Abstract
Charcot–Marie–Tooth disease is a genetically heterogeneous group of motor and sensory neuropathies associated with mutations in more than 30 genes. Charcot–Marie–Tooth disease type 4J (OMIM 611228) is a recessive, potentially severe form of the disease caused by mutations of the lipid phosphatase FIG4. We provide a more complete view of the features of this disorder by describing 11 previously unreported patients with Charcot–Marie–Tooth disease type 4J. Three patients were identified from a small cohort selected for screening because of their early onset disease and progressive proximal as well as distal weakness. Eight patients were identified by large-scale exon sequencing of an unselected group of 4000 patients with Charcot–Marie–Tooth disease. In addition, 34 new FIG4 variants were detected. Ten of the new CMT4J cases have the compound heterozygous genotype FIG4I41T/null described in the original four families, while one has the novel genotype FIG4L17P/null. The population frequency of the I41T allele was found to be 0.001 by genotyping 5769 Northern European controls. Thirty four new variants of FIG4 were identified. The severity of Charcot–Marie–Tooth disease type 4J ranges from mild clinical signs to severe disability requiring the use of a wheelchair. Both mild and severe forms have been seen in patients with the same genotype. The results demonstrate that Charcot–Marie–Tooth disease type 4J is characterized by highly variable onset and severity, proximal as well as distal and asymmetric muscle weakness, electromyography demonstrating denervation in proximal and distal muscles, and frequent progression to severe amyotrophy. FIG4 mutations should be considered in Charcot–Marie–Tooth patients with these characteristics, especially if found in combination with sporadic or recessive inheritance, childhood onset and a phase of rapid progression.
Keywords: Charcot–Marie–Tooth disease, neurodegenerative disorders, clinical characteristics, demyelinating disease, molecular genetics
Introduction
Charcot–Marie–Tooth disease is a genetically heterogeneous motor and sensory peripheral neuropathy caused by mutations in more than 30 genes (Barisic et al., 2008; Saporta et al., 2011). Recessive inheritance is observed in ∼1–4% of cases (Saporta et al., 2011), although the true number may be higher, since sporadic cases may carry recessive mutations and in some populations increased consanguinity is associated with a higher rate of recessive disorders. Charcot–Marie–Tooth disease typically affects distal muscles with slow progression and little effect on lifespan. We recently reported that mutations in the lipid phosphatase FIG4 cause a severe recessive form of the disorder designated Charcot–Marie–Tooth disease type 4J (CMT4J) (Chow et al., 2007). Three of the initial families had early onset and one had adult onset with progression to wheelchair confinement (Chow et al., 2007; Zhang et al., 2008). In this article, we describe the clinical and genetic characterization of 11 previously unreported patients with CMT4J.
CMT4J is caused by mutations in FIG4, a phosphatase that removes the 5-phosphate from the low abundance signalling phosphoinositide PI(3,5)P2 that is localized on the cytoplasmic surface of vesicles of the endosome/lysosome pathway. The original four patients with CMT4J were compound heterozygotes carrying one unique null allele of FIG4 in combination with the shared ancestral missense allele FIG4-I41T (Chow et al., 2007). FIG4-I41T retains partial function in yeast (Chow et al., 2007), and overexpression of FIG4-I41T as a transgene in Fig4 null mice rescues neurodegeneration and lethality (Lenk et al., 2011). The I41T substitution is located near the surface of the non-catalytic domain of FIG4 (Manford et al., 2010) and impairs interaction with the scaffold protein VAC14, rendering the FIG4 protein unstable in vivo (Lenk et al., 2011). In cultured cells, co-transfection with VAC14 increases the half-life of a green fluorescent protein-FIG4 hybrid protein but the I41T mutation prevented this effect (Ikonomov et al., 2010). The very low abundance of FIG4-I41T protein in patient fibroblasts can be partially restored by treatment with the proteasome inhibitor MG-132 (Lenk et al., 2011).
At the cellular level, the reduced abundance of PI(3,5)P2 in Fig4 null mice leads to accumulation of enlarged vesicles derived from the endosome/lysosome pathway in fibroblasts and neurons (Chow et al., 2007; Ferguson et al., 2009). Similar vesicle accumulation is seen in fibroblasts from a patient with CMT4J (Zhang et al., 2008). The Fig4 null mice also exhibit impaired autophagy with accumulation of ubiquitinated proteins and p62 in astrocytes and neurons (Ferguson et al., 2009). The data are consistent with a defect in the regeneration of lysosomes from the mature autolysosome.
Two mouse models with Fig4 deficiency have been studied. Homozygous Fig4 null mice survive for 4–6 weeks and exhibit extensive spongiform neurodegeneration in brain and peripheral ganglia, and loss of large diameter-myelinated neurons in sciatic nerve (Chow et al., 2007). A transgenic model expressing the CMT4J variant Fig4-I41T on a Fig4 null background exhibits dose-dependent rescue of mutant phenotypes, including neurodegeneration in the brain and dorsal root ganglia, and myelination of the sciatic nerve (Lenk et al., 2011).
Here, we extend the clinical characterization of CMT4J by describing the clinical and genetic characteristics of 11 additional patients. We also describe the frequency of the pathogenic FIG4-I41T allele in the Northern European control population and the overall contribution of FIG4 to Charcot–Marie–Tooth disease.
Materials and methods
Mutation detection
Genomic DNA was isolated from peripheral blood. The 23 exons of FIG4 were amplified by polymerase chain reaction and analysed by automated sequencing in the University of Michigan Sequencing Core or as part of a diagnostic panel for patients with Charcot–Marie–Tooth disease at Athena Diagnostics (Worcester). Novel variants are listed in the Supplementary Tables and in the Inherited Peripheral Neuropathies database at the University of Antwerp (www.molgen.ua.ac.be/CMTMutations/Default.cfm).
Deletion of exon 2
The genomic DNA of Patient A7 was analysed with the TaqMan real-time polymerase chain reaction assay using primers flanking exon 2 (Hs02702611_cn) with internal reference RNase P (4403326). Samples were assayed at two different concentrations in quadruplicate using the StepOne Plus machine at University of Michigan Microarray Core. ΔCt values (Ct of FIG4 – Ct of RNase P) were calculated.
Yeast functional assay
Rescue of enlarged vacuoles in Fig4p null yeast was assayed as previously described (Chow et al., 2007; Jin et al., 2008).
Genotyping the I41T mutation in Northern European controls
Thirteen heterozygotes were identified among 5769 individuals (11 538 alleles) by genotyping the nucleotide substitution c.122T>C as described below.
Nine I41T heterozygotes were detected among 4414 unaffected control subjects from the National Institute of Mental Health Schizophrenia Genetics Initiative (NIMH-GI), collected by the Molecular Genetics of Schizophrenia II (MGS-2) collaboration, using Sequenom matrix-assisted laser desorption/ionization-time of flight mass spectrometry (MALDI-TOF; MassARRAY), iPLEX chemistry and mass-spectrometric detection (Sequenom Inc.). Custom single nucleotide polymorphism genotyping was performed in 24-plex polymerase chain reaction and primer-extension reaction format. Assays were designed utilizing Sequenom Assay Designer 3.1 software; primer sequences are available from the authors. Known heterozygous samples served as positive controls. Genotyping calls were made using SpectroAnalyser 3.4 software. Heterozygotes were confirmed by direct sequencing in forward and reverse directions.
Sequencing exon 2 from 764 unaffected adults of Northern European ancestry identified 1 heterozygote in 192 neurologically normal controls in Coriell panels NDPT006 and NDPT009, 1 heterozygote in 163 individual Coriell controls, 1 heterozygote in 206 ClinSeq controls (Biesecker et al., 2009) and 0 heterozygotes in 20 neurologically normal adults and 92 amyotrophic lateral sclerosis spouses (Chow et al., 2009).
One heterozygote was identified among 591 unaffected LOAD controls from the National Cell Repository for Alzheimer's Disease (NCRAD) using the TaqMan Allelic Discrimination Assay on an ABI 7900HT Fast Real-Time PCR system (Applied Biosystems) with custom assay AHY9HWJ containing polymerase chain reaction primers and fluorescent-labelled probes (Applied Biosystems). Polymerase chain reaction was performed in 5 µl volumes containing 10 ng of genomic DNA, 2.5 µl of TaqMan Genotyping Mastermix, 0.25 µl of primer/probe mix and 1.25 µl of water with forward primer GGGAG CAATA ATGCA GAAAC GAAAT and reverse primer TGGAG GATAC TTACC CTGTC ATCAA T. Ten minutes at 95°C was followed by 40 cycles of 15 s at 92°C and 1 min at 60°C. The assay endpoint was read after the polymerase chain reaction reactions; allelic discrimination was accomplished using the SDS v2.3 software (Applied Biosystems).
Results
FIG4 mutations in Australian patients with early-onset progressive Charcot–Marie–Tooth disease
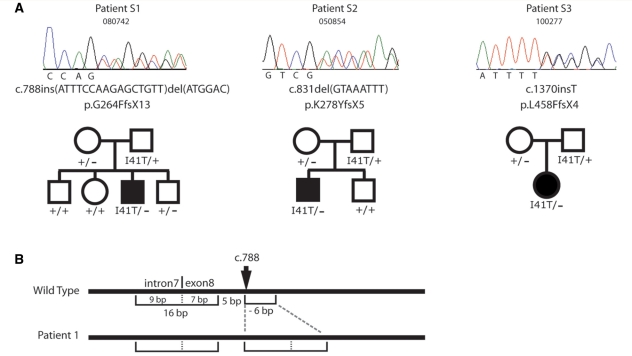
Six patients were selected for specific sequencing of FIG4 based on early disease onset with severe progressive proximal as well as distal weakness; these patients were negative for mutations in the Charcot–Marie–Tooth disease genes PMP22, MPZ and GJB1 (males). The 23 exons of FIG4 were amplified from genomic DNA and sequenced. Three patients were found to be FIG4 compound heterozygotes carrying the I41T missense mutation and a protein truncation mutation (Fig. 1). The truncation mutation in Patient S1 is a complex indel resulting from duplication of a 16-bp fragment spanning the junction between intron 7 and exon 8, combined with a 6-bp deletion (Fig. 1). The net effect is insertion of 10 bp into exon 8, causing a change in reading frame and the premature truncation mutation p.G264FfsX13. The structure of this rearrangement is consistent with the recently described fork stalling, template switching mechanism (FOSTES) (Lee et al., 2007). Patient S2 carries an 8-bp deletion in exon 8 that produces the protein truncation mutation p.K278YfsX5 (Fig. 1). Patient S3 carries a 1-bp insertion in exon 12 resulting in the protein truncation mutation p.L458FfsX4 (Fig. 1). Thus, each patient has the compound heterozygous genotype FIG4I41T/− that characterized the first four families with CMT4J (Chow et al., 2007).
Figure 1.
Autosomal recessive inheritance of FIG4 mutations in three Australian families with CMT4J. (A) DNA chromatographs demonstrating the null mutations in three patients who also carry the FIG4-I41T allele. Autosomal recessive inheritance in evident in these families. (B) Structure of the mutant allele in Patient S1.
Autosomal recessive inheritance of FIG4 mutations in Australian patients with CMT4J
Parents and unaffected siblings of Patients S1, S2 and S3 with CMT4J were genotyped for the patient mutations. None of the parents had signs or symptoms suggestive of disease and five parents had nerve conduction studies that were normal. All of the parents were heterozygous carriers of patient mutations, and the unaffected sibs were heterozygous or had the wild-type reference sequence (Fig. 1B). The data demonstrate autosomal recessive inheritance of the disease in these families.
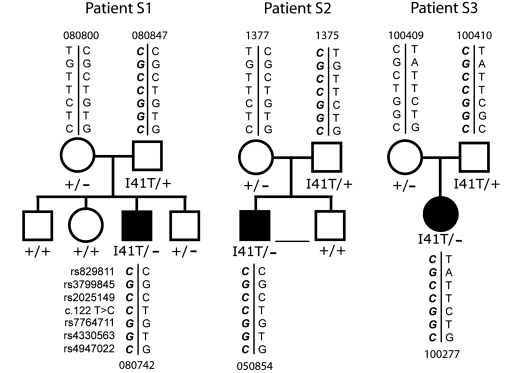
FIG4 haplotypes in Australian families with CMT4J
Parents and affected offspring were genotyped for three single nucleotide polymorphisms used previously to define a 15 kb ancestral haplotype for the I41T mutant allele, rs3799845 (G), rs2025429 (C) and rs7764711 (G) (Chow et al., 2007). Three additional single nucleotide polymorphisms extended the haplotype to 126 kb. The genotypes were consistent with inheritance of I41T on a shared haplotype (Fig. 2). The extended haplotype includes the alleles rs829811(C), rs4330563 (G) and rs4947022 (C). The data support inheritance of the same ancestral mutant allele in these families and the original families (Chow et al., 2007).
Figure 2.
The FIG4-I41T allele is inherited on a shared chromosome 6 haplotype in three unrelated Australian families. The shared alleles are indicated in bold (lower left).
FIG4 mutations identified by large-scale screening of patients with Charcot–Marie–Tooth disease
The 23 exons of FIG4 were sequenced from 4000 consecutive DNA samples submitted for a Charcot–Marie–Tooth disease gene panel to Athena Diagnostics. Eight individuals (0.2%) carrying two mutant alleles of FIG4 were diagnosed as CMT4J. Seven of the eight patients were compound heterozygotes for the I41T allele and a null allele of FIG4. Single nucleotide polymorphism genotypes of these patients were consistent with inheritance of the ancestral haplotype in Fig. 2.
The eighth patient, A9, carried a null allele described below in combination with the missense mutation p.L17P. Leucine 17 is located in the first β sheet at the surface of the protein interaction domain of FIG4 (Manford et al., 2010). The non-conservative substitution of proline for leucine is predicted to alter the conformation of the protein and, like I41T, to affect interaction with other proteins in the PI(3,5)P2 biosynthetic complex (Lenk et al., 2011). Leucine residue 17 is evolutionarily conserved from mammals to yeast. The L17P substitution is predicted to be deleterious by the widely used software programs Polyphen (score 2.3 = probably deleterious), Sift (score 0.00 = pathogenic with confidence of 0.85) and Prophyler (score 7 × 10−6 = high impact on function). These considerations indicate that L17P is a second deleterious variant that is pathogenic when inherited in combination with a null allele.
Null alleles of FIG4 from the large-scale screen
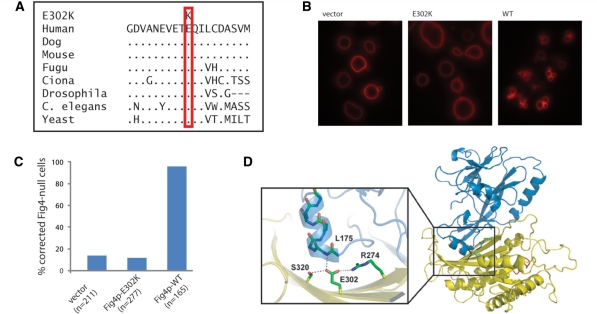
Seven of the null alleles in the patients with CMT4J result in protein truncation, including four frameshift mutations, one nonsense mutation and one deletion of exon 2 (Table 1, A1–A9 and Supplementary Table 2). The eighth null allele is the missense mutation E302K, which alters a residue that is evolutionarily invariant from yeast to human (Fig. 3A). To test the functional consequence of E302K, we introduced the corresponding mutation into the yeast gene and assayed rescue of the enlarged vacuole phenotype of Fig4p null yeast, as previously described (Chow et al., 2007; Jin et al., 2008). Wild-type Fig4p rescues vacuole morphology and generates the normal, multilobed appearance in 95% of cells (Fig. 3B). The E302K mutant did not rescue vacuole enlargement (Fig. 3C) and transformed cells did not differ from the non-transformed controls. Residue E302 is located at the interface between the catalytic domain and the N-terminal protein interaction domain of FIG4 (Fig. 3D) (Manford et al., 2010). Glutamate 302 stabilizes the protein by hydrogen bonding and electrostatic interaction with the conserved residue R274 in the catalytic domain, and formation of a hydrogen bond with the main chain amide group of L175 that stabilizes the last alpha helix dipole in the N-terminal domain. Substitution by lysine is predicted to prevent these interactions and destabilize the protein. The lack of activity in the yeast assay together with the structural considerations indicate that E302K is a functionally null allele of FIG4.
Table 1.
Clinical and genetic features of patients with CMT4J with mutation in FIG4
| Patient | Sex | Genotype (alleles) | Onset | Age at diagnosis | Proximal weakness | Site of weakness | Asymmetry | Diffuse areflexia | Sensory deficits | Comments |
|---|---|---|---|---|---|---|---|---|---|---|
| S1 | M | I41T G264FfsX13 | Early | 14 (2009) | Y | LE, UE | mild, R > L | Y | Vibration, pinprick, proprioception | Gait difficulty, muscle wasting, scoliosis, elevated creatine kinase |
| Wheelchair in late 20 s | ||||||||||
| S2 | M | I41T K278YfsX5 | Early | 5 (1975) | Y | LE, UE | Y | Y | Vibration, pinprick, proprioception | Wheelchair since 20 s and CPAP. |
| Some motor improvement with 0.5 mg/kg prednisolone daily | ||||||||||
| Sural nerve: reduced axon density and size, progressive myelinated fibre loss, onion bulbs (Fig. 6) | ||||||||||
| EMG: active and chronic denervation | ||||||||||
| S3 | F | I41T L458FfsX4 | Early | 5 (1997) | Y | LE, UE | L > R | Y | Proprioception, (intact pinprick and vibration) | Sural nerve: severe loss of myelinated axons, onion bulbs |
| Brain MRI: no central demyelination. Original diagnosis Dejerine–Sottas | ||||||||||
| Wheelchair, modified car controls. | ||||||||||
| Scoliosis (Fig. 5) | ||||||||||
| A1 | M | I41T T556NfsX20 | Late | Unknown | Unknown | Unknown | Unknown | Unknown | ||
| A2 | M | I41T F254SfsX7 | Late | 54 (2008) | Y | LE > UE | L > R | N | Proprioception, light touch | Tingling and numbness in L UE and LE, high arches, hammer toe. |
| Additional diagnosis of Parkinson's disease. | ||||||||||
| Brain MRI: atrophy of frontal and parietal lobes and cerebellum | ||||||||||
| Carbidopa–Levodopa improved cognition and motor control. | ||||||||||
| A3 | M | I41T T556NfsX20 | Late | 47 (2000) | Unknown | LE > UE | L > R | Y | Vibration, pinprick, proprioception | Involuntary jerking in LEs |
| Sural nerve: reduced axon density and size, demyelination | ||||||||||
| EMG: active and chronic denervation | ||||||||||
| Original diagnosis of chronic inflammatory demyelinating polyneuropathy | ||||||||||
| One affected sibling. | ||||||||||
| A4 | F | I41T L458FfsX4 | Early | 4 (2009) | Y | LE > UE | L > R | Y | Intact light touch and pinprick | Normal cognitive development |
| EEG and brain MRI normal | ||||||||||
| Non-epileptic shaking spells and intention tremor in L UE > R UE | ||||||||||
| A6 | F | I41T E302K | Late | 46 (2009) | Y | LE, UE | R > L | Y | Vibration, proprioception | Sudden onset of symptoms |
| Tremor distal UE > LE | ||||||||||
| A7 | M | I41T deletion | Unknown | Unknown | Unknown | Unknown | Unknown | Unknown | ||
| A8 | F | I41T R381X | Early | 10 (2010) | Y | LE > UE | N | N | Intact pinprick, vibration and Proprioception | |
| A9 | – | L17P F254SfsX7 | Unknown | Unknown | Unknown | Unknown | Unknown | Unknown | ||
| C1 | – | I41T F98GfsX4 | Early | <5 | Unknown | Unknown | Unknown | Unknown | Unknown | BAB1079 (Chow et al., 2007) |
| C2 | F | I41T R183X | Late | 37 (1998) | Y | UE > LE | R > L | Y | Vibration, (intact pinprick and proprioception) | BAB1372 (Chow et al., 2007; Zhang et al., 2008) |
| Progressed to quadriplegia. Onset followed physical trauma. | ||||||||||
| Died at 47 of presumed respiratory compromise. | ||||||||||
| Brain MRI unremarkable | ||||||||||
| C3 | M | I41T R183X | Late | 35 (1998) | Y | LE > UE | L > R | Y | Vibration, (intact pinprick and proprioception) | BAB1373, sibling of BAB1372 (Chow et al., 2007; Zhang et al., 2008) |
| Progressed to wheelchair. | ||||||||||
| Sural nerve: reduced axon density and size, demyelination, onion bulbs | ||||||||||
| EMG: active and chronic denervation | ||||||||||
| C4 | – | I41T D348GfsX11 | Early | <5 | Unknown | Unknown | Unknown | Unknown | Unknown | BAB1161 (Chow et al., 2007) |
| Original diagnosis of Dejerine–Sottas Syndrome | ||||||||||
| C5 | – | I41T F254SfsX7 | Early | <5 | Unknown | Unknown | Unknown | Unknown | Unknown | BAB1369 (Chow et al., 2007) |
Each patient carries one null allele and one missense variant. Details in Supplementary material.
S = Sydney, Australia; A = Athena; C = Chow et al., 2007; Y = present; N = absent; UE = upper extremity; LE = lower extremity; R = right; L = left; > indicates greater impairment; CPAP = continuous positive airway pressure.
Figure 3.
The missense mutation E302K is a functionally null allele of FIG4. (A) Residue E302 is evolutionarily conserved in yeast, invertebrates and mammals. Dots represent identity to the amino acid in the human sequence. (B and C) The enlarged vacuole in FIG4 null yeast is rescued by the wild-type (WT) gene but not by the E302K mutant. (D) Location of the E302K mutation between the protein interaction domain and the catalytic domain of FIG4 in the FIG4 protein (courtesy of Yuxin Mao).
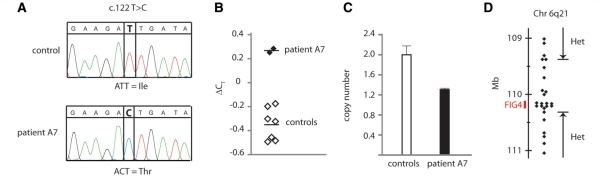
In Patient A7, the sequence chromatogram for exon 2 contains only the mutant nucleotide c.122 T>C encoding p.I41T, and lacks the reference T nucleotide observed in heterozygous patients (Fig. 4A). These data indicate that the patient is either homozygous for the I41T allele or is hemizygous due to a deletion of exon 2 on the other chromosome. The other exons from this patient did not contain a second mutation in FIG4. The low allele frequency of 0.001 for the I41T in controls (described below) suggests that homozygosity is most likely to result from recent consanguinity, which would generate blocks of homozygosity encompassing tens of megabases around the FIG4 gene at 110 Mb on chromosome 6 (www.genome.ucsc.edu, build 36). Genotyping 22 microsatellite markers between 64 and 128 Mb on chromosome 6 demonstrated that Patient A7 is heterozygous throughout this region (Supplementary Table 1), ruling out recent consanguinity. Therefore, to detect a possible deletion of exon 2, we measured the copy number of exon 2 using a quantitative Taqman polymerase chain reaction assay. Comparison with diploid I41T/+ controls demonstrated that the patient’s DNA contains only one copy of exon 2 (Fig. 4B and C). To evaluate the size of the deletion, we genotyped polymorphic single nucleotide polymorphisms with minor allele frequencies >0.3 within and near the FIG4 gene. Patient A7 is homozygous for all of the tested single nucleotide polymorphisms within the FIG4 gene (Fig. 4D). The maximum size of the deletion is 0.94 Mb, based on the location of the closest flanking heterozygous single nucleotide polymorphisms, rs11153138 and rs4245540 (Supplementary Table 1).
Figure 4.
CMT4J patient A7 is a compound heterozygote carrying the I41T mutation in combination with a deletion of FIG4 exon 2. (A) Sequencing of exon 2 from genomic DNA of Patient A7 detected only the c.122C nucleotide encoding threonine 41, and did not detect the reference T allele, indicating that the patient does not carry a wild-type allele. (B and C) Quantitative polymerase chain reaction using intronic primers flanking exon 2 demonstrates reduced copy number for exon 2 compared with diploid controls. The data indicate that the wild-type copy of exon 2 is deleted in this patient. (D) Maximal size of the deletion that includes exon 2. Genotyping polymorphic single nucleotide polymorphisms flanking FIG4 on chromosome 6 identified a region of 0.94 Mb, including the 100 kb FIG4 gene, in which none of the tested markers were heterozygous. This interval, bounded by the heterozygous single nucleotide polymorphisms rs11153138 and rs4245540, defines the maximal size of the deletion. Each symbol represents one genotyped marker. The identities and locations of the single nucleotide polymorphisms and microsatellites are presented in Supplementary Table 1 (www.genome.ucsc.edu, human genome build 38).
Heterozygous variants of FIG4
Eighty heterozygotes carrying a single variant allele were identified among the 4000 screened patients with Charcot–Marie–Tooth disease. Eighteen individuals were I41T/+ heterozygotes (0.5%), approximately twice the frequency in controls described below. Thirteen individuals were +/− heterozygotes carrying protein truncation (null) mutations (0.3%) (Supplementary Table 2). The pathogenicity of heterozygosity for null mutations of FIG4 is currently uncertain, due to the lack of complete FIG4 sequence data from sufficient numbers of controls. Thirty-six patients with Charcot–Marie–Tooth disease were heterozygous for novel missense variants of unknown functional consequence that are listed in Supplementary Table 3. Carriers of the polymorphic coding single nucleotide polymorphisms rs9885672 (V654A) and rs2295837 (M364L) were present at predicted frequencies (Supplementary Table 3). The clinical implications of heterozygosity for these FIG4 variants is not known and familial segregation studies were not possible.
Clinical features of patients with FIG4 mutations
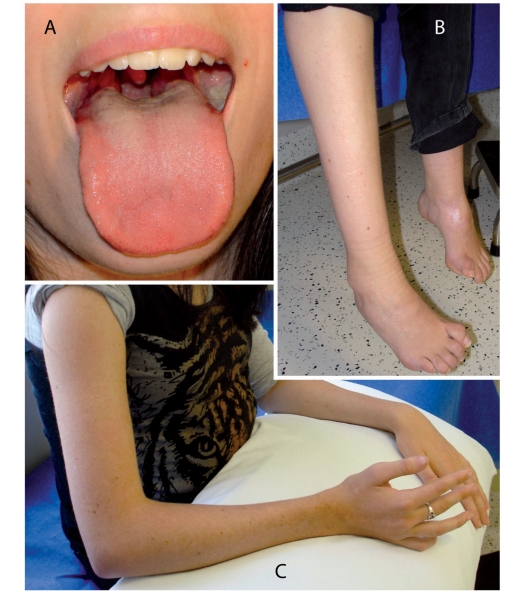
We identified 11 new patients who are compound heterozygotes for FIG4 mutations. Genotypes and clinical features are summarized in Table 1 and described in detail in the Supplementary material (clinical descriptions). Disease onset varies widely from early childhood to the sixth decade. Proximal and distal weakness was approximately equal in those cases with examination data. Signs of weakness in upper and lower extremities and the tongue are shown for Patient S3 at 19 years of age (Fig. 5). Proximal weakness in patients with CMT4J contrasts with typical Charcot–Marie–Tooth disease that is predominantly distal. Most patients had asymmetric weakness and in several cases trauma, such as a fall, appeared to trigger the initial symptoms or cause a period of rapid progression. Most patients were also areflexic. Sensory findings were mild or absent, and were uniformly less severe than motor findings (Table 1 and Supplementary material). There is evidence of cranial nerve involvement affecting right eye abduction in Patient S2 (Supplementary material) and tongue in Patient S3 (Fig. 5).
Figure 5.
Proximal and distal muscle weakness in a patient with CMT4J. At the age of 19 years, Patient S3 demonstrated several features of proximal muscle wasting. (A) Wasted tongue with ‘triple furrow’ sign and (B) foot deformity and cocked up toes. The difference in leg length follows a left femoral fracture. (C) Proximal and distal wasting of the upper limb.
Neurophysiological studies for seven patients are tabulated in Table 2. The general picture is one of severe combined axonal and demyelinating neuropathy. There is major reduction of motor nerve conduction velocity affecting both upper and lower extremities. Several cases demonstrate conduction velocities that are much slower than expected from denervation alone (Van Asseldonk et al., 2005). In several cases, nerve conduction velocity and amplitude are both affected, consistent with prior observations (Chow et al., 2007; Zhang et al., 2008). All but one patient lacked detectable response from the sural nerve, although clinical examination showed relative preservation of sensory function. The one patient with residual sural potential had intermediate median motor nerve conduction velocities of 41 and 37 m/s, indicating that the neurophysiological phenotype is not always demyelinating. Needle EMG for Patients S2 and A3 revealed features of active denervation, including fibrillation potentials, positive sharp waves and reduced recruitment patterns. Evidence of chronic denervation included polyphasic large amplitude motor unit action potentials of long duration. Sites of muscle atrophy correspond to the sites of weakness. Clinical, morphological and neurophysiological observations are consistent with severe demyelinating neuropathy.
Table 2.
Nerve conduction studies
| Patient | Age at testing (years) | Sural SNAP (µV) | Median SNAP (µV) | Median DML (ms) | Median CMAP (mV) | Median MNCV (m/s) | Peroneal CMAP (mV) | Peroneal MNCV (m/s) |
|---|---|---|---|---|---|---|---|---|
| Normal valuesa | – | ≥6 | ≥20 | ≤4.4 | ≥4 | ≥49 | ≥2 | ≥44 |
| S1 | 14 | 3 | 7 | 7 | 5.1 | 41 | 0.8 | 29 |
| 15 | Undetect. | 4 | 13 | 4.3 | 37 | 2.2 | Undetect. | |
| S2 | 7 | – | – | 14 | – | 12 | – | – |
| 19 | – | Undetect. | 18 | – | 4 | – | – | |
| 35 | – | – | 20 | 1 | 4 | – | – | |
| 39 | – | Undetect. | 19 | 0.6 | 3 | – | Undetect. | |
| 41 | Undetect. | Undetect. | 25 | 0.5 | 3 | Undetect. | Undetect. | |
| 41 | – | – | 35 | 0.2 | 3 | Undetect. | 25 | |
| S3 | 4 | Undetect. | – | 14 | 0.5 | 11 | Undetect. | Undetect. |
| 18 | – | Undetect. | 16 | 0.6 | 7 | Undetect. | Undetect. | |
| A2 | 54 | Undetect. | – | – | – | – | Undetect. | Undetect. |
| A3 | 48 | Undetect. | – | – | – | – | 0.3 | Undetect. |
| 56 | Undetect. | 6 | 17 | 1.7 | 23 | Undetect. | Undetect. | |
| A4 | 5 | Undetect. | Undetect. | 13u | 1.9u | 12u | 0.1 | 16 |
| A8 | 11 | Undetect. | 22 | 15 | 5 | 32 | 0.4 | 14 |
The median nerve sensory nerve action potential was measured in digit 2. The median distal motor latency and compound motor action potential were recorded from the abductor pollicis brevis muscle. The median motor nerve conduction velocity was measured from elbow to wrist. The ulnar nerve distal motor latency and compound motor action potential were measured below the elbow, and the ulnar motor nerve conduction velocity was measured across the elbow. The peroneal compound motor action potential was measured in the extensor digitorum brevis muscle, and the peroneal motor nerve conduction velocity was measured from the fibular head to ankle.
aNormal values from Preston and Shapiro, 2005.
CMAP = compound motor action potential; DML = distal motor latency; MNCV = motor nerve conduction velocity; undetect. = no response detected; – = not done; SNAP = sensory nerve action potential; u = Ulnar nerve.
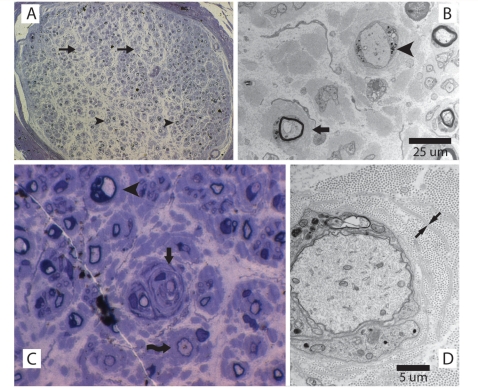
Patient S2 had early onset and progressed to wheelchair dependence over a period of 30 years (Table 1). This patient had two sural nerve biopsies >30 years apart (Fig. 6). Demyelinating features such as onion bulbs were present from the first biopsy at the age of 7 years, as was extensive loss of large diameter myelinated fibres, a feature also seen in sciatic nerve of the Fig4 null mouse (Chow et al., 2007). Progressive loss of myelinated fibres and reduction in axon number is evident from comparison of histology at 7 and 41 years of age. The ultrastructure of one demyelinated axon demonstrates the double basement membrane and surrounding onion bulb lamellae (Fig. 6C). At 41 years of age, there were few residual myelinated fibres (Fig. 6B). Patient S2 retains head and neck function and some function in his left arm. His clinical course represents the most severe end of the CMT4J spectrum, and is quite similar to patients C2 and C3 (Table 1).
Figure 6.
Abnormal sural nerve myelination in early stage disease in a patient with CMT4J. Right sural nerve biopsy at 7 years of age. A later biopsy of the left sural nerve, at the age of 41 years, revealed further loss of axons (data not shown). (A) Cross section of a sural nerve fascicle with extensive loss of large myelinated fibres. There is relative sparing of moderate to small diameter fibres, which range from thinly myelinated (arrowheads) to absent myelin (arrows). Stained with toluidine blue; ×20 magnification. (B) Electron microscopy demonstrating one naked axon with myelin breakdown products (arrowhead) and one thinly myelinated axon with surrounding onion bulb remnants and no clear Schwann cell processes (arrow). (C) Thinly myelinated axon with multiple thin myelin bands within the surrounding onion bulb (arrow). Also visible, a dysmyelinated axon (arrowhead) and a thinly myelinated axon (curving arrow). (D) Electron microscopy demonstrating a minimally myelinated axon with myelin breakdown products. The double basal lamina of a remnant Schwann cell process is visible (arrows).
Frequency of the FIG4-I41T allele in control Northern European population
To measure the population frequency of the pathogenic FIG4-I41T variant responsible for most cases of CMT4J, we genotyped the c.122T>C single nucleotide polymorphism in 5769 control samples, described in the ‘Materials and methods’ section, using three techniques. Sequencing of exon 2 from 764 individuals identified three I41T heterozygotes. Genotyping of 591 controls with a Taqman assay identified one heterozygote. Genotyping 4414 controls with a Sequenom assay identified nine heterozygotes. The overall frequency of I41T heterozygotes was 13/5769 and the calculated allele frequency was 0.001. Thus, FIG4-I41T is a moderately rare allele with frequency lower than polymorphic variants (>0.01), but considerably higher than ‘private’ or rare variants that may be restricted to a single family and have a population frequency of <1/106. The observed allele frequency of I41T can account for its presence in 14 unrelated families with CMT4J.
Complete sequence of FIG4 from unaffected controls
To completely ascertain FIG4 variants and assess the frequency of null alleles in an unaffected population, we carried out complete sequencing of the 23 exons of FIG4 in 206 control individuals from the ClinSeq collection (Biesecker et al., 2009). No deleterious alleles were identified in this relatively small population of controls. The common variants described above were present at expected allele frequencies of 0.05 for M364L (rs2295837) and 0.2 for V654A (rs9885672). One control was heterozygous for the I41T allele. The missense variants M781T, T873A and I902L were each detected in a single individual. No protein truncation mutations were observed, suggesting that their frequency in the general population is low.
Discussion
Our findings expand the clinical and genetic spectrum of patients with FIG4-related neuropathy and indicate that CMT4J is a clinically distinct form of Charcot–Marie–Tooth disease. The genotypes and clinical findings for the 11 previously unreported patients are summarized in Table 1. At the bottom of Table 1 we have added the known features of the five previously reported patients (C1 to C5) to provide a complete view. The unbiased screen described here indicates that the frequency of CMT4J among patients presenting with Charcot–Marie–Tooth disease is ∼8/4000 (0.2%). This is equivalent to ∼20% of recessive Charcot–Marie–Tooth disease and 0.4% of all Charcot–Marie–Tooth disease.
Early-onset CMT4J
The clinical features of the four new early-onset patients (S1, S2, S3 and A4) expands the brief descriptions of three previously published cases (Chow et al., 2007). FIG4 mutations were found in three of the six Australian patients selected for childhood onset with severe progressive proximal and distal weakness, demonstrating that CMT4J is greatly enriched in cases with these features. A key factor for inclusion in the study was proximal weakness, which implies risk of subsequent wheelchair dependence that is rarely required even with marked distal weakness. The Australian patients were negative for mutations in MFN2, MPZ, PMP22 or GAN1, which can also cause severe Charcot–Marie–Tooth disease, and for mutations in GDAP1 and MTMR2.
Patients with early onset lacked symptoms in infancy but exhibited gait abnormalities from the time of walking, with asymmetric involvement of the extremities. No cases of congenital or infantile onset CMT4J have yet been demonstrated. Nerve conduction velocities were within the classical demyelination range for Patients S1, S2 and A4, consistent with sural nerve histology. Conduction amplitudes were consistent with additional significant axonal loss. In contrast to the length-dependent weakness and wasting typical of Charcot–Marie–Tooth disease, the early proximal weakness accompanied by preservation of some distal motor units is distinctive and reflects the amyotrophic denervation in this condition. Needle EMG in Patient S2 provided evidence for chronic, active denervation in the extremities, but the motor nerve conduction velocity in Patients S3 and A4 was much slower than would be expected from denervation alone and is suggestive of significant demyelination. This is in agreement with the results of sural nerve biopsies for four patients (Fig. 6 and Table 1). The combination of generalized demyelinating neuropathy and proximal amyotrophic features emphasized here is highly unusual.
In the earliest stages, minimal sensory abnormalities were present, but patients developed reduced vibration and light touch sensation distally that progressed to total loss of sharp sensation and reduced joint position sense. Sensory nerve action potentials are reduced, becoming unobtainable with time. In summary, early-onset CMT4J affects sensory and motor nerves, proximal and distal muscles, lower and upper extremities and axons and Schwann cells.
Late-onset CMT4J
Screening of unselected patients with Charcot–Marie–Tooth disease revealed a broad range of severity, including one patient in whom the clinical features of Charcot–Marie–Tooth disease were minimally symptomatic. Signal features of late-onset cases include asymmetric presentation, a degree of proximal weakness that is unusual for Charcot–Marie–Tooth disease, and progression to severe motor dysfunction resembling amyotrophic lateral sclerosis in some cases. Physical examination, neurophysiology, needle EMG and peripheral nerve biopsy results were similar to those in the patients with early onset.
Age of onset does not differ between males and females and is not correlated with the position of protein truncation in the null allele (Table 2). Patients A2 and C5 have identical FIG4 genotypes, with a stop codon at residue 261 in the null allele, yet onset was early in C5 and late in A2. An important contribution of genetic background to clinical course is suggested by the siblings C2 and C3, one male and one female, both with onset at 32 years of age and progression within a few years to wheelchair dependence (Zhang et al., 2008). These siblings may share genetic variants at other loci that modify the course of CMT4J. Recent data from human exome sequencing has revealed an unexpectedly large amount of genetic variation in the human population, with each individual carrying 200–400 rare amino acid substitutions (Durbin et al., 2010). It is known from the study of mouse mutants that mutations in other genes affecting PI(3,5)P2 metabolism result in neurodegeneration similar to Fig4 mutants (Bolino et al., 2000; Zhang et al., 2007; Jin et al., 2008). Strain background can influence age of onset in the Fig4 null mouse (our unpublished data). It will be worthwhile to compare the sequences of these and other predicted modifier genes in patients with early- and late-onset CMT4J.
Relationship to Dejerine–Sottas syndrome and chronic inflammatory demyelinating polyneuropathy
Dejerine–Sottas syndrome (OMIM 14 590) is a severe childhood motor and sensory demyelinating neuropathy with elevation of protein in CSF. Three patients with CMT4J were originally diagnosed with Dejerine–Sottas (Table 1), demonstrating clinical overlap. Testing for FIG4 mutations may be indicated in patients with this diagnosis. Four other genes have been implicated in both Dejerine–Sottas and Charcot–Marie–Tooth disease (www.ncbi.nlm.nih.gov/omim/145900).
Late-onset Patient A3 was first diagnosed with chronic inflammatory demyelinating polyneuropathy. Treatment with intravenous immunoglobulin did not improve symptoms in Patients A3 or S2. Sural nerve biopsies from patients with CMT4J lacked inflammatory infiltrates characteristic of chronic inflammatory demyelinating polyneuropathy. Patient S2 has apparently benefitted from oral corticosteroids, which can have transcriptional and psychological effects in addition to their anti-inflammatory and immunosuppressive properties.
The I41T mutant allele of FIG4
Fifteen of the 16 known patients with CMT4J are compound heterozygotes carrying the missense allele I41T in combination with a null allele of FIG4 (Table 1). The I41T mutation is inherited on the same haplotype in all the patients who have been tested, demonstrating segregation of an ancestral mutation. I41T has reached an allele frequency of 0.001 in the Northern European population based on the observations of 13 heterozygotes among 5769 control individuals. This moderately high frequency can account for the presence of the same allele in multiple unrelated families. It is not surprising that FIG4-I41T homozygotes have not been observed, since their predicted frequency is one per million. Using a mouse model of CMT4J, we demonstrated that the FIG4-I41T protein can support normal function if present in sufficiently high abundance, which can be generated by 5-fold overexpression of the messenger RNA (Lenk et al., 2011). In the same study, expression at twice the normal level was insufficient to prevent disease. Since homozygous FIG4I41T/I41T patients would express at a level equivalent to 1-fold, the mouse studies indicate that they are likely to be severely affected.
A small number of patients with Charcot–Marie–Tooth disease (18/4000, 0.45%) were heterozygous carriers of the I41T allele (I41T/+). Some of these patients might carry an undetected deletion or intronic mutation that was missed by our exonic sequencing strategy. If so, the frequency of CMT4J could approach 1% of all cases of Charcot–Marie–Tooth disease. The frequency of I41T/+ heterozygotes in the control population, 13/5769 (0.2%), and predicts that eight heterozygotes are expected by chance in the patient population of 4000. We consider it unlikely that heterozygosity for I41T is a major risk for Charcot–Marie–Tooth disease.
Null alleles of FIG4
Thirteen patients with Charcot–Marie–Tooth disease (13/4000, 0.3%) were heterozygous carriers of null alleles of FIG4. FIG4+/− heterozygosity was previously observed in 3/600 (0.5%) of patients with amyotrophic lateral sclerosis (Chow et al., 2009). The clinical significance of the FIG4+/− genotype remains uncertain, because the frequency of null alleles in control populations is unknown. Complete sequencing of 206 controls did not detect any null alleles, but sequencing of several thousand controls will be needed to determine whether null heterozygotes have an increased rate of disease. In the mouse, Fig4+/− heterozygotes are unaffected up to 2.5 years of age (Lenk et al., 2011). Homozygous null mice do not survive beyond 2 months of age, indicating that this would be a lethal genotype.
The complex indel in Patient S1 could have arisen by the fork stalling, template switching mutation mechanism (FOSTES) (Lee et al., 2007). We also observed three recurrent frameshift mutations caused by single nucleotide indels within homopolymer runs of 3–5 nt (Supplementary Table 2). The known susceptibility of homopolymers to frameshift mutation (Denver et al., 2004) is consistent with independent re-mutation of these sites. CpG demethylation of arginine codons is another source of recurrent mutations (Kearney et al., 2006) and could account for the premature stop codon R183X found in two unrelated patients (Supplementary Table 2).
Overall, 1% of the 4000 patients with Charcot–Marie–Tooth disease whose FIG4 exons were sequenced are heterozygous carriers of rare FIG4 variants not found in the single nucleotide polymorphism database. It is possible that some of these FIG4 variants might act as genetic modifiers contributing to disease severity in patients with primary mutations in other Charcot–Marie–Tooth disease genes. However, the observed 1% frequency of FIG4 variants in patients is not substantially higher than might be expected in the population at large. Recent results from genome sequencing indicate that each human genome carries between 200 and 400 rare variants affecting protein sequence (Ng et al., 2009; Durbin et al., 2010; Roach et al., 2010). Assessing the role of rare variants in human disease will require measurements of variant frequencies in large populations combined with functional assessment in biological assays (Davis et al., 2011).
Our findings indicate that CMT4J is a rare, clinically distinct form of Charcot–Marie–Tooth disease. Patients with sporadic or recessive inheritance of severe progressive distal and proximal weakness should be tested for FIG4 mutations, beginning with genotyping the I41T variant that is present in 15/16 known cases. FIG4 mutations are enriched among patients with early-onset disease with proximal weakness. Demyelination can be severe, with marked sensory axonal loss and an amyotrophic pattern of motor axonal loss. Asymmetry and deterioration after minor injury may provide additional indications. Clinical manifestation ranges from early to adult onset and from mild impairment to fatal outcome. It is striking that overexpression of the causal FIG4-I41T mutant can rescue neurodegeneration in a mouse model (Lenk et al., 2011). Development of methods for upregulation of the I41T allele in patients with CMT4J could lead to treatment for this rare but potentially devastating disorder.
Funding
National Institutes of Health research (grant R01 GM24872) (to M.H.M.); the Charcot–Marie–Tooth disease Association of Australia (to G.N.); the NHMRC of Australia (Grant number APP1007705 to G.N.); MDA USA (grant number 158509 to G.N.); Hartwell Foundation (to G.M.L., postdoctoral fellowship); Medical Scientist Training Program at the University of Michigan (NIH T32 GM07863 to C.J.F.). The National Cell Repository for Alzheimer's Disease (NCRAD) receives government support under a cooperative agreement grant (U24 AG21886) awarded by the National Institute on Aging (NIA).
Supplementary material
Supplementary material is available from Brain online.
Acknowledgements
We thank Dr Min-Xia Wang, Dr Sue Bremmer and Prof. John Pollard for sural nerve histology and imaging. We are grateful to Prof. Robert Ouvrier for clinical information and assistance. We acknowledge Dr Yuxin Mao (Cornell University) for the depiction of protein structure in Fig. 3D, and Dr E. A. Otto (University of Michigan) for genotyping of chromosome 6 microsatellites in Patient A7 (Supplementary Table 1). We thank Eric Klein for assistance with large-scale genotyping of the FIG4-I41T mutation in control samples. We are grateful to Drs Anthony Antonellis, William Dauer, James Dowling and Andrew Lieberman for critical reading of the manuscript.
For the population screen of I41T frequency, DNA from normal controls was obtained from the following sources. We thank contributors, including the Alzheimer's Disease Centres who collected samples used in this study, as well as patients and their families, whose help and participation made this work possible. The National Institute of Mental Health Schizophrenia Genetics Initiative (NIMH-GI) data and biomaterials were collected by the Molecular Genetics of Schizophrenia II (MGS-2) collaboration. The investigators and co-investigators are: ENH/Northwestern University, Evanston, IL, MH059571, Pablo V. Gejman, M.D. (Collaboration Coordinator; PI), Alan R. Sanders, MD; Emory University School of Medicine, Atlanta, GA,MH59587, Farooq Amin, MD (PI); Louisiana State University Health Sciences Centre; New Orleans, Louisiana, MH067257, Nancy Buccola APRN, BC, MSN (PI); University of California-Irvine, Irvine, CA,MH60870, William Byerley, MD (PI); Washington University, St Louis, MO, U01, MH060879, C. Robert Cloninger, MD (PI); University of Iowa, Iowa, IA,MH59566, Raymond Crowe, MD (PI), Donald Black, MD; University of Colorado, Denver, CO, MH059565, Robert Freedman, MD (PI); University of Pennsylvania, Philadelphia, PA, MH061675, Douglas Levinson MD (PI); University of Queensland, Queensland, Australia, MH059588, Bryan Mowry, MD (PI); Mt. Sinai School of Medicine, New York, NY,MH59586, Jeremy Silverman, PhD (PI). In addition, cord blood samples were collected by V L Nimgaonkar's group at the University of Pittsburgh, as part of a multi-institutional collaborative research project with J Smoller, MD DSc and P Sklar, MD PhD (Massachusetts General Hospital) (grant MH 63420).
Glossary
Abbreviations
- CMT4J
Charcot–Marie–Tooth disease type 4J
References
- Barisic N, Claeys KG, Sirotkovic-Skerlev M, Lofgren A, Nelis E, De Jonghe P, et al. Charcot-Marie-Tooth disease: a clinico-genetic confrontation. Ann Hum Genet. 2008;72:416–41. doi: 10.1111/j.1469-1809.2007.00412.x. [DOI] [PubMed] [Google Scholar]
- Biesecker LG, Mullikin JC, Facio FM, Turner C, Cherukuri PF, Blakesley RW, et al. The ClinSeq Project: piloting large-scale genome sequencing for research in genomic medicine. Genome Res. 2009;19:1665–74. doi: 10.1101/gr.092841.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chow CY, Landers JE, Bergren SK, Sapp PC, Grant AE, Jones JM, et al. Deleterious variants of FIG4, a phosphoinositide phosphatase, in patients with ALS. Am J Hum Genet. 2009;84:85–88. doi: 10.1016/j.ajhg.2008.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chow CY, Zhang Y, Dowling JJ, Jin N, Adamska M, Shiga K, et al. Mutation of FIG4 causes neurodegeneration in the pale tremor mouse and patients with CMT4J. Nature. 2007;448:68–72. doi: 10.1038/nature05876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis EE, Zhang Q, Liu Q, Diplas BH, Davey LM, Hartley J, et al. TTC21B contributes both causal and modifying alleles across the ciliopathy spectrum. Nat Genet. 2011;43:189–96. doi: 10.1038/ng.756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denver DR, Morris K, Kewalramani A, Harris KE, Chow A, Estes S, et al. Abundance, distribution, and mutation rates of homopolymeric nucleotide runs in the genome of Caenorhabditis elegans. J Mol Evol. 2004;58:584–95. doi: 10.1007/s00239-004-2580-4. [DOI] [PubMed] [Google Scholar]
- Durbin RM, Abecasis GR, Altshuler DL, Auton A, Brooks LD, Gibbs RA, et al. A map of human genome variation from population-scale sequencing. Nature. 2010;467:1061–73. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferguson CJ, Lenk GM, Meisler MH. Defective autophagy in neurons and astrocytes from mice deficient in PI(3,5)P2. Hum Mol Genet. 2009;18:4868–78. doi: 10.1093/hmg/ddp460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikonomov OC, Sbrissa D, Fligger J, Delvecchio K, Shisheva A. ArPIKfyve regulates Sac3 protein abundance and turnover: disruption of the mechanism by Sac3I41T mutation causing Charcot-Marie-Tooth 4J disorder. J Biol Chem. 2010;285:26760–4. doi: 10.1074/jbc.C110.154658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin N, Chow CY, Liu L, Zolov SN, Bronson R, Davisson M, et al. VAC14 nucleates a protein complex essential for the acute interconversion of PI3P and PI(3,5)P(2) in yeast and mouse. EMBO J. 2008;27:3221–34. doi: 10.1038/emboj.2008.248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kearney JA, Wiste AK, Stephani U, Trudeau MM, Siegel A, RamachandranNair R, et al. Recurrent de novo mutations of SCN1A in severe myoclonic epilepsy of infancy. Pediatr Neurol. 2006;34:116–20. doi: 10.1016/j.pediatrneurol.2005.07.009. [DOI] [PubMed] [Google Scholar]
- Lee JA, Carvalho CM, Lupski JR. A DNA replication mechanism for generating nonrecurrent rearrangements associated with genomic disorders. Cell. 2007;131:1235–47. doi: 10.1016/j.cell.2007.11.037. [DOI] [PubMed] [Google Scholar]
- Lenk GM, Ferguson CJ, Chow CY, Jin N, Jones JM, Grant AE, et al. Rescue of neurodegeneration by transgenic expression of a pathogenic variant of FIG4 responsible for Charcot-Marie-Tooth Disease. PLOS Genetics. 2011;7:e1002104. doi: 10.1371/journal.pgen.1002104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manford A, Xia T, Saxena AK, Stefan C, Hu F, Emr SD, et al. Crystal structure of the yeast Sac1: implications for its phosphoinositide phosphatase function. EMBO J. 2010;29:1489–98. doi: 10.1038/emboj.2010.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng SB, Turner EH, Robertson PD, Flygare SD, Bigham AW, Lee C, et al. Targeted capture and massively parallel sequencing of 12 human exomes. Nature. 2009;461:272–6. doi: 10.1038/nature08250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preston DC, Shapiro BE. Electromyography and neuromuscular disorders. 2nd edn. Philadelphia PA: Elsevier; 2005. [Google Scholar]
- Roach JC, Glusman G, Smit AF, Huff CD, Hubley R, Shannon PT, et al. Analysis of genetic inheritance in a family quartet by whole-genome sequencing. Science. 2010;328:636–9. doi: 10.1126/science.1186802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saporta AS, Sottile SL, Miller LJ, Feely SM, Siskind CE, Shy ME. Charcot-marie-tooth disease subtypes and genetic testing strategies. Ann Neurol. 2011;69:22–33. doi: 10.1002/ana.22166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Asseldonk JT, Van den Berg LH, Kalmijn S, Wokke JH, Franssen H. Criteria for demyelination based on the maximum slowing due to axonal degeneration, determined after warming in water at 37 degrees C: diagnostic yield in chronic inflammatory demyelinating polyneuropathy. Brain. 2005;128:880–91. doi: 10.1093/brain/awh375. [DOI] [PubMed] [Google Scholar]
- Zhang X, Chow CY, Sahenk Z, Shy ME, Meisler MH, Li J. Mutation of FIG4 causes a rapidly progressive, asymmetric neuronal degeneration. Brain. 2008;131:1990–2001. doi: 10.1093/brain/awn114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Zolov SN, Chow CY, Slutsky SG, Richardson SC, Piper RC, et al. Loss of Vac14, a regulator of the signaling lipid phosphatidylinositol 3,5-bisphosphate, results in neurodegeneration in mice. Proc Natl Acad Sci USA. 2007;104:17518–23. doi: 10.1073/pnas.0702275104. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.