Abstract
Recent studies have shown that ampicillin resistance has increased steadily over the past 3 decades within U.S. Enterococcus faecium isolates. Analysis of the predicted PBP5 protein of 41 isolates showed a consensus PBP5 pattern for the 9 isolates with MICs of <4 μg/ml that is distinctly different from the PBP5 consensus of the 32 isolates with MICs of >4 μg/ml with ∼5% difference between these; however, there were no consistent amino acid changes that correlated with specific increases in the MICs of ampicillin within the latter group. Analysis of three other genes encoding cell wall/surface proteins also showed that there are two distinct evolutionary groups for each gene, but with occasional mixing of genes, consistent with a species that evolves by recombination.
INTRODUCTION
Decreased ampicillin susceptibility in Enterococcus faecium is due to the expression of the low-affinity class B penicillin-binding protein 5 (PBP5fm) (7, 28). PBP5fm has been implicated as being responsible for both intrinsic low-level ampicillin resistance and higher-level resistance and is also responsible for cell wall synthesis function when the other PBPs are saturated by ampicillin (7, 17, 24, 28). Previous studies have shown that clinical strains with intermediate levels of ampicillin resistance overproduce PBP5; however, strains that exhibit much higher MICs, such as H80721 and C68 (MICs of 256 μg/ml), contain a very low affinity PBP5 (at least 10-times-lower affinity) which is not overproduced (6, 12, 22, 30). In addition, it has been shown that the low-affinity pbp5 gene of strain C68 (and some other clinical strains) can be transferred horizontally, suggesting circulation of a low-affinity pbp5 among clinical isolates (21). Many of the changes found in the pbp5 gene of highly resistant clinical isolates have been proposed to lower the affinity for β-lactam antibiotics, resulting in higher MICs, although those implicated have varied among different studies (11, 13, 16, 18, 19, 22, 30). A specific impact of an individual or multiple pbp5 mutations has been hard to determine in these studies, due to the fact that they are mostly correlation studies in nonisogenic clinical isolates. However, a study using pbp5 cloned onto a shuttle plasmid in a pbp5 deletion mutant showed that specific substitutions seen in the previous studies conferred modest levels of resistance when present as single point mutations and that resistance was increased when some of these mutations were present in combination (19).
The progression of resistance of E. faecium to ampicillin in the United States was originally reported in a 22-year study (1968 to 1990) using isolates from a single Boston hospital; Grayson et al. suggested an alteration in PBP5 as a possible mechanism for increased resistance (9). In a recent publication by Galloway-Peña et al. (8), we found that ampicillin MICs of clinical E. faecium isolates from around the United States have also increased steadily over the past 3 decades. Using multilocus sequence typing (MLST), these isolates were usually ST17, a single-locus variant, or a double-locus variant of ST17; i.e., most of them are part of the hospital-associated CC17 genogroup. This genetic lineage is responsible for the worldwide emergence of multiresistant E. faecium (14, 25, 27). The goal of the current study was to determine if specific amino acid variations could be correlated with increasing MICs displayed by these strains.
MATERIALS AND METHODS
Bacterial isolates and pbp5 gene sequencing.
Twenty-nine isolates from our previous study (8) were chosen to represent each major outbreak, ampicillin MIC, and sequence type by MLST; these isolates were collected between 1971 and 1993. The complete pbp5 gene (GenBank accession no. ZP_00603984) of the 29 isolates was sequenced using the primers found in Table 1. Many of these isolates were known to be in the CC17 genogroup. Twelve additional strains isolated between 1985 and 2006 were studied because they have sequenced genomes or a sequenced pbp5 region (http://www.ncbi.nlm.nih.gov/genome and http://www.broadinstitute.org/annotation/genome/enterococcus_faecalis/GenomesIndex.html).
Table 1.
Primers used in this study to sequence the complete pbp5 gene of Enterococcus faecium
| Primer name | Primer sequence (5′–3′) | Amplicon size (nucleotides) |
|---|---|---|
| Efmpbp5-1outsideF | GGAATGACAAGCAAGAGAAGGAGG | 522 |
| Efmpbp5-1F | ATGAAAAGAAGTGACAAGCACGGC | 493 |
| Efmpbp5-1R | CTTACTTTGTCATTTCCTTC | |
| Efmpbp5-2F | GAAGGAAATGACAAAGTAAG | 466 |
| Efmpbp5-2R | GTCCCACGAAGATCCTTATCAAAAGCC | |
| Efmpbp5-3F | GGCTTTTGATAAGGATCTTCGTGGGAC | 563 |
| Efmpbp5-3R | CCCATTTTCAACGTTTCTTGTGCC | |
| Efmpbp5-4F | GGCACAAGAAACGTTGAAAATGGG | 584 |
| Efmpbp5-4R | TTATTGATAATTTTGGTTGAGGTATTG | |
| Efmpbp5-4outsideR | CGCCACAGTCCTTTTACTGTAC | 619 |
| Rpbp5_1F | GCAAAGATGAATACCTCATTAGG | 357 |
| Rpbp5_1R | CAAAGTAATCGGGTTGTACCCAGC | |
| Rpbp5_2F | CAGAACTTCCAGCTGGAGCTAC | 413 |
| Rpbp5_2R | GATCATAGCTTGGAGAGCTAGC | |
| Rpbp5_3F | GCGACAGGTTATGCTCCTGG | 423 |
| Rpbp5_3R | GAATACATTGCTGCTTGCTGGATAGG |
MICs.
Ampicillin MICs were determined using the agar dilution method according to CLSI guidelines (5). The MICs for previously published strains were found to be the same (8).
MLST.
PCR and MLST were performed as described previously (10). Fragments of 7 housekeeping genes (atpA, ddl, gdh, purk, gyd, pstS, and adk) were sequenced, allelic profiles were obtained, and a sequence type (ST) was designated for each unique allelic profile based on the MLST website (http://efaecium.mlst.net).
Other gene comparisons and phylogenetic studies.
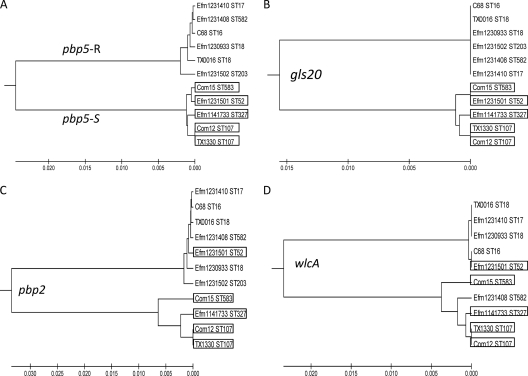
Three other genes were studied to determine if the variability found in pbp5 was comparable to the variability of other genes that encode putative surface proteins within the same genomes and if the isolates with these genes grouped the same with these genes as with pbp5. The additional genes analyzed were pbp2 (GenBank accession no. ZP_03980638), gls20 (GenBank accession no. ZP_03981586) (4), and wlcA (GenBank accession no. ZP_03980840) (a gene that we are currently studying which is related to the Enterococcus faecalis WxL genes [1, 2]). Using the sequence from E. faecium strain TX1330, we blasted the accessible genomes of E. faecium isolated in the United States available on the Broad Institute or NCBI website for each gene. Each of the genes was aligned using ClustalW, and the percent difference between the nucleotide and amino acid sequences was analyzed. The nucleotide sequence for each gene was then analyzed using MEGA 4.0.2 software, and phylogenetic UPGMA (unweighted-pair group method using average linkages) trees were constructed for each gene (Fig. 1 and 2). Only strains with sequenced genomes isolated in the United States were used, as this began as a study to compare the sequenced U.S. isolates. In addition, we also did not include any sequences of genes that were ambiguous, that had poor sequence quality, or were incomplete.
Fig. 1.
PBP5 phylogeny coincides with MIC distribution. An UPGMA tree of PBP5 was constructed using the amino acid sequences from all strains analyzed in this study. PBP5 splits isolates into two groups using the amino acid sequence, and these two groups coincide with the MIC distribution. Isolates with an ampicillin MIC of <4 μg/ml all group together in the lower branch of the tree (pbp5-S group) (except for TX2050, which has an MIC of 4 μg/ml), while all isolates with an MIC of >4 μg/ml group together in the top branch of the tree (pbp5-R group). The MIC in micrograms per milliliter and the multilocus sequence type are listed next to the strain name. STND designates sequence typing that was not done.
Fig. 2.
UPGMA trees of four E. faecium genes show that isolates generally fall into two distinct lineages. UPGMA trees of pbp5 (A), gls20 (B), pbp2 (C), and wlcA (D) are depicted using the sequenced U.S. isolates and available on the Broad Institute or NCBI website. In order to determine if other genes showed the same two groupings as pbp5 (designated pbp5-S and pbp5-R) (A), UPGMA trees of gls20 (B), pbp2 (C), and wlcA (D) were made. The boxes indicate isolates that share the ampicillin pbp5-S (MIC of <4 μg/ml) consensus. The sequence for wlcA of Efm 1231502 was unavailable.
RESULTS AND DISCUSSION
Up to 98 single nucleotide polymorphisms (see Table S1 in the supplemental material), up to 73 silent polymorphisms (see Table S2), and up to 20 amino acid variations (Table 2 ) were found when comparing isolates with an ampicillin MIC of <4 μg/ml and those with an MIC of ampicillin of >4 μg/ml, resulting in an approximately 5% nucleotide difference. All strains with MICs of <4 μg/ml (pbp5-S group) had almost identical amino acid profiles, while all strains with MICs of >4 μg/ml (pbp5-R group) had amino acid profiles which were different from the first group and which were almost identical to each other and to the transferable low-affinity PBP5 found in the E. faecium strain C68 (∼99 to 100% sequence identity) (3, 20). The consensus sequence (Table 2) of the amino acids that differed between the two groups was derived for the strains with an MIC of <4 μg/ml and those with an MIC of >4 μg/ml and is consistent with the sequences previously reported (16, 20, 22).
Table 2.
Amino acid changes between ampicillin-susceptible and -resistant Enterococcus faecium strains from the United States
| Ampicillin MIC, μg/ml (no. of isolates) | Strain | Multilocus sequence type | Amino acid at position: |
||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 24 | 27 | 34 | 66 | 68 | 85 | 100 | 144 | 172 | 177 | 204 | 216 | 324 | 466′ | 485 | 496 | 499 | 525 | 586 | 629 | 667 | |||
| MICs of ≤4 μg/ml | |||||||||||||||||||||||
| Consensus sequence | V | S | R | G | A | E | E | K | T | L | D | A | T | —d | M | N | A | E | V | E | P | ||
| 0.5–1 (3) | 1231501 | 52 | |||||||||||||||||||||
| Com15 | 583 | ||||||||||||||||||||||
| Com12 | 107 | A | |||||||||||||||||||||
| 2 (1) | TX1401a | NDb | L | V | S | ||||||||||||||||||
| 1141733 | 327 | S | |||||||||||||||||||||
| 4 (4) | TX2050 | 296 | V | ||||||||||||||||||||
| TX2042a | 19c | A | G | Q | E | Q | Q | A | I | S | A | K | T | D | V | ||||||||
| TX2043a | 25 | A | G | Q | E | Q | Q | A | I | S | A | K | T | D | |||||||||
| D366a | 25 | A | G | Q | E | Q | Q | A | I | G | S | A | K | I | D | ||||||||
| MICs of >4 μg/ml | |||||||||||||||||||||||
| Consensus sequence | A | G | Q | E | T | D | Q | Q | A | I | G | S | A | —d | T | K | T | D | L | V | S | ||
| 8 (2) | TX1399 | 475c | |||||||||||||||||||||
| TX2053 | 112 | V | |||||||||||||||||||||
| 16 (5) | TX0016 | 18c | M | V | E | ||||||||||||||||||
| TX2058 | 476 | D | A | ||||||||||||||||||||
| TX2062 | 92 | D | A | ||||||||||||||||||||
| TX2063 | 280c | A | |||||||||||||||||||||
| TX2008 | 10 | S | V | ||||||||||||||||||||
| 32 (5) | TX2061 | 92 | D | A | |||||||||||||||||||
| TX2029 | 17c | ||||||||||||||||||||||
| TX2067 | 17c | ||||||||||||||||||||||
| TX2016 | 18c | ||||||||||||||||||||||
| TX1371 | 17c | ||||||||||||||||||||||
| 64 (9) | TX2033 | 17c | A | ||||||||||||||||||||
| TX2034 | 17c | ||||||||||||||||||||||
| TX2038 | 17c | ||||||||||||||||||||||
| TX2024 | 16c | ||||||||||||||||||||||
| TX2025 | 16c | ||||||||||||||||||||||
| TX2027 | 17c | ||||||||||||||||||||||
| TX2068 | 17c | A | |||||||||||||||||||||
| TX2069 | 17c | A | |||||||||||||||||||||
| TX1405 | 17c | L | |||||||||||||||||||||
| 128 (7) | 1230933 | 18c | A | ||||||||||||||||||||
| 1231408 | 582c | S | A | V | |||||||||||||||||||
| 1231410 | 17c | S | A | V | |||||||||||||||||||
| TX2436 | 16c | A | |||||||||||||||||||||
| TX2416 | 20c | L | |||||||||||||||||||||
| TX2420 | 17c | ||||||||||||||||||||||
| TX2022 | ND | ||||||||||||||||||||||
| 256 (4) | 1231502 | 203c | S | A | V | ||||||||||||||||||
| TX2070 | 17c | D | A | ||||||||||||||||||||
| C68 | 16c | S | A | V | |||||||||||||||||||
| H80721 | ND | S | A | V | |||||||||||||||||||
“Hybrid-type” pbp5 gene.
ND, the multilocus sequence type was not determined.
ST17, single-locus variant of ST17, or double-locus variant of ST17 and therefore considered to be in the CC17 genogroup.
—, absence of 466 in the consensus sequence.
By analyzing the silent polymorphism sites found when comparing pbp5-S and pbp5-R strains, we found that pbp5-R strains use the same codon at all silent polymorphism sites whereas the pbp5-S and hybrid strains may use two different codons for an amino acid at a particular position (18 out of 73 silent polymorphisms) (see Tables S1 and S2 in the supplemental material).
Interestingly, some strains (e.g., TX2042) with an MIC of 4 μg/ml had a “hybrid-like” PBP5 (indicated in Table 2) with some of the amino acids matching the consensus of the more susceptible strains, while most of the protein sequence matched the consensus of the resistant strains. One strain with an MIC of 2 μg/ml (TX1401) had the consensus sequence of susceptible strains, except for the last three consensus sequence amino acids, 586, 629, and 667, which coincided with the consensus of the resistant strains. TX1401 was the only strain predicted to have derived its sequence via homologous recombination using the maximum chi-square test with putative recombination sites at bases 1888 (P = 0.009) and 1587 (P = 0.016) with approximately the first three-fourths of the sequence resembling the pbp5-S strains and the last part resembling pbp5-R strains.
Amino acid changes near the active site predicted in previous publications (11, 13, 16, 18, 19, 22, 24, 30) that were thought to be the cause of high-level resistance include an additional serine after amino acid position 466 (Ser-466′) and Met-485-Ala/Thr. Ser-466′ was found in three of the four most highly resistant isolates (MIC of ampicillin, 256 μg/ml) and two isolates with an ampicillin MIC of 128 μg/ml but also in one low-level-resistant strain (e.g., TX2008) with an MIC of 16 μg/ml, indicating that there is not an absolute association between these changes and high-level resistance. Interestingly, some strains (both low-level and high-level ampicillin resistant) also had an insertion of an aspartic acid residue instead of a serine at position 466′ (e.g., TX2058 and TX2070), which is located in the loop that forms the outer edge of the active site. Both residues are polar, so they may have the same effect, although aspartic acid is negatively charged, while serine is neutral. It was theorized by Sauvage et al. that the insertion at residue 466′ may displace the Val-465 in the active site, reducing accessibility for penicillin, affecting substrate specificity (23). Although these variations were found only in resistant isolates, whether there was a serine or aspartic acid at this position did not seem to correlate with specific increases in MICs. However, the presence of a Ser-466′ or Asp-466′ in combination with Ala-485 was found in all four isolates with an MIC of 256 μg/ml, and therefore, while it may not be sufficient, it may be required to achieve this high level of ampicillin resistance. It is thought that removal of the bulky methionine at position 485 would allow for conformational freedom near the Ser-480-Asp-Asn active site, resulting in a less efficient acylation process (23). Although there are a number of amino acid differences between pbp5-S and pbp5-R strains (as well as silent polymorphisms) in the membrane anchor (positions 1 to 45), N-terminal extension (positions 46 to 162), and N-terminal domain (positions 163 to 345), these are unlikely to be of great significance since the transpeptidase domain is at the C terminus (positions 346 to 680), although we cannot rule out this possibility. Of note, however, all of the amino acid differences among the resistant strains do occur in the transpeptidase domain.
Outside of Ser/Asp-466′ and Ala-485, no single amino acid variation, or combination thereof, correlated with specific ampicillin MICs or the progressive increase in MICs seen in U.S. outbreak isolates with an ampicillin MIC of >4 μg/ml. Table 2 groups the isolates by MICs and shows how each isolate's PBP5 differs from the consensus sequence. Interestingly, strain background did not seem to correlate with the differences seen in the pbp5 sequence, as multiple variations were found within the same ST, and the same sequence variation was found in multiple STs. However, the more resistant strains are of STs found in the hospital-associated CC17 genogroup, as expected (8) (CC17 genogroup isolates are footnoted in Table 2).
The finding that most E. faecium strains with an MIC of >4 μg/ml had a primary sequence highly divergent from the sequences of those that had an MIC of <4 μg/ml is similar to what has been found in Streptococcus pneumoniae, except that the pbp1, pbp2b, and pbp2x sequences in resistant clinical isolates revealed mosaic genes, with blocks of sequences that differed up to 23% (29). To determine if the 5% variation and division of isolates into two groups (Fig. 1) were restricted to pbp5, or if this was a difference that could be found on a larger genomic scale, we looked at three other genes, including pbp2, encoding proteins thought to be on the cell surface and, thus, possibly more likely to change compared to more conserved housekeeping genes. The sequences of gls20, pbp2, and wlcA from the 11 sequenced U.S. isolates available on the Broad Institute and NCBI websites were analyzed (Fig. 2). The isolates also fell into two distinct lineages based on each of the other three genes, with up to a 3% nucleotide difference for gls20, up to a 7% difference for pbp2, and up to a 10% difference for wlcA between the two groups. These two lineages generally corresponded with the pbp5-S and pbp5-R groups, which also corresponded to a CC17 genogroup (ST17, plus single-locus variants, and/or double-locus variants of it) and a non-CC17 genogroup. While most of the time the isolates in the pbp5-S and pbp5-R groups also grouped together using the other three genes, this was not always the case (Fig. 2). For example, Efm 1231501 groups with the pbp5-R group using pbp2 and wlcA genes, despite being a non-CC17 isolate, and with the pbp5-S group for pbp5 and gls20. Efm 1231408 grouped with the pbp5-S group using wlcA, even though it is a CC17 genogroup isolate, and with the pbp5-R for pbp5, pbp2, and gls20. This apparent exchange of genes is as expected with an organism that appears to evolve by recombination and which has been shown to demonstrate extensive interstrain genomic diversity linked to the presence of phages, plasmids, pathogenicity islands, and conjugative elements (26). This observation supports the idea of at least two distinct E. faecium lineages, probably representative of commensal and hospital-associated clades (15), horizontally transferring certain genes between individual isolates of the two lineages.
Conclusion.
Although there is a conserved amino acid sequence corresponding to a more resistant PBP5, there must be other factors such as regulation, expression, translational modifications, or other genes leading to the differences in MICs seen among strains with the resistant pbp5 sequence. Together, these results suggest that the high MICs of ampicillin of hospital-associated E. faecium strains are due not only to the presence of a more resistant PBP5 but also to other changes that remain to be determined. In addition, the genomic variability seen between the pbp5 genes of the available U.S. E. faecium genomes was also seen in three other genes studied. The genomic variability appears to represent distinct lineages, although the other genes were not always associated with the same pbp5 gene, consistent with a species that shares and exchanges its genes by homologous recombination. These findings suggest that there are two distinct evolutionary clades of E. faecium.
Supplementary Material
ACKNOWLEDGMENTS
B.E.M. is supported in part by NIH grant R01 AI067861 from the National Institute of Allergy and Infectious Diseases (NIAID). J.G.-P. was supported by T32AI55449 during the time of this work and is currently supported by F31AI092891 from the NIAID. L.B. is supported by a merit review from the Department of Veterans Affairs and by NIH grant R01 AI045626 from the NIAID.
Footnotes
Supplemental material for this article may be found at http://aac.asm.org/.
Published ahead of print on 16 May 2011.
REFERENCES
- 1. Brinster S., Furlan S., Serror P. 2007. C-terminal WxL domain mediates cell wall binding in Enterococcus faecalis and other gram-positive bacteria. J. Bacteriol. 189:1244–1253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Brinster S., et al. 2007. Enterococcal leucine-rich repeat-containing protein involved in virulence and host inflammatory response. Infect. Immun. 75:4463–4471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Carias L. L., Rudin S. D., Donskey C. J., Rice L. B. 1998. Genetic linkage and cotransfer of a novel, vanB-containing transposon (Tn5382) and a low-affinity penicillin-binding protein 5 gene in a clinical vancomycin-resistant Enterococcus faecium isolate. J. Bacteriol. 180:4426–4434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Choudhury T., Singh K. V., Sillanpää J., Nallapareddy S. R., Murray B. E. 2011. Importance of two Enterococcus faecium loci encoding Gls-like proteins for in vitro bile salts stress response and virulence. J. Infect. Dis. 203:1147–1154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Clinical and Laboratory Standards Institute 2006. Performance standards for antimicrobial susceptibility testing; 16th informational supplement M100-S16. Clinical and Laboratory Standards Institute, Wayne, PA [Google Scholar]
- 6. Fontana R., et al. 1994. Overproduction of a low-affinity penicillin-binding protein and high-level ampicillin resistance in Enterococcus faecium. Antimicrob. Agents Chemother. 38:1980–1983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Fontana R., Grossato A., Rossi L., Cheng Y. R., Satta G. 1985. Transition from resistance to hypersusceptibility to beta-lactam antibiotics associated with loss of a low-affinity penicillin-binding protein in a Streptococcus faecium mutant highly resistant to penicillin. Antimicrob. Agents Chemother. 28:678–683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Galloway-Pena J. R., Nallapareddy S. R., Arias C. A., Eliopoulos G. M., Murray B. E. 2009. Analysis of clonality and antibiotic resistance among early clinical isolates of Enterococcus faecium in the United States. J. Infect. Dis. 200:1566–1573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Grayson M. L., et al. 1991. Increasing resistance to beta-lactam antibiotics among clinical isolates of Enterococcus faecium: a 22-year review at one institution. Antimicrob. Agents Chemother. 35:2180–2184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Homan W. L., et al. 2002. Multilocus sequence typing scheme for Enterococcus faecium. J. Clin. Microbiol. 40:1963–1971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Jureen R., Mohn S. C., Harthug S., Haarr L., Langeland N. 2004. Role of penicillin-binding protein 5 C-terminal amino acid substitutions in conferring ampicillin resistance in Norwegian clinical strains of Enterococcus faecium. APMIS 112:291–298 [DOI] [PubMed] [Google Scholar]
- 12. Klare I., Rodloff A. C., Wagner J., Witte W., Hakenbeck R. 1992. Overproduction of a penicillin-binding protein is not the only mechanism of penicillin resistance in Enterococcus faecium. Antimicrob. Agents Chemother. 36:783–787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Klibi N., et al. 2008. Polymorphism in pbp5 gene detected in clinical Enterococcus faecium strains with different ampicillin MICs from a Tunisian hospital. J. Chemother. 20:436–440 [DOI] [PubMed] [Google Scholar]
- 14. Leavis H. L., Bonten M. J., Willems R. J. 2006. Identification of high-risk enterococcal clonal complexes: global dispersion and antibiotic resistance. Curr. Opin. Microbiol. 9:454–460 [DOI] [PubMed] [Google Scholar]
- 15. Leavis H. L., et al. 2007. Insertion sequence-driven diversification creates a globally dispersed emerging multiresistant subspecies of E. faecium. PLoS Pathog. 3:e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ligozzi M., Pittaluga F., Fontana R. 1996. Modification of penicillin-binding protein 5 associated with high-level ampicillin resistance in Enterococcus faecium. Antimicrob. Agents Chemother. 40:354–357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Lleo M. M., Canepari P., Cornaglia G., Fontana R., Satta G. 1987. Bacteriostatic and bactericidal activities of beta-lactams against Streptococcus (Enterococcus) faecium are associated with saturation of different penicillin-binding proteins. Antimicrob. Agents Chemother. 31:1618–1626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Poeta P., et al. 2007. Polymorphisms of the pbp5 gene and correlation with ampicillin resistance in Enterococcus faecium isolates of animal origin. J. Med. Microbiol. 56:236–240 [DOI] [PubMed] [Google Scholar]
- 19. Rice L. B., et al. 2004. Impact of specific pbp5 mutations on expression of beta-lactam resistance in Enterococcus faecium. Antimicrob. Agents Chemother. 48:3028–3032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Rice L. B., et al. 2001. Penicillin-binding protein 5 and expression of ampicillin resistance in Enterococcus faecium. Antimicrob. Agents Chemother. 45:1480–1486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Rice L. B., et al. 2005. Enterococcus faecium low-affinity pbp5 is a transferable determinant. Antimicrob. Agents Chemother. 49:5007–5012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Rybkine T., Mainardi J. L., Sougakoff W., Collatz E., Gutmann L. 1998. Penicillin-binding protein 5 sequence alterations in clinical isolates of Enterococcus faecium with different levels of beta-lactam resistance. J. Infect. Dis. 178:159–163 [DOI] [PubMed] [Google Scholar]
- 23. Sauvage E., et al. 2002. The 2.4-A crystal structure of the penicillin-resistant penicillin-binding protein PBP5fm from Enterococcus faecium in complex with benzylpenicillin. Cell. Mol. Life Sci. 59:1223–1232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Sifaoui F., Arthur M., Rice L., Gutmann L. 2001. Role of penicillin-binding protein 5 in expression of ampicillin resistance and peptidoglycan structure in Enterococcus faecium. Antimicrob. Agents Chemother. 45:2594–2597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Top J., Willems R., Bonten M. 2008. Emergence of CC17 Enterococcus faecium: from commensal to hospital-adapted pathogen. FEMS Immunol. Med. Microbiol. 52:297–308 [DOI] [PubMed] [Google Scholar]
- 26. van Schaik W., Willems R. J. 2010. Genome-based insights into the evolution of enterococci. Clin. Microbiol. Infect. 16:527–532 [DOI] [PubMed] [Google Scholar]
- 27. Willems R. J., et al. 2005. Global spread of vancomycin-resistant Enterococcus faecium from distinct nosocomial genetic complex. Emerg. Infect. Dis. 11:821–828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Williamson R., le Bouguenec C., Gutmann L., Horaud T. 1985. One or two low affinity penicillin-binding proteins may be responsible for the range of susceptibility of Enterococcus faecium to benzylpenicillin. J. Gen. Microbiol. 131:1933–1940 [DOI] [PubMed] [Google Scholar]
- 29. Zapun A., Contreras-Martel C., Vernet T. 2008. Penicillin-binding proteins and beta-lactam resistance. FEMS Microbiol. Rev. 32:361–385 [DOI] [PubMed] [Google Scholar]
- 30. Zorzi W., et al. 1996. Structure of the low-affinity penicillin-binding protein 5 PBP5fm in wild-type and highly penicillin-resistant strains of Enterococcus faecium. J. Bacteriol. 178:4948–4957 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.