatchewan was quite high but specific single nucleotide polymorphisms and amino acid polymorphisms corresponded with the placement of 90 SK in Clade 2 and 42 SK and 43 SK in Clade 1. The sequences from the Alberta pools are also of interest. Two of the nucleotide sequences (10 and 95) were virtually identical to the 52–48 sequence from Manitoba and one (27) appeared most similar to those found in British Columbia. Although there was clustering of similar CLBOVs in various provinces or regions, the results also show evidence of co-circulation of distinct genotypes within the same general area from which mosquitoes were obtained.
Although CLBOV is now added to the rapidly increasing list of insect-specific flaviviruses,3,4 it exhibits only 60–70% NS5 amino acid similarity with other members of the complex. Further analysis of genomic regions encoding the NS3 gene indicated similar phylogenetic grouping (Drebot MA, unpublished data). On the basis of these observations and the recent isolation of this virus from mosquitoes obtained in California and Colorado (Bolling and others19 and Brault AC, unpublished data), it is likely that CLBOV will be recognized as a new species within the genus Flavivirus.
This study and other recent publications indicated that CLBOV is associated primarily with Cx. tarsalis mosquitoes.18,19 Real time RT-PCR testing of 400 pools of Cx. pipiens from the province of Quebec provided no evidence for CLBOV RNA. However, preliminary studies using CxFV-specific primers indicated that a related virus may be present in eastern Canada (Drebot MA, unpublished data), as observed in Cx. p. pipiens and Cx. p. quinquefasciatus from the United States and other countries.4,9,12–16 In addition, Aedes and other genera of mosquitoes obtained in Canada and the United States should be assayed for viruses similar to Aedes flavivirus, which was recently identified in Japan.11 The ever widening geographic range of these viruses is an interesting finding and should be explored further.
Our study also demonstrates that infection rates for CLBOV virus are variable across geographic areas. For example, we detected CLBOV RNA in more than 40% of Cx. tarsalis pools obtained from sites in southern Alberta (minimum infection rate = 16.5). However, mosquitoes were obtained in Alberta during 2003 and 2005, and Cx. tarsalis from other western provinces in Canada were obtained in 2007. Seasonal and yearly prevalence and infection rates of CLBOV in Cx. tarsalis at various sites or regions in Canada warrant further study. Recent studies in Colorado indicate that there can be differences in the infection rates of mosquitoes obtained from the same site at various times during the spring and summer.19 Similar changes in virus prevalence may occur in western Canada and other geographic areas that could be influenced by factors such as climate, environmental conditions, and ecosystems.
A number of questions remain regarding the evolutionary origins of these viruses, the mechanism for their transmission, the extent of arthropod taxa for which these virus–host relationships exist, and the specificity of these associations. For example, some of the insect-specific flaviviruses appear to be associated with Aedes spp. mosquitoes, and the CxFVs have been linked with the Cx. pipiens complex.3 The CLBOV agent described in our study has been associated with Cx. tarsalis mosquitoes from western Canada and the western United States in California and Colorado. This geographic pattern is paralleled by the distribution of Cx. tarsalis mosquitoes in western North America and in conjunction with the virus only being identified in this one species could indicate that a highly specific virus–host association has evolved between these mosquitoes and CLBOV.
Wider surveillance testing for CLBOV and other insect-specific viruses will be needed to test this hypothesis further. However, it is interesting that CxFVs have been identified in primarily Cx. pipiens complex mosquito pools from Asia and Central and North America. More restricted geographic distribution of Cx. tarsalis mosquitoes compared with that of the Cx. pipiens complex could indicate restricted gene flow and consequently enhanced evolutionary rates of these viruses compared with that of the CxFV complex. Close genetic identity of CxFVs from Texas with sequence data generated from CxFV detected in Cx. pipiens mosquitoes from Japan indicate the potential for exchange of these CxFV genotypes through large ports such as Houston by which Cx. pipiens mosquitoes are likely imported. The limited distribution of Cx. tarsalis mosquitoes could preclude such global gene flow of CLBOV genotypes. However, it should also be noted that studies using microsatellite markers have indicated extensive gene flow between Cx. tarsalis populations in North America.24,25 This finding could explain why similar CLBOV genotypes have been identified in Manitoba and Colorado. Detailed genetic studies of CLBOVs from geographically isolated Cx. tarsalis mosquitoes will need to be assessed over time to fully evaluate this hypothesis.
It should also be determined whether infection of a mosquito with CLBOV alters the susceptibility for superinfection with an alternative human disease flavivirus.3 During the screening of Cx. tarsalis mosquitoes from Manitoba, we observed that a significant proportion (30%) of pools containing CLBOV RNA also contained WNV RNA (Drebot MA, unpublished data). However, this number was reduced (1.6%) in Cx. tarsalis pools from California 2008 (Brault AC and others, unpublished data). Co-infection of Cx. quinquefasciatus with CxFV and WNV indicated that overall there was little impact on the infectivity or transmissibility of WNV in these mosquitoes.4 However, it was noted that the level of transmission of WNV could be enhanced depending on the origin of the mosquito colony used.
Recently, a second study characterizing co-infections of Culex spp. with WNV and CxFV in Chicago implied a positive ecological association between the two viruses.26 Further studies are required to characterize possible interference or perhaps augmentation of arbovirus co-infection that may be conferred by persistent infection with these viruses. Co-infections of mosquitoes by various combinations of medically important flaviviruses and insect-specific flaviviruses such as CLBOV will shed further light on the role these agents may play in arboviral ecology, prevalence, vector competence, and vertical transmission of other flaviviruses.3 Studies are also warranted with respect to how insect-specific flaviviruses might affect the biology of the arthropod host, including reproductive patterns, longevity, sensitivity to environmental conditions such as weather and climate changes, and other characteristics that might be artificially manipulated to interrupt or modulate arthropod breeding or reproductive capacity. Also, the study of molecular determinants of these viruses that encode altered mosquito tropism, mosquito transmission mechanisms, and loss of vertebrate infectivity will provide fundamental insights into virus–host interactions.
ACKNOWLEDGMENTS
We thank Connie Blakeston, Matthew Walker, Antonia Dibernardo, Robbin Lindsay, Melissa Rabb, Katarina Bernat, and Kimberly Holloway, Chet Moore, and Lars Eisen for expert technical assistance and advice. We also thank Mohammed Morshed (British Columbia Centers for Disease Control, Vancouver, British Columbia), Philip Curry (Saskatchewan Health, Regina, Saskatchewan), Ryan McDonald (Saskatchewan Disease Control Laboratory, Regina, Saskatchewan), Jock Macintosh (Alberta Environment, Calgary, Alberta), Magdy Dawood and Paul Van Caeseele (Cadham Provincial Laboratory, Winnipeg, Manitoba), and Hugues Charest (Laboratoire de Sante Publique du Quebec, Montreal, Quebec) for providing mosquito pools used in this study. We also thank the Kern County Mosquito and Vector Control District for collecting mosquito pools used in this study.
Footnotes
Financial support: Support for genetic characterization of CLBOV-positive pools from Kern County, California was provided through a contract with the Sacramento–Yolo County Vector Control Association with the University of California, Davis, CA. This study at Colorado State University was supported in part by contract N01-AI-25489 from the National Institutes of Allergy and Infectious Diseases.
Authors' addresses: Shaun Tyler and Michael A. Drebot, National Microbiology Laboratory, Public Health Agency of Canada, Winnipeg, Manitoba, Canada, E-mails: shaun.tyler@phac-aspc.gc.ca and mike.drebot@phac-aspc.gc.ca. Bethany G. Bolling, Carol D. Blair, and Charles H. Calisher, Arthropod-borne and Infectious Diseases Laboratory, Department of Microbiology, Immunology and Pathology, Colorado State University, Fort Collins, CO, E-mails: bolling@ColoState.EDU, Carol.Blair@ColoState.EDU and calisher@cybersafe.net. Aaron C. Brault, Division of Vector-borne Diseases, Centers for Disease Control and Prevention, Fort Collins, CO, E-mail: acbrault1@mac.com. Kanti Pabbaraju, Provincial Laboratory for Public Health, Calgary, Alberta, Canada, E-mail: Kanti.Pabbaraju2@albertahealthservices.ca. M. Veronica Armijos and David C. Clark, Department of Pathology, Microbiology and Immunology, School of Veterinary Medicine and Center for Vector-Borne Diseases, University of California, Davis, CA, E-mails: mvarmijos@ucdavis.edu and dcclark09@gmail.com.
References
- 1.Lindenbach BD, Thiel HJ, Rice CM. In: Fields Virology. 5th edition. Knipe DM, Howley PM, editors. Philadelphia, PA: Lippincott-Raven Publishers; 2007. pp. 1101–1152. (Flaviviridae: the viruses and their replication). [Google Scholar]
- 2.Cook S, Holmes EC. A multigene analysis of the phylogenetic relationships among the flaviviruses (family: Flaviviridae) and the evolution of vector transmission. Arch Virol. 2006;151:309–325. doi: 10.1007/s00705-005-0626-6. [DOI] [PubMed] [Google Scholar]
- 3.Firth AE, Blitvich BJ, Willis NM, Miller CL, Atkins JF. Evidence for ribosomal frameshifting and a novel overlapping gene in the genomes of insect-specific flaviviruses. Virology. 2010;399:153–166. doi: 10.1016/j.virol.2009.12.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kent RJ, Crabtree MB, Miller BR. Transmission of West Nile virus by Culex quinquefasciatus say infected with Culex flavivirus Izabal. PLoS Negl Trop Dis. 2010;4:e671. doi: 10.1371/journal.pntd.0000671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stollar V, Thomas VL. An agent in the Aedes aegypti cell line (Peleg) which causes fusion of Aedes albopictus cells. Virology. 1975;64:367–377. doi: 10.1016/0042-6822(75)90113-0. [DOI] [PubMed] [Google Scholar]
- 6.Cook S, Bennett SN, Holmes EC, De Chesse R, Moureau G, de Lamballerie X. Isolation of a new strain of the flavivirus cell fusing agent virus in a natural mosquito population from Puerto Rico. J Gen Virol. 2006;87:735–748. doi: 10.1099/vir.0.81475-0. [DOI] [PubMed] [Google Scholar]
- 7.Crabtree MB, Sang RC, Stollar V, Dunster LM, Miller BR. Genetic and phenotypic characterization of the newly described insect flavivirus, Kamiti River virus. Arch Virol. 2003;148:1095–1118. doi: 10.1007/s00705-003-0019-7. [DOI] [PubMed] [Google Scholar]
- 8.Sang RC, Gichogo A, Gachoya J, Dunster MD, Ofula V, Hunt AR, Crabtree MB, Miller BR, Dunster LM. Isolation of a new flavivirus related to cell fusing agent virus (CFAV) from field-collected flood-water Aedes mosquitoes sampled from a dambo in central Kenya. Arch Virol. 2003;148:1085–1093. doi: 10.1007/s00705-003-0018-8. [DOI] [PubMed] [Google Scholar]
- 9.Hoshino K, Isawa H, Tsuda Y, Yano K, Sasaki T, Yuda M, Takasaki T, Kobayashi M, Sawabe K. Genetic characterization of a new insect flavivirus isolated from Culex pipiens mosquito in Japan. Virology. 2007;359:405–414. doi: 10.1016/j.virol.2006.09.039. [DOI] [PubMed] [Google Scholar]
- 10.Crabtree MB, Nga PT, Miller BR. Isolation and characterization of a new mosquito flavivirus, Quang Binh virus, from Vietnam. Arch Virol. 2009;154:857–860. doi: 10.1007/s00705-009-0373-1. [DOI] [PubMed] [Google Scholar]
- 11.Hoshino K, Isawa H, Tsuda Y, Sawabe K, Kobayashi M. Isolation and characterization of a new insect flavivirus from Aedes albopictus and Aedes flavopictus mosquitoes in Japan. Virology. 2009;391:119–129. doi: 10.1016/j.virol.2009.06.025. [DOI] [PubMed] [Google Scholar]
- 12.Morales-Betoulle ME, Monzón Pineda ML, Sosa SM, Panella N, López MR, Cordón-Rosales C, Komar N, Powers A, Johnson BW. Culex flavivirus isolates from mosquitoes in Guatemala. J Med Entomol. 2008;45:1187–1190. doi: 10.1603/0022-2585(2008)45[1187:cfifmi]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 13.Farfan-Ale JA, Loroño-Pino MA, Garcia-Rejon JE, Hovav E, Powers AM, Lin M, Dorman KS, Platt KB, Bartholomay LC, Soto V, Beaty BJ, Lanciotti RS, Blitvich BJ. Detection of RNA from a novel West Nile-like virus and high prevalence of an insect-specific flavivirus in mosquitoes in the Yucatan Peninsula of Mexico. Am J Trop Med Hyg. 2009;80:85–95. [PMC free article] [PubMed] [Google Scholar]
- 14.Kim DY, Guzman H, Bueno R, Jr, Dennett JA, Auguste AJ, Carrington CV, Popov VL, Weaver SC, Beasley DW, Tesh RB. Characterization of Culex flavivirus (Flaviviridae) strains isolated from mosquitoes in the United States and Trinidad. Virology. 2009;386:154–159. doi: 10.1016/j.virol.2008.12.034. [DOI] [PubMed] [Google Scholar]
- 15.Blitvich BJ, Lin M, Dorman KS, Soto V, Hovav E, Tucker BJ, Staley M, Platt KB, Bartholomay LC. Genomic sequence and phylogenetic analysis of Culex flavivirus, an insect-specific flavivirus, isolated from Culex pipiens (Diptera:Culicidae) in Iowa. J Med Entomol. 2009;46:934–941. doi: 10.1603/033.046.0428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cook S, Moureau G, Harbach RE, Mukwaya L, Goodger K, Ssenfuka F, Gould E, Holmes EC, de Lamballerie X. Isolation of a novel species of flavivirus and a new strain of Culex flavivirus (Flaviviridae) from a natural mosquito population in Uganda. J Gen Virol. 2009;90:2669–2678. doi: 10.1099/vir.0.014183-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Crochu S, Cook S, Attoui H, Charrel RN, De Chesse R, Belhouchet M, Lemasson JJ, de Micco P, de Lamballerie X. Sequences of flavivirus-related RNA viruses persist in DNA form integrated in the genome of Aedes spp. mosquitoes. J Gen Virol. 2004;85:1971–1980. doi: 10.1099/vir.0.79850-0. [DOI] [PubMed] [Google Scholar]
- 18.Pabbaraju K, Ho KC, Wong S, Fox JD, Kaplen B, Tyler S, Drebot M, Tilley PA. Surveillance of mosquito-borne viruses in Alberta using reverse transcription polymerase chain reaction with generic primers. J Med Entomol. 2009;46:640–648. doi: 10.1603/033.046.0332. [DOI] [PubMed] [Google Scholar]
- 19.Bolling BG, Eisen L, Moore CG, Blair CD. Insect-specific flaviviruses from Culex mosquitoes in Colorado, with evidence of vertical transmission. Am J Trop Med Hyg. 2011;85:169–177. doi: 10.4269/ajtmh.2011.10-0474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Scaramozzino N, Crance JM, Jouan A, DeBriel A, Stoll F, Garin D. Comparison of flavivirus universal primer pairs and development of a rapid, highly sensitive heminested reverse transcription-PCR assay for detection of flaviviruses targeted to a conserved region of the NS5 gene sequences. J Clin Microbiol. 2001;39:1922–1927. doi: 10.1128/JCM.39.5.1922-1927.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ayers M, Adachi D, Johnson G, Andonova M, Drebot M, Tellier R. A single tube RT-PCR assay for the detection of mosquito-borne flaviviruses. J Virol Methods. 2006;135:235–239. doi: 10.1016/j.jviromet.2006.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Higgins DG, Thompson JD, Gibson TJ. Using CLUSTAL for multiple sequence alignments. Methods Enzymol. 1996;266:383–402. doi: 10.1016/s0076-6879(96)66024-8. [DOI] [PubMed] [Google Scholar]
- 23.Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- 24.Venkatesan M, Westbrook CJ, Hauer MC, Rasgon JL. Evidence for a population expansion in the West Nile virus vector Culex tarsalis. Mol Biol Evol. 2007;24:1208–1218. doi: 10.1093/molbev/msm040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Venkatesan M, Rasgon JL. Population genetic data suggest a role for mosquito-mediated dispersal of West Nile virus across the western United States. Mol Ecol. 2010;19:1573–1584. doi: 10.1111/j.1365-294X.2010.04577.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Newman CM, Cerutti F, Anderson TK, Hamer GL, Walker ED, Kitron UD, Ruiz MO, Brawn JD, Goldberg TL. Vector Borne Zoonotic Dis. 2011. (Culex flavivirus and West Nile virus mosquito coinfection and positive ecological association in Chicago, United States). Jan 22 [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Darsie RF, Jr, Ward RA. Identification and Geographical Distribution of the Mosquitoes of North America, North of Mexico. Gainesville, FL: University Press of Florida; 2005. [Google Scholar]


