Abstract
This article reports the spread of blaKPC-2 in the Sao Paulo and Rio de Janeiro states, facilitated by globally spread K. pneumoniae clonal complex 258 (CC258) clones (ST258, ST11, and ST437) and a diversity of plasmids (IncFII, IncN, and IncL/M, two untypeable plasmids carrying Tn4401a or Tn4401b) successfully disseminated among species of the Enterobacteriaceae (Enterobacter cloacae, Serratia marcescens, and Citrobacter freundii). It also constitutes the first description of sequence type 258 (ST258) in Brazil, which was associated with a nosocomial hospital outbreak in Ribeirao Preto city.
TEXT
KPC (Klebsiella pneumoniae carbapenemase) enzymes are globally spread β-lactamases of Ambler class A, comprising 10 variants, KPC-2 and KPC-3 being predominant (26; http://www.lahey.org/studies/). They are mainly associated with K. pneumoniae, although KPC producers of other Enterobacteriaceae species, Pseudomonas and Acinetobacter, are increasingly reported (26, 39). The blaKPC genes are part of Tn4401, which is often linked to mosaic platforms derived from Tn1331, a transposon containing blaOXA-9 and blaTEM-1 (24, 33). Expansion of clones and plasmids that have acquired Tn4401 seems to have fuelled the recent pandemic dissemination of blaKPC genes (8, 15, 20, 21, 34). To date, only sporadic cases of K. pneumoniae, Enterobacter cloacae, Serratia marcescens, and Escherichia coli KPC-2 producers have been documented in Brazil (6, 9, 11, 19, 23, 28, 35, 40).
We analyzed 64 carbapenem-resistant isolates: 57 K. pneumoniae isolates, 5 E. cloacae isolates, 1 S. marcescens isolate, and 1 Citrobacter freundii isolate from different patients at 6 hospitals in two distant Brazilian regions (Table 1 and Fig. 1). Identification and antimicrobial susceptibility testing were accomplished by using semiautomatic systems and standard methods (7, 13). Clonal relatedness was established by pulsed-field gel electrophoresis (PFGE) and also by multilocus sequence typing in the case K. pneumoniae isolates (http://www.pasteur.fr/recherche/genopole/PF8/mlst/Kpneumoniae.html). Characterization of KPC-2 producers included phenotypic assays, PCR, and further sequencing as reported previously (8).
Table 1.
Plasmids carrying blaKPC-2 genes in Enterobacteriaceae from Brazila
| Inc group | Size (kb)b | Tn4401 variant | Other beta-lactamases | Species | ST | PFGE patternc | Susceptibility phenotyped | No. of isolates | Datee | City/state | Sample |
|---|---|---|---|---|---|---|---|---|---|---|---|
| FIIs | 50, 130, 240 | a | OXA-9, TEM-1, SHV-1 | K. pneumoniae | ST258 | KpA | GEN | 34 | 03/2007–06/2008 | Ribeirao Preto/SP | Several samplesf |
| FIIs | 50, 130, 240 | a | OXA-9, TEM-1, SHV-1 | K. pneumoniae | ST258 | KpA1 | GEN | 1 | 06/2007 | Ribeirao Preto/SP | Surgical wound |
| FIIs | 50, 130, 240 | a | OXA-9, TEM-1, SHV-1 | K. pneumoniae | ST258 | KpA2 | GEN | 2 | 02–06/2008 | Ribeirao Preto/SP | Bronco-alveolar lavage; urine |
| FIIs | 130, 150, 270 | a | OXA-9, TEM-1, SHV-1 | K. pneumoniae | ST258 | KpA | GEN | 1 | 04/2007 | Franca/SP | Urine |
| FIIs | 130 | a | OXA-9, TEM-1, SHV-1 | K. pneumoniae | ST258 | KpA | GEN | 7 | 07–08/2009 | Ribeirao Preto/SP | Rectal swab |
| FIIs | 130 | a | OXA-9, TEM-1, SHV-1 | K. pneumoniae | ST258 | KpA1′ | GEN | 3 | 07–08/2009 | Ribeirao Preto/SP | Rectal swab |
| FIIs | 130 | a | OXA-9, TEM-1, SHV-1 | K. pneumoniae | ST258 | KpA4 | GEN | 2 | 08/2009 | Ribeirao Preto/SP | Rectal swab |
| FIIs | 130 | a | OXA-9, TEM-1, SHV-1 | K. pneumoniae | ST258 | KpA4′ | GEN | 1 | 08/2009 | Ribeirao Preto/SP | Rectal swab |
| FIIs | 130, 240 | a | OXA-9, TEM-1, SHV-1 | K. pneumoniae | ST11g | KpA6 | GEN, AMI, TOB | 1 | 07/2009 | Ribeirao Preto/SP | Rectal swab |
| FIIs | 130, 140, 160 | a | OXA-9, TEM-1, SHV-1, CTX-M-2 | K. pneumoniae | ST48 | KpE | AMI, CIP, LEV, NIT, STX | 1 | 08/2009 | Ribeirao Preto/SP | Rectal swab |
| ut | 50, 110 | b | OXA-9, TEM-1, SHV-1, CTX-M-2 | K. pneumoniae | ST44 | KpC | GEN, AMI, TOB | 2 | 2008–2009 | Rio de Janeiro/RJ | Urine |
| N | 40, 100, 150 | b | OXA-9, TEM-1, SHV-1, CTX-M-2 | K. pneumoniae | ST327 | KpB | GEN, AMI, TOB | 1 | 01/2008 | Rio de Janeiro/RJ | Urine |
| N | 40, 140 | b | OXA-9, TEM-1, SHV-1, CTX-M-2 | K. pneumoniae | ST437 | KpD | GEN, AMI, TOB | 1 | 2009 | Rio de Janeiro/RJ | Urine |
| N | 40 | b | OXA-9, TEM-1 | E. cloacae | NA | Eclα | —j | 2 | 11/2007–10/2008 | Porto Alegre; Lageado/RS | Fragment of foot amputated; urine |
| ut | 20, 100, 250, 300 | ut | OXA-9, TEM-1 | E. cloacaeh | NA | Eclβ | — | 3 | 01–04/2009 | Porto Alegre/RS | Venous catheter; urine; blood |
| L/M | 60 | b | OXA-9, TEM-1 | S. marcescensi | NA | Sm | GEN | 1 | 04/2010 | Duque de Caxias/RJ | Blood |
| L/M | 50 | b | OXA-9, TEM-1 | C. freundii | NA | Cf | AMI | 1 | 06/2010 | Duque de Caxias/RJ | Bile secretion |
Abbreviations: GEN, gentamicin; AMI, amikacin; TOB, tobramycin; CIP, ciprofloxacin; LEV, levofloxacin; NIT, nitrofurantoin; STX, trimethoprim-sulfamethoxazole; NA, not applicable; ut = untypeable.
Underlining indicates size of the plasmid carrying blaKPC-2.
PFGE types are defined by capital letters. Subtypes are designated by a number (indicating the number of bands that differed from those of the index strain) and primes when necessary (to distinguish among subtypes differing in the same number of bands, with such bands being different).
The following antibiotics were tested in this study: ampicillin, ampicillin-sulbactam, piperacillin, piperacillin-tazobactam, cefazolin, cefuroxime, cefuroxime-axetil, cefotetan, ceftazidime, ceftriaxone, cefepime, aztreonan, imipenen, meropenen, AMI, GEN, TOB, CIP, LEV, NIT, STX, colistin, and tigecycline.
Dates are given as month/year.
This clone was recovered from samples of blood (n = 10), peritoneal cavity (n = 1), oropharynx (n = 2), urine (n = 7), venous catheter (n = 7), rectal swab (n = 3), broncoalveolar lavage (n = 1), sputum (n = 1), and wound/abscess (n = 2).
ST11 and ST258 show highly related PFGE profiles, as has also been noted in other studies (37).
Strains initially described in reference 40.
Strain initially described in reference 11.
—, susceptible only to colistin and tigecycline according to EUCAST breakpoints (13).
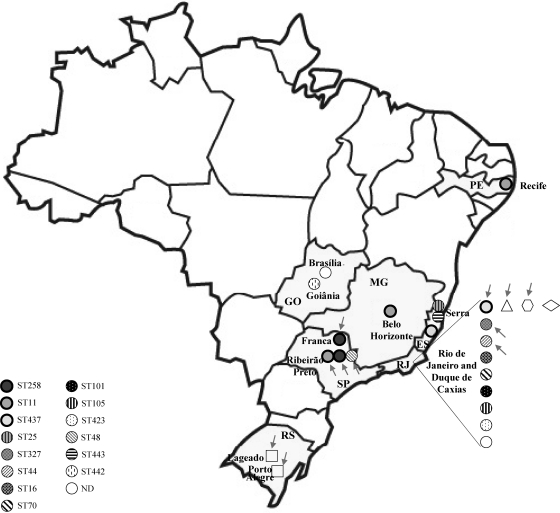
Fig. 1.
Geographic distribution of KPC-2-producing Enterobacteriaceae in Brazil. Abbreviations: ES, Espirito Santo; GO, Goias; DF, Distrito Federal; MG, Minas Gerais; PE, Pernambuco; RJ, Rio de Janeiro; RS, Rio Grande do Sul; SP, Sao Paulo; ND, not determined. States where KPC producers were isolated appear shaded in gray. K. pneumoniae isolates are represented by circles, E. cloacae isolates by squares, S. marcescens by a triangle, C. freundii by a hexagon, and E. coli isolates by a rhombus. Bold circles represent STs belonging to CC258 (ST258, ST11, and ST437). Clones analyzed in this study are indicated by arrows. ST25 and ST327 are double-locus variants of each other.
The K. pneumoniae isolates corresponded to 5 PFGE types linked to 6 sequence types (STs): KpA-ST258 (n = 51, comprising 6 subtypes: 42 KpA, 1 KpA1, 3 KpA1′, 2 KpA2, 2 KpA4, and 1 KpA4′), KpA6-ST11 (n = 1), KpB-ST327 (n = 1), KpC-ST44 (n = 2), KpD-ST437 (n = 1), and KpE-ST48 (n = 1). All isolates were resistant to β-lactams, susceptible to colistin and tigecycline, and showed a variable phenotype against aminoglycosides, quinolones, nitrofurantoin, and trimethoprim-sulfamethoxazole (Table 1). Heteroresistance to carbapenems was observed for K. pneumoniae and C. freundii isolates, as previously reported for KPC and VIM producers (29, 36).
The detection of isolates belonging to the multidrug-resistant K. pneumoniae clonal complex 258 (CC258) (ST258 and its single-locus variants ST11 and ST437) is of special concern (37). The ST258 clone is now globally spread and is mostly associated with the emergence and dissemination of KPC producers in different countries of North America, Europe, and Asia (2, 3, 14, 16, 20, 25, 26, 34). The ST11-K. pneumoniae lineage, first reported in France in 1997, is currently predominant in China, South Korea, and Hungary and has also been detected in the Netherlands, Norway, Poland, Portugal, Spain, and Brazil (2, 9, 17, 27, 30, 32, 35, 37; Luísa Peixe, personal communication). This clone has been extensively associated with different extended-spectrum beta-lactamases (ESBLs), mainly CTX-M-15 and CTX-M-14 (17, 27, 37) and more recently the KPC-2 (2, 30, 34) and VIM enzymes (18). Only sporadic cases of ST11 have been identified in Brazil; however, its recovery in different Brazilian cities might mirror a wider distribution than that reported (9, 35) (http://www.pasteur.fr/recherche/genopole/PF8/mlst/Kpneumoniae.html) (Fig. 1). Even though only one ST437 K. pneumoniae isolate, recovered in Rio de Janeiro in 2009, was included in our study, one recent study by Seki et al. (35) has shown the recovery of ST437 isolates producing KPC-2 in this city from 2007 to 2009, and a degree of epidemicity similar to that reported for other CC258 members cannot be discarded. Other clones recovered in this study are not well represented in the databases, such as ST327, previously detected in Israel linked to KPC-2 (21), or ST44 and ST48, which have not previously been associated with KPC production. Although most known KPC-2 producers of Serratia, Enterobacter, and Citrobacter have acquired highly transmissible plasmids from Klebsiella isolates, the spread of selected clones of these species might also be happening (31, 38).
The presence of genes encoding carbapenemases (GES, OXA, SPM, IMP, and VIM), ESBLs (CTX-M, TEM, and SHV), and cefamicinases (CMY) was analyzed as described previously (1, 10, 12). The genes blaOXA-9 and blaTEM-1 were detected in all K. pneumoniae isolates, while blaCTX-M-2 was identified in K. pneumoniae clones KpB, KpC, KpD, and KpE (Table 1). The frequent association of ESBLs or metallo-beta-lactamases (MBLs) with K. pneumoniae KPC producers in this and other studies seems to reflect acquisition of transmissible plasmids carrying blaKPC-2 by local endemic strains or transmissible plasmids carrying blaESBL by epidemic clones of KPC producers (8). The presence of CTX-M-2 among K. pneumoniae clones other than ST258 or ST11 suggests different origins, importation of globally spreading strains, and/or acquisition of KPC plasmids by endemic CTX-M-2-producing K. pneumoniae.
The analysis of the genetic environment of blaKPC by amplification of Tn4401, comparison of the restriction fragment length polymorphism (RFLP) patterns obtained by digestion of amplicons with PstI, HindIII, and BamHI, and sequencing of variants representative of distinct types revealed the presence of Tn4401 variants known as “a” (53 K. pneumoniae isolates) and “b” (4 K. pneumoniae isolates, 1 S. marcescens isolate, 1 C. freundii isolate, and 2 E. cloacae isolates) and also an untypeable platform (3 E. cloacae isolates). They were associated with a diversity of plasmids which were identified by typing the replication region (PCR and hybridization of S1-digested DNA) and comparison of fingerprintings corresponding to XhoI and HindIII-digested plasmid DNA as described previously (4, 8). While Tn4401a was located on a 130-kb IncFII plasmid, Tn4401b was detected in plasmids belonging to the incompatibility groups IncN (40 kb), IncL/M (50 to 60 kb), and two untypeable plasmids (20 and 50 kb). Only those of IncFII were transferred by conjugation.
The IncFII plasmids from ST258 K. pneumoniae isolates harboring a copy of Tn4401a were recovered in Ribeirao Preto city for 2 years. The IncN plasmids carrying the isoform Tn4401b were detected in KpB-ST327, KpD-ST437, and E. cloacae (Eclα) isolated from unrelated patients in Rio de Janeiro, Porto Alegre, and Lageado, cities sited 1,500 km apart in the south/southwest area of Brazil. IncN plasmid replicons were identical to that of pNL194, a 70-kb plasmid widely disseminated in Greece, which carries an MBL gene (22) (GenBank accession no. GU585907). A diversity of IncN plasmids with similar backbones but slightly different replicon sequences are increasingly associated with genes encoding carbapenemases of class A (KPC) or class B (VIM), reflecting the spread of IncN evolving from a common precursor (5, 15, 22). The IncL/M plasmids that also contained Tn4401b were recovered from S. marcescens and C. freundii. Cuzon et al. have recently described the location of the blaKPC-2 gene in 12-kb IncL/M plasmids from K. pneumoniae isolates collected in northeast Brazil (9), and the national dissemination of highly transferable IncL/M plasmids carrying blaKPC-2 cannot be discarded.
This article reports the spread of blaKPC-2 in Sao Paulo and Rio de Janeiro states facilitated by globally spread K. pneumoniae CC258 clones (ST258, ST11, and ST437) and a diversity of plasmids successfully disseminated among Enterobacteriaceae species. It also constitutes the first description of ST258 in Brazil associated with a nosocomial outbreak in a university hospital of Ribeirao Preto city. Although only sporadic cases of KPC producers have been documented in South America (Argentina, Colombia, and Brazil) (9, 11, 24, 40), this work has pointed out the high diversity of available genetic platforms carrying blaKPC-2. This might greatly amplify the dissemination of KPC genetic elements in this continent.
Acknowledgments
This study was funded by research grants from Conselho Nacional de Desenvolvimento Científico e Tecnológico in Brazil (CNPq, Projeto Universal, no. 480848/2008-4), the European Commission (LSHM-CT-2009-227258 HEALTH-F3-2008-223031), and the CIBERESP Network for Biomedical Research in Epidemiology and Public Health (Instituto Carlos III, Ministerio de Ciencia e Innovación of Spain, reference number CB06/02/0053). Initial identification and characterization of the strains were supported with laboratory equipment by Fundação de Amparo à Pesquisa do Estado de São Paulo (FAPESP). L.N.A. is supported by a CNPQ fellowship (Doutorado no País, no. 142736/2008-2) and CNPQ fellowship (Doutorado sanduíche, no. 201191/2009-1). T.C. is supported by a fellow research contract grant, FI09/00901, from Instituto Carlos III, Spanish Ministry of Science and Innovation.
We thank the staff of the Laboratory of Microbiology of Hospital das Clínicas of Faculty of Medicine of Ribeirão Preto, University of São Paulo (HC-FMRP-USP), for providing most of the isolates included in this study. We also thank personnel belonging to the infection control committee and to the intensive care unit (ICU) for supporting the investigation of intestinal colonization by carbapenemase-producing members of the Enterobacteriaceae among inpatients at the ICU of HC-FMRP-USP.
The clonal data are publicly available at http://www.pasteur.fr/mlst.
Footnotes
Published ahead of print on 16 May 2011.
REFERENCES
- 1. Andrade L. N., et al. 2010. Determinants of beta-lactam resistance in meningitis-causing Enterobacteriaceae in Brazil. Can. J. Microbiol. 56:399–407 [DOI] [PubMed] [Google Scholar]
- 2. Baraniak A., et al. 2009. Emergence of Klebsiella pneumoniae ST258 with KPC-2 in Poland. Antimicrob. Agents Chemother. 53:4565–4567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bogaerts P., et al. 2010. Emergence of clonally related Klebsiella pneumoniae isolates of sequence type 258 producing KPC-2 carbapenemase in Belgium. Antimicrob. Agents Chemother. 65:361–362 [DOI] [PubMed] [Google Scholar]
- 4. Carattoli A., et al. 2005. Identification of plasmids by PCR-based replicon typing. J. Microbiol. Methods 63:219–228 [DOI] [PubMed] [Google Scholar]
- 5. Carattoli A., et al. 2010. Complete nucleotide sequence of the IncN plasmid pKOX105 encoding VIM-1, QnrS1and SHV-12 proteins in Enterobacteriaceae from Bolzano, Italy compared with IncN plasmids encoding KPC enzymes in the USA. J. Antimicrob. Chemother. 65:2070–2075 [DOI] [PubMed] [Google Scholar]
- 6. Carvalho-Assef A. P. D., et al. 2010. Escherichia coli producing KPC-2 carbapenemase: first report in Brazil. Diagn. Microbiol. Infect. Dis. 68:337–338 [DOI] [PubMed] [Google Scholar]
- 7. Clinical Laboratory Standards Institute 2011. Performance standards for antimicrobial susceptibility testing. CLSI M100-S21. Clinical and Laboratory Standards Institute, Wayne, PA.
- 8. Curiao T., et al. 2010. Emergence of blaKPC-3-Tn4401a associated with a pKPN3/4-like plasmid within ST384 and ST388 Klebsiella pneumoniae clones in Spain. J. Antimicrob. Chemother. 65:1608–1614 [DOI] [PubMed] [Google Scholar]
- 9. Cuzon G., et al. 2010. Worldwide diversity of Klebsiella pneumoniae that produces β-lactamase blaKPC-2 gene. Emerg. Infect. Dis. 16:1349–1356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. D'Andrea M. M., et al. 2006. CMY-16, a novel acquired AmpC-type beta-lactamase of the CMY/LAT lineage in multifocal monophyletic isolates of Proteus mirabilis from northern Italy. Antimicrob. Agents Chemother. 50:618–624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Del Peloso P. F., Barros M. F. L., Santos F. A. 2010. Sepse por Serratia marcescens KPC. J. Bras. Patol. Med. Lab. 46:365–367 [Google Scholar]
- 12. Ellington M. J., Kistler J., Livermore D. M., Woodford N. 2007. Multiplex PCR for rapid detection of genes encoding acquired metallo-β-lactamases. J. Antimicrob. Chemother. 59:321–322 [DOI] [PubMed] [Google Scholar]
- 13. European Committee on Antimicrobial Susceptibility Testing 5 January 2011. Breakpoint tables for interpretation of MICs and zone diameters. EUCAST clinical breakpoint table v. 1.3 2011-01-05. European Committee on Antimicrobial Susceptibility Testing, Vaxjo, Sweden. http://www.eucast.org/fileadmin/src/media/PDFs/EUCAST_files/Disk_test_documents/EUCAST _breakpoints_v1.3_pdf.pdf
- 14. Giani T., et al. 2009. Emergence in Italy of Klebsiella pneumoniae sequence type 258 producing KPC-3 carbapenemase. J. Clin. Microbiol. 47:3793–3794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Gootz T. D., et al. 2009. Genetic organization of transposase regions surrounding blaKPC carbapenemase genes on plasmids from Klebsiella strains isolated in a New York city hospital. Antimicrob. Agents Chemother. 53:1998–2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kitchel J., et al. 2009. Molecular epidemiology of KPC-producing Klebsiella pneumoniae isolates in the United States: clonal expansion of multilocus sequence type 258. Antimicrob. Agents Chemother. 53:3365–3370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ko K. S., et al. 2010. Predominance of an ST11 extended-spectrum beta-lactamase-producing Klebsiella pneumoniae clone causing bacteraemia and urinary tract infections in Korea. J. Med. Microbiol. 59:822–828 [DOI] [PubMed] [Google Scholar]
- 18. Kristóf K., et al. 2010. Identification of a blaVIM-4 gene in the internationally successful Klebsiella pneumoniae ST11 clone and in a Klebsiella oxytoca strain in Hungary. J. Antimicrob. Chemother. 65:1303–1305 [DOI] [PubMed] [Google Scholar]
- 19. Leão R. S., et al. 2011. KPC-2 producing Klebsiella pneumoniae and Escherichia coli co-infection in a catheter related infection. Clin. Microbiol. Infect. 17:380–382 [DOI] [PubMed] [Google Scholar]
- 20. Leavitt A., Chmelnitsky I., Carmeli Y., Navon-Venezia S. 2010. Complete nucleotide sequence of KPC-3-encoding plasmid pKpQIL in the epidemic Klebsiella pneumoniae sequence type 258. Antimicrob. Agents Chemother. 54:4493–4496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Leavitt A., et al. 2010. Molecular epidemiology, sequence types, and plasmid analyses of KPC-producing Klebsiella pneumoniae strains in Israel. Antimicrob. Agents Chemother. 54:3002–3006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Miriagou V., et al. 2010. Sequence of pNL194, a 79.3-kilobase IncN plasmid carrying the blaVIM-1 metallo-β-lactamase gene in Klebsiella pneumoniae. Antimicrob. Agents Chemother. 54:4497–4502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Monteiro J., Santos A. F., Asensi M. D., Peirano G., Gales A. C. 2009. First report of KPC-2-producing Klebsiella pneumoniae in Brazil. Antimicrob. Agents Chemother. 53:333–334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Naas T., et al. 2008. Genetic structures at the origin of acquisition of the β-lactamase blaKPC gene. Antimicrob. Agents Chemother. 52:1257–1263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Navon-Venezia S., et al. 2009. First report on a hyperepidemic clone of KPC-3-producing Klebsiella pneumoniae in Israel genetically related to a strain causing outbreaks in the United States. Antimicrob. Agents Chemother. 53:818–820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Nordmann P., Cuzon G., Naas T. 2009. The real threat of Klebsiella pneumoniae carbapenemase-producing bacteria. Lancet Infect. Dis. 9:228–236 [DOI] [PubMed] [Google Scholar]
- 27. Oteo J., et al. 2009. Emergence of CTX-M-15-producing Klebsiella pneumoniae of multilocus sequence types 1, 11, 14, 17, 20, 35 and 36 as pathogens and colonizers in newborns and adults. J. Antimicrob. Chemother. 64:524–528 [DOI] [PubMed] [Google Scholar]
- 28. Peirano G., et al. 2009. Carbapenem hydrolysing β-lactamase KPC-2 in Klebsiella pneumoniae isolated in Rio de Janeiro, Brazil. J. Antimicrob. Chemother. 63:265–568 [DOI] [PubMed] [Google Scholar]
- 29. Pournaras S., et al. 2010. Characteristics of meropenem heteroresistance in Klebsiella pneumoniae carbapenemase (KPC)-producing clinical isolates of K. pneumoniae. J. Clin. Microbiol. 48:2601–2604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Qi Y., et al. 2011. ST11, the dominant clone of KPC-producing Klebsiella pneumoniae in China. J. Antimicrob. Chemother. 66:307–312 [DOI] [PubMed] [Google Scholar]
- 31. Rasheed J. K., et al. 2008. Detection of the Klebsiella pneumoniae carbapenemase type 2 carbapenem-hydrolyzing enzyme in clinical isolates of Citrobacter freundii and Klebsiella oxytoca carrying a common plasmid. J. Clin. Microbiol. 46:2066–2069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Rhee J. Y., et al. 2010. KPC-producing extreme drug-resistant Klebsiella pneumoniae isolate from a patient with diabetes mellitus and chronic renal failure on hemodialysis in South Korea. Antimicrob. Agents Chemother. 54:2278–2279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Rice L. B., et al. 2008. The KQ element, a complex genetic region conferring transferable resistance to carbapenems, aminoglycosides, and fluoroquinolones in Klebsiella pneumoniae. Antimicrob. Agents Chemother. 52:3427–3429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Samuelsen Ø., et al. 2009. Emergence of clonally related Klebsiella pneumoniae isolates of sequence type 258 producing plasmid-mediated KPC carbapenemase in Norway and Sweden. J. Antimicrob. Chemother. 63:654–658 [DOI] [PubMed] [Google Scholar]
- 35. Seki L. M., et al. 10 March 2011. Molecular epidemiology of KPC-2-producing Klebsiella pneumoniae isolates in Brazil: the predominance of sequence type 437. Diagn. Microbiol. Infect. Dis. [Epub ahead of print.] [DOI] [PubMed] [Google Scholar]
- 36. Tato M., et al. 2010. Carbapenem heteroresistance in VIM-1-producing Klebsiella pneumoniae isolates belonging to the same clone: consequences for routine susceptibility testing. J. Clin. Microbiol. 48:4089–4093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Tóth A., et al. 2010. Emergence of a colistin-resistant KPC-2-producing Klebsiella pneumoniae ST258 clone in Hungary. Eur. J. Clin. Microbiol. Infect. Dis. 29:765–769 [DOI] [PubMed] [Google Scholar]
- 38. Tsakris A., et al. 2010. In vivo acquisition of a plasmid-mediated bla(KPC-2) gene among clonal isolates of Serratia marcescens. J. Clin. Microbiol. 48:2546–2549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Walsh T. R. 2010. Emerging carbapenemases: a global perspective. Int. J. Antimicrob. Agents 36S3:S8–S14 [DOI] [PubMed] [Google Scholar]
- 40. Zavascki A. P., et al. 2009. KPC-2-producing Enterobacter cloacae in two cities from Southern Brazil. Int. J. Antimicrob. Agents 34:286–298 [DOI] [PubMed] [Google Scholar]