Abstract
Azithromycin at clinically relevant doses does not inhibit planktonic growth of the opportunistic pathogen Pseudomonas aeruginosa but causes markedly reduced formation of biofilms and quorum-sensing-regulated extracellular virulence factors. In the Gac/Rsm signal transduction pathway, which acts upstream of the quorum-sensing machinery in P. aeruginosa, the GacA-dependent untranslated small RNAs RsmY and RsmZ are key regulatory elements. As azithromycin treatment and mutational inactivation of gacA have strikingly similar phenotypic consequences, the effect of azithromycin on rsmY and rsmZ expression was investigated. In planktonically growing cells, the antibiotic strongly inhibited the expression of both small RNA genes but did not affect the expression of the housekeeping gene proC. The azithromycin treatment resulted in reduced expression of gacA and rsmA, which are known positive regulators of rsmY and rsmZ, and of the PA0588-PA0584 gene cluster, which was discovered as a novel positive regulatory element involved in rsmY and rsmZ expression. Deletion of this cluster resulted in diminished ability of P. aeruginosa to produce pyocyanin and to swarm. The results of this study indicate that azithromycin inhibits rsmY and rsmZ transcription indirectly by lowering the expression of positive regulators of these small RNA genes.
INTRODUCTION
The macrolide azithromycin (AZM) is a widely used antibiotic that blocks translation in susceptible bacteria by binding to the 50S ribosomal subunit. Although AZM does not inhibit planktonic growth of the opportunistic pathogen Pseudomonas aeruginosa at therapeutically relevant doses, the antibiotic has nevertheless been found to have beneficial effects in the treatment of chronic P. aeruginosa lung infections in cystic fibrosis patients. The observed improvement of pulmonary function may result from a combination of several mechanisms, among which the anti-inflammatory effects and the antibiofilm and antivirulence properties of AZM appear to be most important (10, 16, 29, 30, 39, 51). Beneficial effects of AZM have also been seen in experimental urinary tract infections caused by P. aeruginosa (1). Numerous studies have documented that AZM at low concentrations inhibits the formation of a range of extracellular virulence factors in P. aeruginosa in vitro (24, 25, 44).
The mode of action of AZM in P. aeruginosa is not entirely clear. The drug accumulates intracellularly and inhibits quorum sensing and biofilm formation in a dose-dependent manner (8, 11, 44, 45). Moreover, AZM appears to kill biofilm cells selectively (29). Quorum sensing in P. aeruginosa consists of a network of regulatory mechanisms that are activated by chemical signals. These signal molecules—mainly N-(3-oxododecanoyl)-homoserine lactone (3-oxo-C12-HSL) and N-butanoyl-homoserine lactone (C4-HSL)—are produced and released by the bacteria in response to cell population densities, as well as to environmental and nutritional factors (6, 38, 49). 3-Oxo-C12-HSL activates the master regulator of quorum sensing, the LasR transcription factor. LasR, together with the additional regulators VqsR and QscR, positively controls the expression of the RhlR transcription factor and its cognate signal, C4-HSL (49). Several studies have shown that AZM (at 2 μg/ml) downregulates the formation of 3-oxo-C12-HSL and C4-HSL in P. aeruginosa and that the resulting inhibition of the quorum-sensing machinery leads to diminished formation of the blue pigment pyocyanin, extracellular lytic enzymes, and biofilm matrix polymers, whereas the type III secretion system and the type IV pili are expressed at enhanced levels (16, 19, 31, 40). Furthermore, AZM improves clearing of P. aeruginosa from infected mouse lungs (16).
The Gac/Rsm signal transduction pathway positively controls quorum sensing in P. aeruginosa. In this regulatory cascade, the GacS/GacA two-component system activates transcription of two genes, rsmY and rsmZ, encoding small RNAs (sRNAs), which have high affinity for the RsmA protein. RsmA is a translational regulator; it can repress translation of target mRNAs by preventing ribosome binding. When the RsmY and RsmZ sRNAs are present in high concentrations, they sequester RsmA and thus allow translation of target mRNAs, including those in charge of quorum-sensing regulation (3, 20, 22, 33, 35). Mutational inactivation of gacA or overexpression of rsmA results in decreased formation of the quorum-sensing signal molecules (mainly C4-HSL), pyocyanin, and exoenzymes, as well as in perturbed biofilm-forming capacity, whereas the type III secretion system and type IV pili are upregulated (2, 4, 20, 27, 32, 33, 35, 41). Furthermore, a gacA mutant is deficient in colonizing the mouse lung (5), whereas an rsmA mutant colonizes the lung more persistently (28). Overall, the phenotypic effects of a gacA mutation are strikingly similar to those of AZM in the wild type of P. aeruginosa, suggesting that AZM might inhibit the Gac/Rsm signal transduction pathway. Here, we report that AZM blocks rsmY and rsmZ expression, and we find that the expression of several activators of rsmY and rsmZ transcription is sensitive to inhibition by AZM.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
The bacterial strains, plasmids, and oligonucleotides used are listed in Table 1. Strains of Escherichia coli and P. aeruginosa PAO1 were routinely grown in nutrient yeast broth (NYB) (42) or Luria broth (LB) (36) with shaking or on nutrient agar (42) plates amended with the following antibiotics when required: ampicillin at 100 μg/ml, tetracycline (Tc) at 25 μg/ml (125 μg/ml for selection of P. aeruginosa), and carbenicillin (Cb) at 300 μg/ml. Chloramphenicol was used at a concentration of 10 μg/ml to counterselect E. coli in mating experiments. AZM was used at 2 μg/ml. When relevant, 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal) was added to plates at a final concentration of 0.02%. Swarming was evaluated on plates containing 0.5% (wt/vol) agar, 8 g/liter of nutrient broth (Oxoid), and 5 g/liter of glucose (15).
Table 1.
Strains, plasmids, and primers used in the study
| Strain, plasmid, or primer | Relevant characteristics or sequence (5′→ 3′)a | Reference or source |
|---|---|---|
| E. coli | ||
| DH5α | λ− φ80dlacZΔM15 Δ(lacZYA-argF)U169 recA1 endA1 hsdR17(rK− mK−) supE44 thi-1 gyrA relA1 | 36 |
| HB101 | proA2hsdS20 (rb−mb−) recA13ara-14 lacY1 galK2 rpsL20 supE44 xyl-5 mtl-1 F− | 36 |
| P. aeruginosa | ||
| PAO1 | Wild type | ATCC 15692 |
| PAO1-retS | PAO1 containing a deletion in the retS gene | 26 |
| PAO6281 | gacA::ΩSm/Sp; Sm/Spr | 35 |
| PAO6327 | gacS::ΩSm/Sp; Sm/Spr | C. Reimmann, unpublished data |
| PAO6733 | PAO1 containing a 8,320-bp deletion in the PA0588-PA0584 genes | This study |
| Plasmids | ||
| pME3087 | Suicide vector for allelic replacement; ColE1 replicon; Tcr | 46 |
| pME3331 | Transcriptional rsmZ-lacZ fusion; Tcr | 15 |
| pME3641 | Plasmid carrying a translational proC′-′lacZ fusion, Cbr | 37 |
| pME3859 | Plasmid carrying a translational rsmA′-′lacZ fusion; Tcr | 33 |
| pME6013 | Broad-host-range vector for translational ′lacZ fusions; Tcr | 14 |
| pME6119 | Translational gacA′-′lacZ fusion; Cbr | 35 |
| pME7311 | Transcriptional rsmY-lacZ fusion; Tcr | 20 |
| pME9902 | Suicide construct derived from pME3087 used for deletion of the PA0588-PA0584 locus; Tcr | This study |
| pME9920 | pME6013 derivative carrying a translational PA0588′-′lacZ fusion; Tcr | This study |
| pRK2013 | Helper plasmid; ColE1 replicon; Tra+ Kmr | 9 |
| Primers | ||
| PRSMZ1 | CGTACAGGGAACACGCAACC | 15 |
| PRSMZ2 | AAAAAAAGGGGCGGGGTATT | 15 |
| PAOTRR1 | GTCAGGACATTGCGCAGGAAGC | 47 |
| PAOTRR2 | AAAACCCCGCCTTTTGGGCG | 47 |
| UPPA0588 | CGAAAGCTTCACCCGCATGCGT | This study |
| PA0588-78 | GCTGACGTCGAGATATTCCTGGAG | This study |
| PA0584-1226 | AGACTGCAGGCCTGAAGCAGCGC | This study |
| DOWNPA0584 | TATGAATTCGCCCGGGAATC | This study |
| TranslfusPA0588-F | CAGGGATCCTATTTCATCCAGCAG | This study |
| TranslfusPA0588-R | TCCCTGCAGAAAATGCTCATGACGTC | This study |
| 5SrRNA-1 | GCCTAAGCTTTTGGACAGGATGGGGTTGGA | 13 |
| 5SrRNA-2 | CAAAGAATTCGACGATTGTGTGTTGTAAGG | 13 |
Underlined sequences in primers indicate restriction sites as specified in Materials and Methods.
Construction of plasmids and of a gene replacement mutant.
DNA cloning and plasmid preparations were performed according to standard methods (36). Large-scale preparations of plasmid DNA were performed using JETstar 2.0 (Genomed). To construct strain PAO6733 (ΔPA0588-0584), an upstream 1,057-bp fragment and a downstream 627-bp fragment were amplified by PCR using the primer pairs UPPA0588F/PA0588-78 and PA0584-1226/DOWNPA0584, respectively. These products were digested with HindIII plus PstI and PstI plus EcoRI, respectively, and cloned into the corresponding sites of the suicide vector pME3087, yielding plasmid pME9902. This construct, carried by E. coli DH5α, was then introduced into P. aeruginosa strain PAO1 by triparental mating, using the helper strain E. coli HB101(pRK2013). Merodiploids were resolved as described previously (50). The resulting P. aeruginosa strain PAO6733 carried a PA0588-PA0584 deletion. The deletion was confirmed by PCR, and the PCR fragment obtained was checked by sequencing.
A translational PA0588′-′lacZ fusion, in which the ′lacZ reporter gene was fused to the 4th codon of the PA0588 open reading frame, was constructed by cloning a 380-bp fragment containing the PA0588 promoter and translation initiation region into the BamHI and PstI sites of pME6013, yielding pME9920. This fragment was generated by PCR using the P. aeruginosa PAO1 genome as the template and primers TranslfusPA0588-F and TranslfusPA0588-R.
β-Galactosidase assay.
P. aeruginosa cells were grown in 20 ml of NYB, with or without AZM, in 50-ml Erlenmeyer flasks. Triton X-100 (0.1% [wt/vol]) was added to the cultures to avoid cell aggregation. Samples were taken during various growth phases and permeabilized with 5% (vol/vol) toluene. β-Galactosidase activities were then measured according to the Miller method (23).
Pyocyanin assay.
The pyocyanin assay is based on the absorbance of pyocyanin at 520 nm in acidic solution (7). Overnight cultures of the strains to be tested were diluted to an optical density at 600 nm (OD600) of 0.05 in 25 ml of LB and grown at 37°C until they reached an OD600 of 2 to 3 (after 8 h). A 5-ml sample of each strain was centrifuged, and the supernatant was extracted with 3 ml of chloroform. After being mixed with a vortex, the lower phase was taken and mixed with 1 ml of 0.2 M HCl in glass tubes until the upper phase showed a pink to deep-red color (approximately 30 min). The absorbance of this solution was measured at 520 nm (A520). Concentrations, expressed as μg of pyocyanin produced per ml of culture supernatant, were determined by multiplying the A520 value by 17.072 (7).
RNA extraction and Northern blots.
P. aeruginosa PAO1 was inoculated at an OD600 of 0.04 into 20 ml of NYB medium, with or without 2 μg/ml of AZM. The cultures were grown at 37°C with vigorous shaking until they reached an OD600 of approximately 3, and then cells were harvested and RNAProtect Bacteria (Qiagen) was added. Total RNA was isolated by the hot-phenol method as described previously (13). The quality of total RNA was checked by agarose gel electrophoresis, and the quantity was measured in a NanoDrop spectrophotometer. Digoxigenin (DIG)-labeled probes were obtained with DNA-labeling mix (Roche) as follows: PCR fragments were synthesized with P. aeruginosa PAO1 genomic DNA as the template and primer pairs PRSMZ1-PRSMZ2 and PAOTRR1-PAOTRR2 for rsmZ and rsmY probes, respectively. RNAs (∼3 μg per lane) were separated on a denaturing urea-polyacrylamide gel and analyzed by Northern blotting as previously described (47), with minor modifications: RNAs were transferred by electroblotting (30 min at 150 mA) onto a charged nylon membrane (Hybond-N+; GE-Amersham) and revealed by hybridization with the DIG-labeled probes and exposure to a light-sensitive film (Super RX; Fujifilm). 5S rRNA served as a loading control. For this purpose, a 5S rRNA gene probe was synthesized with primers 5SrRNA-1 and 5S-rRNA-2 (Table 1) and DIG labeled. The membranes could be used a second time for Northern blot detection of 5S rRNA. To completely detach the probe previously hybridized, the membranes were rinsed with diethylpyrocarbonate-treated H2O and immersed twice in 0.2 M NaOH containing 0.1% SDS for 15 min. The membranes were then rinsed with 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) solution (36) and used in the second Northern blot.
RESULTS
AZM blocks induction of rsmZ and rsmY expression in P. aeruginosa.
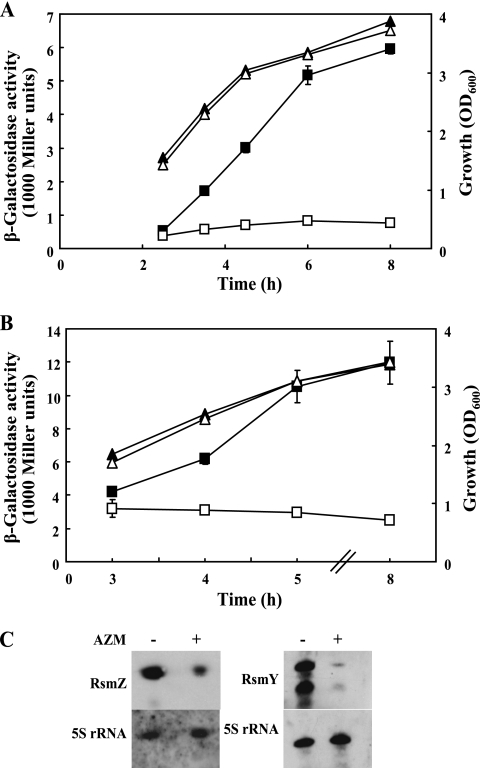
We initially verified that AZM at 2 μg/ml had no effect on planktonic growth of P. aeruginosa PAO1 in NYB but inhibited the formation of quorum-sensing-dependent exoproducts (data not shown), in agreement with published data (8, 16, 31, 40, 45). The expression of transcriptional rsmZ-lacZ and rsmY-lacZ fusions was strongly inhibited by AZM at 2 μg/ml; AZM prevented cell population density-dependent induction of both rsmZ and rsmY during postexponential growth (Fig. 1A and B). This result was confirmed by Northern blotting: the intracellular concentrations of RsmZ and RsmY sRNAs were reduced about 5- and 10-fold, respectively, after AZM had been administered (Fig. 1C). As noted previously (20), RsmY sRNA was present as a full-length molecule and as a shortened form, which possibly lacks the terminator structure. In a control experiment, the expression of a translational lacZ fusion to a constitutively expressed housekeeping gene, proC (37), was unaffected by the addition of AZM (Fig. 2A). In conclusion, AZM strongly inhibits the immediate output of the GacS-GacA system, i.e., the synthesis of the RsmZ and RsmY sRNAs, and this mode of action occurs upstream of the quorum-sensing machinery (20, 33, 35).
Fig. 1.
Regulation of rsmZ and rsmY expression in the presence or absence of AZM. (A) The β-galactosidase activities of an rsmZ-lacZ transcriptional fusion carried by pME3331 (squares) and growth (triangles) were determined in P. aeruginosa PAO1 in the absence (filled symbols) or presence (empty symbols) of 2 μg/ml of AZM. The experiments were done in triplicate; average values ± standard deviations are shown. (B) The β-galactosidase activities of an rsmY-lacZ transcriptional fusion carried by pME7311 (squares) and growth (triangles) were determined in P. aeruginosa PAO1 in the absence (filled symbols) and presence (empty symbols) of 2 μg/ml of AZM. The experiments were done in triplicate; average values ± standard deviations are shown. (C) Northern blot analysis of RsmZ and RsmY RNA in the absence (−) and presence (+) of AZM. Total RNA (3 μg) was prepared from cultures in the late exponential phase (OD600 ≥ 3). 5S rRNA was used as a loading control.
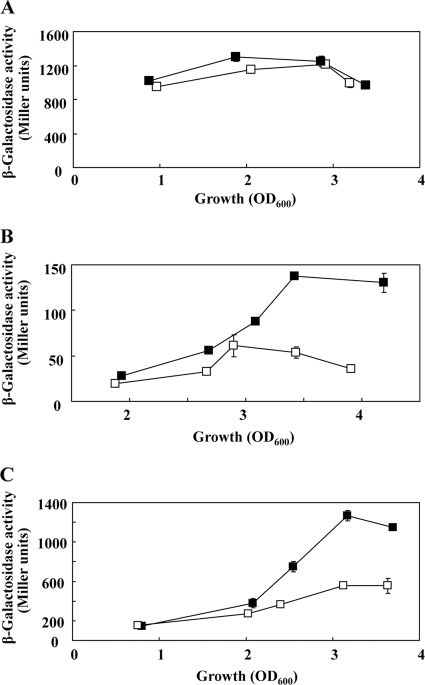
Fig. 2.
Regulation of proC, gacA, and rsmA expression in the presence or absence of AZM. The β-galactosidase activities of translational proC′-′lacZ, gacA′-′lacZ, and rsmA′-′lacZ fusions carried by pME3641 (A), pME6119 (B), and pME3859 (C), respectively, were determined in P. aeruginosa PAO1 in the absence (filled symbols) or presence (empty symbols) of 2 μg/ml of AZM. The experiments were done in triplicate; average values ± standard deviations are shown.
AZM treatment results in diminished expression of GacA and RsmA.
The response regulator GacA binds to an upstream activating sequence in the rsmZ and rsmY promoters, and this interaction is essential for transcriptional induction of both genes (3, 17, 22). Furthermore, the RsmA protein has a marked positive effect on rsmZ and rsmY transcription; this regulation occurs via a feedback loop, whose components are not known (15, 20). Addition of AZM led to reduced expression of translational gacA′-′lacZ and rsmA′-′lacZ fusions in the wild-type PAO1 compared to the uninhibited control (Fig. 2B and C), suggesting that AZM may curtail the availability of both GacA and RsmA.
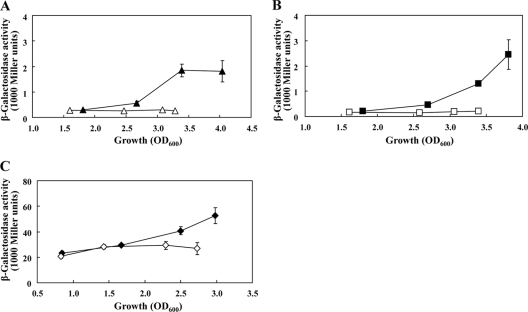
Although rsmZ-lacZ expression was low in a gacA mutant, AZM still lowered the residual levels (Fig. 3A), implying that, besides gacA, there are additional targets of AZM. In theory, AZM might interact with the GacS sensor or its antagonist, the RetS sensor (12), and such interference with signaling might account for the observed decrease in rsmZ and rsmY expression (Fig. 1A and B). However, in both a gacS and a retS background, AZM remained able to reduce rsmZ expression (Fig. 3B, C). Taken together, these data suggest that the pronounced effect of AZM on rsmZ and rsmY transcription involves several potentially synergistic interactions with activators of rsmZ and rsmY.
Fig. 3.
Expression of an rsmZ-lacZ transcriptional fusion (pME3331) in the P. aeruginosa gacA mutant PAO6281 (A), in the gacS mutant PAO6327 (B), and in the retS mutant PAO1-retS (C) in the absence (filled symbols) or presence (empty symbols) of 2 μg/ml of AZM. The experiments were done in triplicate; average values ± standard deviations are shown.
The PA0588-PA0584 gene cluster contributes to rsmZ and rsmY expression.
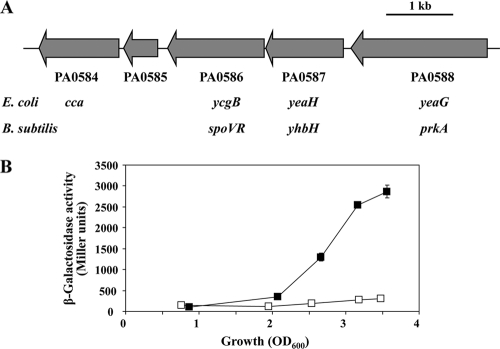
A recent transcriptomic study (19) shows that AZM at 2 μg/ml has a particularly strong (about 50-fold) negative effect on the abundance of the PA0586 and PA0587 transcripts in P. aeruginosa. Little is currently known about the functions of the PA0588-PA0584 gene cluster from which these transcripts emanate, except that some genes have homologues in other bacteria (Fig. 4A). The proximal PA0588 gene appears to encode a Ser/Thr protein kinase, which has been detected as a phosphorylated protein in P. aeruginosa (34). The distal PA0584 gene is annotated as cca, which is likely to code for a tRNA-nucleotidyltransferase adding a CCA trinucleotide to the 3′ ends of tRNAs (48). The PA0587-PA0585 genes specify hypothetical proteins (43). We verified that the AZM treatment resulted in strongly diminished expression of a translational PA0588′-′lacZ fusion carried by pME9920 (Fig. 4B).
Fig. 4.
(A) Schematic representation of the PA0588-PA0584 gene cluster (43). Homologous genes found in E. coli and Bacillus subtilis are indicated below the corresponding PA numbers. Mutant PAO6733 was constructed in P. aeruginosa PAO1 by deletion of all five genes, as described in Materials and Methods. (B) Influence of AZM on the expression of a translational PA0588′-′lacZ fusion. β-Galactosidase activities were determined in the absence (filled symbols) or presence (empty symbols) of AZM. The experiments were done in triplicate; average values ± standard deviations are shown.
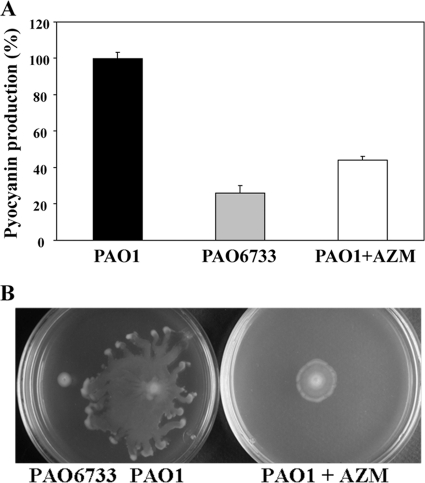
We constructed a deletion mutant, PAO6733, lacking the entire PA0588-PA0584 gene cluster. Production of pyocyanin was reduced in this mutant at the end of growth in comparison with the wild-type PAO1 (Fig. 5A). A similar reduction of pyocyanin formation was seen upon AZM treatment of the wild type (Fig. 5A). Furthermore, the deletion mutant PAO6733 had a swarming deficiency resembling that of the AZM-treated wild type (21) (Fig. 5B). Thus, the functions of the PA0588-PA0584 cluster overlap with those of the quorum-sensing machinery.
Fig. 5.
Influence of the PA0588-PA0584 gene cluster and AZM on pyocyanin production and swarming motility in P. aeruginosa. (A) Pyocyanin levels were determined in P. aeruginosa PAO1 (black bar), PAO6733 (ΔPA0588-0584; grey bar), and in P. aeruginosa PAO1 in the presence of AZM (white bar). The value obtained for PAO1 was set at 100%. Each value is the average of three different LB cultures grown for 8 h, and the error bars show standard deviations. (B) Swarming ability was tested on semisolid medium after 16 h of incubation at 37°C for PAO1 and PAO6733 in the absence or presence of AZM.
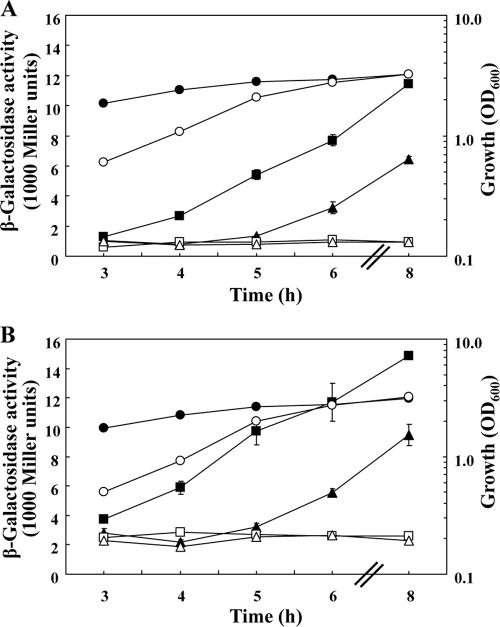
We then asked whether deletion of the PA0588-PA0584 cluster would affect rsmZ and rsmY expression. In the deletion mutant PAO6733, expression of both rsmZ-lacZ and rsmY-lacZ was reduced about 2-fold at the end of growth, and AZM caused further lowering of expression (Fig. 6A and B). Although the mutant was handicapped during an early growth phase in NYB because of a delayed exit from stationary phase and a longer doubling time (60 min instead of 40 min for the wild type), the final cell population densities of the mutant were similar to those of the wild type (Fig. 6A and B). Thus, one function of the PA0588-PA0584 cluster is to sustain full rsmZ and rsmY expression, and this positive-control effect on rsmZ and rsmY expression adds to the known positive effects of GacA and RsmA. As none of the five genes in the PA0588-PA0584 cluster encodes a transcription factor, the effect that deletion of the cluster has on rsmZ and rsmY transcription is probably indirect.
Fig. 6.
Effect of a PA0588-PA0584 deletion on the expression of the rsmZ and rsmY genes. (A) The growth (circles) and β-galactosidase activities of an rsmZ-lacZ transcriptional fusion carried by pME3331 were determined in the P. aeruginosa wild type (squares) and in the deletion mutant PAO6733 (triangles) in the absence (filled symbols) or presence (empty symbols) of 2 μg/ml of AZM. The experiments were done in triplicate; average values ± standard deviations are shown. (B) Growth (circles) and β-galactosidase activities of an rsmY-lacZ transcriptional fusion carried by pME7311 were determined in the P. aeruginosa wild type (squares) and in the mutant PAO6733 (triangles) in the absence (filled symbols) and presence (empty symbols) of 2 μg/ml of AZM. The experiments were done in triplicate; average values ± standard deviations are shown.
DISCUSSION
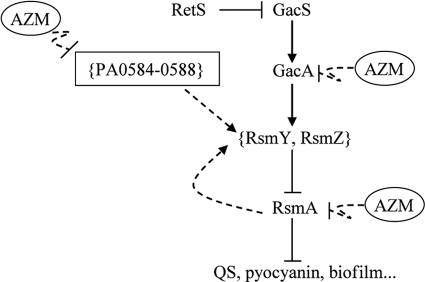
The strong inhibitory effect of AZM on the expression of the sRNA genes rsmY and rsmZ in P. aeruginosa is striking, especially as growth of the organism and expression of a housekeeping gene (proC) were unaffected under the same conditions (Fig. 1 and 2A). Given that AZM is an inhibitor of translation in susceptible bacteria, including P. aeruginosa (21), it is likely that AZM acts indirectly on rsmY and rsmZ transcription. This could occur via AZM-mediated inhibition of translational expression of regulatory proteins that activate rsmY and rsmZ transcription. The evidence obtained in this study supports this view. GacA and RsmA, both known to be crucial for induction of rsmY and rsmZ transcription (15, 20), were expressed at significantly reduced levels upon AZM addition (Fig. 2B and C). Moreover, we newly identified an AZM-sensitive gene cluster (PA0588-PA0584) that makes a positive contribution to rsmY and rsmZ expression (Fig. 4 and 6). While more work is needed to elucidate the roles of individual genes in this cluster, we conclude from our work and from a recent independent study (19) that AZM downregulates the expression of a number of genes that exert positive-control functions upstream of the quorum-sensing machinery. A model summarizing the mechanisms investigated in this work is presented in Fig. 7. In the same vein, Kai and collaborators (19) have pointed out that AZM downregulates the expression of several genes that are needed to supply the metabolic precursors of the quorum-sensing signals 3-oxo-C12-HSL and C4-HSL.
Fig. 7.
Model of the influence of AZM on the expression of the untranslated sRNAs RsmY and RsmZ in P. aeruginosa PAO1. Expression of the sRNA genes rsmY and rsmZ depends on the positive control exerted by the response regulator GacA (directly) and by the RNA-binding protein RsmA (indirectly). In addition, the PA0588-PA0584 cluster has an indirect positive effect, although the mechanism is not yet known. The combined negative influences of AZM on the expression of GacA, RsmA, and the cluster PA0588-PA0584 could explain the strongly reduced levels of the RsmY and RsmZ sRNAs in the presence of the antibiotic. ⊥ represents negative control; → represents positive control; dashed lines indicate indirect effects.
Although in theory the strong negative effect of AZM on the RsmY and RsmZ sRNAs might involve some interference with signal transduction from the membrane-bound sensors RetS and GacS to the response regulator GacA, two observations do not support such a hypothesis. First, AZM is known to accumulate intracellularly and to inhibit translation of susceptible mRNAs in P. aeruginosa (21, 44). Second, AZM continued to have inhibitory effects on rsmZ expression even in gacA, gacS, and retS mutants (Fig. 3). We therefore conclude, as have other authors (21, 44), that AZM basically acts on translating ribosomes in the cytoplasm. It still remains to be understood which features an mRNA needs to have for translation to be AZM sensitive in P. aeruginosa.
Among the multiple effects that AZM has been reported to exert in P. aeruginosa, not all may necessarily depend on interference with rsmYZ expression and hence with quorum sensing. For instance, AZM-mediated killing of stationary-phase and biofilm cells and changes in membrane permeability (16, 18, 29, 40) may involve other mechanisms. However, to our knowledge, the observation that AZM inhibits the expression of RsmY and RsmZ is the first example of an antibiotic inhibiting the expression of sRNAs in a bacterium. This highlights the possibility that RsmY and RsmZ could be used as sensitive reporters of AZM action in P. aeruginosa, which may be helpful in future clinical studies evaluating the efficacy of AZM as a treatment of P. aeruginosa infections.
ACKNOWLEDGMENTS
We thank A. Goodman for making a retS mutant available to us. We are grateful to Cornelia Reimmann for supplying a gacS mutant and for critically reading the manuscript.
This work was supported by the Velux Foundation and the Swiss National Science Foundation.
Footnotes
Published ahead of print on 2 May 2011.
REFERENCES
- 1. Bala A., Kumar R., Hajai K. 2011. Quorum sensing inhibition of Pseudomonas aeruginosa by azithromycin and its effectiveness in urinary tract infections. J. Med. Microbiol. 60:300–306 [DOI] [PubMed] [Google Scholar]
- 2. Brencic A., Lory S. 2009. Determination of the regulon and identification of novel mRNA targets of Pseudomonas aeruginosa RsmA. Mol. Microbiol. 72:612–632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Brencic A., et al. 2009. The GacS/GacA signal transduction system of Pseudomonas aeruginosa acts exclusively through its control over the transcription of the RsmY and RsmZ regulatory small RNAs. Mol. Microbiol. 73:434–445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Burrowes E., Abbas A., O'Neill A., Adams C., O'Gara F. 2005. Characterisation of the regulatory RNA RsmB from Pseudomonas aeruginosa PAO1. Res. Microbiol. 156:7–16 [DOI] [PubMed] [Google Scholar]
- 5. Coleman F. T., et al. 2003. Hypersusceptibility of cystic fibrosis mice to chronic Pseudomonas aeruginosa oropharyngeal colonization and lung infection. Proc. Natl. Acad. Sci. U. S. A. 100:1949–1954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Duan K., Surette M. G. 2007. Environmental regulation of Pseudomonas aeruginosa PAO1 Las and Rhl quorum-sensing systems. J. Bacteriol. 189:4827–4836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Essar D. W., Eberly L., Hadero A., Crawford I. P. 1990. Identification and characterization of genes for a second anthranilate synthase in Pseudomonas aeruginosa: interchangeability of the two anthranilate synthases and evolutionary implications. J. Bacteriol. 172:884–900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Favre-Bonté S., Köhler T., van Delden C. 2003. Biofilm formation by Pseudomonas aeruginosa: role of the C4-HSL cell-to-cell signal and inhibition by azithromycin. J. Antimicrob. Chemother. 52:598–604 [DOI] [PubMed] [Google Scholar]
- 9. Figurski D. H., Helinski D. R. 1979. Replication of an origin-containing derivative of plasmid RK2 dependent on a plasmid function provided in trans. Proc. Natl. Acad. Sci. U. S. A. 76:1648–1652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Florescu D. F., Murphy P. J., Kalil A. C. 2009. Effects of prolonged use of azithromycin in patients with cystic fibrosis: a meta-analysis. Pulm. Pharmacol. Ther. 22:467–472 [DOI] [PubMed] [Google Scholar]
- 11. Gillis R. J., Iglewski B. H. 2004. Azithromycin retards Pseudomonas aeruginosa biofilm formation. J. Clin. Microbiol. 42:5842–5845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Goodman A. L., et al. 2009. Direct interaction between sensor kinase proteins mediates acute and chronic disease phenotypes in a bacterial pathogen. Genes Dev. 23:249–259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. González N., et al. 2008. Genome-wide search reveals a novel GacA-regulated small RNA in Pseudomonas species. BMC Genomics 9:167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Heeb S., et al. 2000. Small, stable shuttle vectors based on the minimal pVS1 replicon for use in gram-negative, plant-associated bacteria. Mol. Plant Microbe Interact. 13:232–237 [DOI] [PubMed] [Google Scholar]
- 15. Heurlier K., et al. 2004. Positive control of swarming, rhamnolipid synthesis, and lipase production by the posttranscriptional RsmA/RsmZ system in Pseudomonas aeruginosa PAO1. J. Bacteriol. 186:2936–2945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hoffmann N., et al. 2007. Azithromycin blocks quorum sensing and alginate polymer formation and increases the sensitivity to serum and stationary-growth-phase killing of Pseudomonas aeruginosa and attenuates chronic P. aeruginosa lung infection in Cftr−/− mice. Antimicrob. Agents Chemother. 51:3677–3687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Humair B., Wackwitz B., Haas D. 2010. GacA-controlled activation of promoters for small RNA genes in Pseudomonas fluorescens. Appl. Environ. Microbiol. 76:1497–1506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Imamura Y., et al. 2005. Azithromycin exhibits effects on Pseudomonas aeruginosa through interaction with the outer membrane. Antimicrob. Agents Chemother. 49:1377–1380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kai T., et al. 2009. A low concentration of azithromycin inhibits the mRNA expression of N-acyl homoserine lactone synthesis enzymes, upstream of lasI and rhlI, in Pseudomonas aeruginosa. Pulm. Pharmacol. Ther. 22:483–486 [DOI] [PubMed] [Google Scholar]
- 20. Kay E., et al. 2006. Two GacA-dependent small RNAs modulate the quorum sensing response in Pseudomonas aeruginosa. J. Bacteriol. 188:6026–6033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Köhler T., Dumas J. L., van Delden C. 2007. Ribosome protection prevents azithromycin-mediated quorum-sensing modulation and stationary-phase killing of Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 51:4243–4248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lapouge K., Schubert M., Allain F. H. T., Haas D. 2008. Gac/Rsm signal transduction pathway of γ-proteobacteria: from RNA recognition to regulation of social behaviour. Mol. Microbiol. 67:241–253 [DOI] [PubMed] [Google Scholar]
- 23. Miller J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY [Google Scholar]
- 24. Mizukane R., et al. 1994. Comparative in-vitro exoenzyme-suppressing activities of azithromycin and other macrolide antibiotics against Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 38:528–533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Molinari G., Guzman C. A., Pece A., Schito G. C. 1993. Inhibition of Pseudomonas aeruginosa virulence factors by subinhibitory concentrations of azithromycin and other macrolide antibiotics. J. Antimicrob. Chemother. 31:681–688 [DOI] [PubMed] [Google Scholar]
- 26. Mougous J. D., et al. 2006. A virulence locus of Pseudomonas aeruginosa encodes a protein secretion apparatus. Science 312:1526–1530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Mulcahy H., O'Callaghan J., O'Grady E. P., Adams C., O'Gara F. 2006. The posttranscriptional regulator RsmA plays a role in the interaction between Pseudomonas aeruginosa and human airway epithelial cells by positively regulating the type III secretion system. Infect. Immun. 74:3012–3015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Mulcahy H., et al. 2008. Pseudomonas aeruginosa RsmA plays an important role during murine infection by influencing colonization, virulence, persistence, and pulmonary inflammation. Infect. Immun. 76:632–638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Mulet X., et al. 2009. Azithromycin in Pseudomonas aeruginosa biofilms: bactericidal activity and selection of nfxB mutants. Antimicrob. Agents Chemother. 53:1552–1560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Murphy D. M., Forrest I. A., Curran D., Ward C. 2010. Macrolide antibiotics and the airway: antibiotic or non-antibiotic effects? Expert Opin. Investig. Drugs 19:401–414 [DOI] [PubMed] [Google Scholar]
- 31. Nalca Y., et al. 2006. Quorum-sensing antagonistic activities of azithromycin in Pseudomonas aeruginosa PAO1: a global approach. Antimicrob. Agents Chemother. 50:1680–1688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Parkins M. D., Ceri H., Storey D. G. 2001. Pseudomonas aeruginosa GacA, a factor in multihost virulence, is also essential for biofilm formation. Mol. Microbiol. 40:1215–1226 [DOI] [PubMed] [Google Scholar]
- 33. Pessi G., et al. 2001. The global posttranscriptional regulator RsmA modulates production of virulence determinants and N-acylhomoserine lactones in Pseudomonas aeruginosa. J. Bacteriol. 183:6676–6683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Ravichandran A., Sugiyama N., Tomita M., Swarup S., Ishihama Y. 2009. Ser/Thr/Tyr phosphoproteome analysis of pathogenic and non-pathogenic Pseudomonas species. Proteomics 9:2764–2775 [DOI] [PubMed] [Google Scholar]
- 35. Reimmann C., et al. 1997. The global activator GacA of Pseudomonas aeruginosa PAO positively controls the production of the autoinducer N-butyryl-homoserine lactone and the formation of the virulence factors pyocyanin, cyanide, and lipase. Mol. Microbiol. 24:309–319 [DOI] [PubMed] [Google Scholar]
- 36. Sambrook J., Russell D. W. 2001. Molecular cloning: a laboratory manual, 3rd ed., Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 37. Savioz A., Zimmermann A., Haas D. 1993. Pseudomonas aeruginosa promoters which contain a conserved GG-N10-GC motif but appear to be RpoN-independent. Mol. Gen. Genet. 238:74–80 [DOI] [PubMed] [Google Scholar]
- 38. Shrout J. D., et al. 2006. The impact of quorum sensing and swarming motility on Pseudomonas aeruginosa biofilm formation is nutritionally conditional. Mol. Microbiol. 62:1264–1277 [DOI] [PubMed] [Google Scholar]
- 39. Sibley C. D., Grinwis M. E., Rabin H. R., Surette M. G. 2010. Azithromycin paradox in the treatment of cystic fibrosis airway disease. Future Microbiol. 5:1315–1319 [DOI] [PubMed] [Google Scholar]
- 40. Skindersøe M. E., et al. 2008. Effects of antibiotics on quorum sensing in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 52:3648–3663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Soscia C., Hachani A., Bernadac A., Filloux A., Bleves S. 2007. Cross talk between type III secretion and flagellar assembly systems in Pseudomonas aeruginosa. J. Bacteriol. 189:3124–3132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Stanisich V. A., Holloway B. W. 1972. A mutant sex factor of Pseudomonas aeruginosa. Genet. Res. Camb. 19:91–108 [DOI] [PubMed] [Google Scholar]
- 43. Stover C. K., et al. 2000. Complete genome sequence of Pseudomonas aeruginosa PAO1, an opportunistic pathogen. Nature 406:959–964 [DOI] [PubMed] [Google Scholar]
- 44. Tateda K., et al. 1996. Direct evidence for antipseudomonal activity of macrolides: exposure-dependent bactericidal activity and inhibition of protein synthesis by erythromycin, clarithromycin, and azithromycin. Antimicrob. Agents Chemother. 40:2271–2275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Tateda K., et al. 2001. Azithromycin inhibits quorum sensing in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 45:1930–1933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Voisard C., et al. 1994. Biocontrol of root diseases by Pseudomonas fluorescens CHA0: current concepts and experimental approaches, p. 67–89 In O'Gara F., Dowling D., Boesten B. (ed.), Molecular ecology of rhizosphere microorganisms. VCH Publishers, Weinheim, Germany [Google Scholar]
- 47. Valverde C., Heeb S., Keel C., Haas D. 2003. RsmY, a small regulatory RNA, is required in concert with RsmZ for GacA-dependent expression of biocontrol traits in Pseudomonas fluorescens CHA0. Mol. Microbiol. 50:1361–1379 [DOI] [PubMed] [Google Scholar]
- 48. Vörtler S., Mörl M. 2010. tRNA-nucleotidyltransferases: highly unusual RNA polymerases with vital functions. FEBS Lett. 584:297–302 [DOI] [PubMed] [Google Scholar]
- 49. Williams P., Cámara M. 2009. Quorum sensing and environmental adaptation in Pseudomonas aeruginosa: a tale of regulatory networks and multifunctional signal molecules. Curr. Opin. Microbiol. 12:182–191 [DOI] [PubMed] [Google Scholar]
- 50. Ye R. W., et al. 1995. Anaerobic activation of the entire denitrification pathway in Pseudomonas aeruginosa requires Anr, an analog of Fnr. J. Bacteriol. 177:3606–3609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Yousef A. A., Jaffe A. 2010. The role of azithromycin in patients with cystic fibrosis. Paediatr. Respir. Rev. 11:108–114 [DOI] [PubMed] [Google Scholar]