Abstract
The metabolic processing of dehydrophos, a broad-spectrum peptide antibiotic containing an unusual vinyl-phosphonate moiety, was examined by using a panel of Salmonella enterica mutants deficient in peptide uptake and catabolism. Dehydrophos bioactivity is lost in opp tpp double mutants, demonstrating a requirement for uptake via nonspecific oligopeptide permeases. Dehydrophos bioactivity is also abolished in a quadruple Salmonella mutant lacking the genes encoding peptidases A, B, D, and N, showing that hydrolysis of the peptide bond is required for activity. 31P nuclear magnetic resonance spectroscopy was used to assess the fate of dehydrophos following in vitro digestion of the antibiotic with purified PepA. The results suggest that the initial product of peptidase processing is 1-aminovinyl-phosphonate O-methyl ester. This phosphonate analogue of dehydroalanine undergoes rearrangement to the more stable imine, followed by spontaneous hydrolysis to yield O-methyl-acetylphosphonate, a structural analogue of pyruvate. This compound is a known inhibitor of pyruvate dehydrogenase and pyruvate oxidase and is probably the active species responsible for dehydrophos bioactivity.
INTRODUCTION
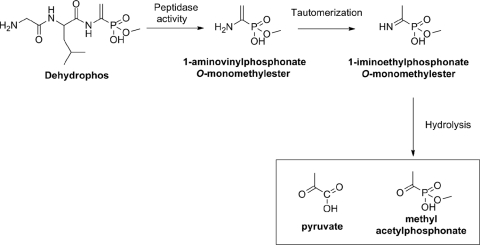
Dehydrophos (formerly A53868) is an unusual antibiotic produced by Streptomyces luridus. This compound has broad spectrum activity in vitro and is effective in vivo in Salmonella challenged chickens (15). Dehydrophos consists of a glycine-leucine dipeptide linked to an O-methylated aminovinylphosphonate (28) (Fig. 1). The cellular target of the compound has yet to be established, and it is unknown whether it acts as a tripeptide or whether it is metabolized by the target cell to release a bioactive product. Structure-activity relationships of several dehydrophos derivatives clearly revealed the importance of the O-methyl and vinyl moieties but did not allow conclusions to be made regarding whether peptide bond cleavage is required for bioactivity (16).
Fig. 1.
Trojan horse model for the bioactivity of dehydrophos. Cleavage of the peptide bonds in dehydrophos releases 1-aminovinylphosphonate O-monomethyl ester, which rearranges to the preferred imine. This imine is then subject to attack by a water molecule to yield methyl acetylphosphonate (MAP), a toxic pyruvate mimic.
The coupling of phosphonic acids to amino acids has been observed in numerous bioactive natural products. In the case of rhizocticin, plumbemycin, bialaphos, and phosalacine, these amino acids convert a toxic phosphonate molecule into an inactive “Trojan horse” that is readily taken up by target organisms. Once inside the cell, peptidase activity serves to release the bioactive phosphonate moiety (6, 17). In other cases the entire peptide is required for bioactivity. For example, the angiotensin converting enzyme inhibitor K-26 binds to its target as a tripeptide consisting of Ile-Tyr coupled to (R)-1-amino-2-(4-hydroxyphenyl)-ethylphosphonic acid (AHEP) (2, 23).
Two mutually exclusive mechanisms for dehydrophos bioactivity can be envisioned based on whether the peptide bonds are cleaved or remain intact. In reporting the revised structure of antibiotic, Whitteck et al. noted that cleavage of the peptide bond would produce an unstable enamine phosphonate, expected to undergo tautomeric rearrangement followed by spontaneous hydrolysis (Fig. 1) (28). The expected product of this transformation would be methyl acetylphosphonate (MAP), a structural analog of pyruvate and a known inhibitor of both pyruvate oxidase and pyruvate dehydrogenase (24). Alternatively, the vinylphosphonate moiety of dehydrophos could undergo a Michael-type addition to sulfhydryl-containing target enzymes in a reaction analogous to that which occurs during the inactivation of cysteine dependent proteases by vinyl sulfones (25). This mechanism would require that the peptide bond remain intact to prevent the rearrangement and hydrolysis reactions described above.
Both peptide transport and catabolism have been extensively characterized in Salmonella enterica, providing a facile experimental system to distinguish between the two models for dehydrophos action. S. enterica contains three distinct systems for peptide uptake encoded the opp, tpp, and dpp operons, which encode oligopeptide, tripeptide, and dipeptide permeases, respectively (10). The Opp permease is the primary peptide uptake system, transporting molecules containing up to 5 amino acids (11–14). The Tpp permease is relatively specific, preferring hydrophobic residues and requiring anaerobic conditions for expression (7, 16), whereas the Dpp permease transports dipeptide substrates and acts as the signal receptor for peptide chemotaxis (1). Mutant strains lacking all three permeases are unable to assimilate most peptides (10). Peptides are converted to free amino acids in S. enterica via the action of eight major peptidases, designated A, B, D, E, N, P, Q, and T, with overlapping specificities. Peptidases A, B, and D possess broad substrate specificities, with A and B belonging to the leucyl aminopeptidase family (19, 21). Peptidase N also has broad substrate specificity but belongs to the lysyl aminopeptidase family and appears subject to a more complex regulation scheme. The remaining peptidases display more restricted substrate ranges and/or are only expressed under certain conditions (20, 21).
We report here the permease- and peptidase-dependent nature of dehydrophos activity in S. enterica. Consistent with the “Trojan horse” model of activity, we also demonstrate in vitro peptidase-dependent generation of the toxic pyruvate mimic MAP and a derived metabolite, 1-hydroxyethylphosphonate monomethyl ester (1-HEP O-Me), following hydrolysis of dehydrophos. These data suggest that dehydrophos functions by generation of a pyruvate analogue capable of inhibiting multiple targets, a useful feature in minimizing bacterial antibiotic resistance.
MATERIALS AND METHODS
Bacterial strains, plasmids, and culture conditions.
The strains used in the present study are listed in Table 1. Streptomyces strains were propagated on ISP4 medium (Difco/Becton Dickinson Microbiology Systems, Sparks, MD) at 30°C unless otherwise noted. Antibiotic production cultures were inoculated with 50 μl of a spore suspension (∼109 CFU) and incubated in 500-ml baffled flasks containing 100 ml of nutrient broth (Difco) agitated at 250 rpm for 48 h prior to analysis of the supernatant. S. enterica was routinely grown in Luria-Bertani broth (LB) or on 1.5% agar LB plates supplemented with appropriate antibiotics. Antibiotics were used at the following concentrations: ampicillin, 100 μg/ml; chloramphenicol, 10 μg/ml; apramycin, 37.5 μg/ml; and kanamycin, 50 μg/ml.
Table 1.
Strains used in the study
| Straina | Genotype/relevant features | Source or reference |
|---|---|---|
| Salmonella enterica | ||
| TN215 | leu-485 pepA1 pepB1 pepD1 pepN1 | 22 |
| TN271 | leu-485 pepA1 pepB1 pepD1 (pepN+) | 22 |
| TN272 | leu-485 pepB1 pepD1 pepN1 (pepA+) | 22 |
| TN273 | leu-485 pepA1 pepD1 pepN1 (pepB+) | 22 |
| TN274 | leu-485 pepA1 pepB1 pepN1 (pepD+) | 22 |
| TN1379 | leuBCD485 | 9 |
| TN1890 | leuBCD485 oppA208 | C. G. Miller, unpublished data |
| TN2216 | leuBCD485 pepB11 pepN90 pepA16 supQ302 (proAB pepD) pepP1 pepQ1 pJG6 (pepA+) | J. Glazebrook and C. G. Miller, unpublished data |
| TN2271 | oppBC250leu::Tn10tppB9::Tn5 | 7 |
| Streptomyces lividans | ||
| 66 | Wild type | ARS culture collection |
| WM4467 | S. lividans 66 with a dehydrophos gene cluster fosmid integrated into the φC31 attB site | 5 |
| Streptomyces luridus NRRL 15101 | Native dehydrophos producer | ARS culture collection |
All of the S. enterica strains are derivatives of S. enterica serovar Typhimurium LT2, except for TN2271, which is a derivative of S. enterica serovar Typhimurium CH379.
Bioassay of dehydrophos growth inhibition.
Lawns of the appropriate Salmonella strain were made by plating 0.1 ml of an exponential-phase culture (optical density at 600 nm of ∼0.1) suspended in molten (∼40°C) E medium (26) top agar (0.5%) onto E medium agar (1.5%) plates containing succinate (40 mM) and leucine (50 μM). Agar cores were then taken from cultures of Streptomyces luridus NRRL 15101 grown for 48 h at 30°C on ISP4 plates. Alternatively, filter disks were spotted with 10 μl of an authentic standard dehydrophos (1 mg/ml). These cores and/or disks were then placed onto the freshly inoculated bioassay plates, incubated at 37°C for 12 h, and then scored for bioactivity.
Overexpression, purification, and activity assay of PepA.
PepA was purified by using a modified version of the Vogt purification of E. coli aminopeptidase I (27). A 500-ml culture of the peptidase A overproducer S. enterica TN2216 was grown for 16 h at 37°C, and the cells were then harvested by centrifugation at 5,000 × g for 10 min. The cell pellet was resuspended in 20 ml of ice-cold buffer 1 (50 mM Tris-HCl [pH 7.8], 200 mM KCl, 10 mM MgCl2, 100 mM EDTA, 100 mM dithiothreitol, and 5% glycerol) and disrupted by a single passage through a chilled French press cell at 20,000 lb/in2. Solid debris was removed by centrifugation at 13,000 × g for 30 min at 4°C. The resuspended cells were then placed into a 70°C water bath and continuously swirled until the temperature reached 65°C. Precipitated proteins were removed by centrifugation at 13,000 × g for 15 min at 4°C. The supernatant was dialyzed (Pierce, 10K MWCO) against 2 liters of 0.1 M phosphate buffer (pH 7.6) for 18 h at 4°C with one change of buffer. After dialysis, the extract was centrifuged again at 13,000 × g for 15 min at 4°C to remove insoluble material. The supernatant was then dialyzed against buffer 2 (20 mM Tris-HCl [pH 8.4], 10 mM KCl, 1 mM magnesium acetate, 0.1 mM EDTA) for 18 h. The precipitate that formed was collected and redissolved in buffer 3 (20 mM KHCO3 [pH 10.3], 50 mM KCl, 1 mM magnesium acetate, 0.1 mM EDTA). This precipitate failed to completely solubilize in buffer 3, leaving the solution turbid. PepA purity was evaluated by visual inspection of an SDS-PAGE gel stained with Coomassie brilliant blue (estimated to be at least 95% pure). PepA action on dehydrophos was assayed at 37°C over 18 h by direct addition of dehydrophos (7 mM) to dissolved PepA. The reaction was monitored by 31P nuclear magnetic resonance (NMR) spectroscopy.
31P NMR spectroscopy-based identification of dehydrophos metabolites and monitoring of the hydrolysis reaction.
Culture supernatant was passed through a 0.2-μm-pore-size filter, concentrated 20-fold by evaporation, and D2O was added to 25% as a lock solvent. 31P NMR spectra were acquired on a Varian Unity Inova 600 spectrophotometer equipped with a 5-mm AutoTuneX probe at the Varian Oxford Instrument Center for Excellence at the University of Illinois Urbana-Champaign. An external standard of 85% phosphoric acid was defined as 0 ppm. Experiments to confirm the identity of proposed metabolites were routinely performed by adding 1 to 5 mM standard to the sample and then reacquiring the spectra.
RESULTS
Dehydrophos is transported into S. enterica via the Opp and Tpp transporters.
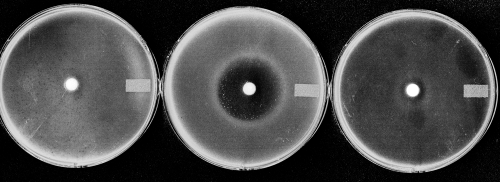
To determine which peptide transport systems are involved in the uptake of dehydrophos, we tested the ability of the antibiotic to inhibit the growth of S. enterica permease mutants. The mutants used are well-characterized strains that have been previously used to characterize peptide uptake in S. enterica (7). TN1379 encodes each of the known peptide permease systems, TN1890 contains both the Dpp and Tpp permease systems (opp mutant) and TN2271 contains only the Dpp system (opp tpp mutant). As expected, dehydrophos inhibits TN1379; however, it fails to inhibit TN2271 and shows only a very small zone of inhibition with TN1890 (Fig. 2). These results indicate that the Opp peptide transporter is the major transporter of dehydrophos, with the Tpp transporter playing a relatively minor role. Dpp is unable to transport dehydrophos. This result also shows that a cytoplasmic target or processing step is required for bioactivity.
Fig. 2.
The oligopeptide permease is required for dehydrophos activity. Bioassays were performed as described by spotting synthetic dehydrophos onto a filter disc and assaying growth inhibition against various Salmonella strains: strain TN1379 (center) contains the Opp, Dpp, and Tpp permease systems; strain TN2271 carries only the Dpp permeases (i.e., opp tpp mutant) (left); and strain TN1890 carries both the Dpp and the Tpp oligopeptide permeases (i.e., opp mutant) (right).
Dehydrophos action requires peptidase activity.
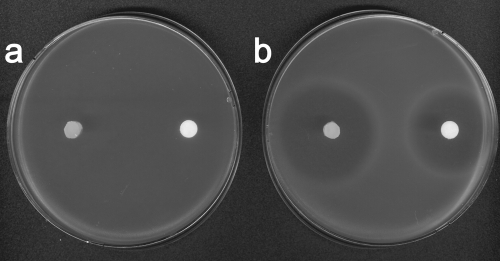
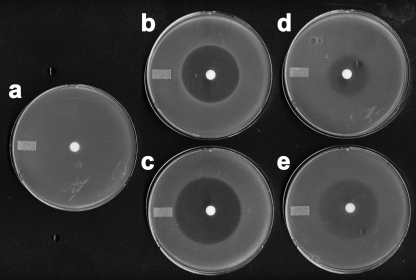
To test whether peptide bond cleavage was required for dehydrophos activity, we examined bioactivity against a series of characterized S. enterica peptidase mutants (22). A quadruple mutant lacking PepA, PepD, PepB, and PepN was resistant to dehydrophos, whereas the isogenic strain containing all of the missing peptidases was sensitive to the antibiotic, indicating that dehydrophos action is dependent on peptidase cleavage (Fig. 3). Strains carrying any one of the deleted peptidases were also sensitive to dehydrophos; however, the severity of the growth inhibition varied depending on the peptidase present. Strains carrying peptidase A or B displayed growth inhibition similar to the wild type, whereas a strain carrying peptidase D had a slightly smaller zone of inhibition, and a strain carrying only peptidase N had a significantly smaller zone of inhibition (Fig. 4).
Fig. 3.
The antibiotic activity of dehydrophos is peptidase dependent. (a) Dehydrophos fails to inhibit S. enterica TN215 lacking peptidases A, B, D, and N when applied as either an agar core from the native producer or as a pure synthetic compound. (b) In contrast, S. enterica TN1379 containing the full complement of peptidases exhibits the typically seen growth inhibition.
Fig. 4.
Reintroduction of any single peptidase allele is sufficient to restore dehydrophos bioactivity. Bioassay plates examining the effects of reintroduction of either PepA (strain TN272 [b]), PepB (strain TN273 [c]), PepN (strain TN271 [d]), PepD (strain TN274 [e]), or to a quadruple mutant of the aforementioned peptidases (strain TN271 [a]).
In vitro peptidase cleavage of dehydrophos by Salmonella peptidase A results in the generation of methyl acetylphosphonate.
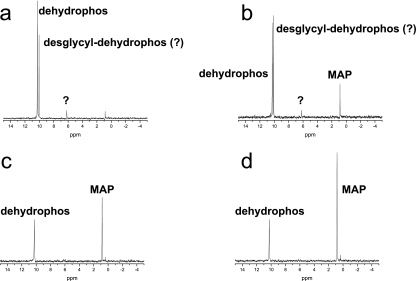
As described above, hydrolysis of the peptide bonds in dehydrophos is expected to yield methyl acetylphosphonate via a series of uncatalyzed chemical reactions. To test this prediction, we incubated synthetically prepared dehydrophos with purified PepA and monitored the products of the cleavage reaction with phosphorus NMR spectroscopy (Fig. 5). Under the conditions used, the reaction proceeded slowly, which allowed for the detection of discrete intermediates. After 1 h of incubation, four separate peaks were observed. Based on a spiking experiment with a synthetic standard, the peak with a chemical shift of 10.2 ppm corresponds to unaltered dehydrophos. The peak at 10 ppm is presumed to be dehydrophos lacking the N-terminal glycine (desglycyldehydrophos); however, this could not be rigorously established due to the lack of an appropriate standard. After 3 h of incubation, the dehydrophos peak decreased in intensity, while the putative desglycyldehydrophos peak increased. After overnight incubation, the desglycyldehydrophos peak disappeared, and the dehydrophos peak was greatly diminished. Two other peaks were seen in the spectra: one located at 6.2 ppm and the other located at 1 ppm. The peak at 6.2 ppm was transiently observed during the first two time points but was not observed after the overnight incubation, whereas the peak at 1 ppm increased in intensity over time and, after overnight incubation, represented the most abundant phosphorus-containing species in the reaction mixture. Spiking of the assay with a synthetic standard identified the 1 ppm peak as MAP. The 6.2-ppm peak was not identified.
Fig. 5.
PepA catalyzed the degradation of dehydrophos. A time course following the fate of dehydrophos, monitored by 31P NMR, incubated with purified S. enterica PepA is shown. (a) After 1 h, a large peak representing unreacted dehydrophos (10.2 ppm) is present, along with a substantial amount of the putative dipeptide (10 ppm). (b) After 3 h, the putative dipeptide peak dominates the spectrum, and the peak at 1 ppm has increased in intensity. (c) After an overnight incubation, only two peaks remain: a large peak at 1 ppm and a peak representing unreacted dehydrophos at 10.2 ppm. (d) The addition of a synthetic standard identifies the peak at 1 ppm as methylacetylphosphonate (MAP).
Dehydrophos bioactivity is not relieved by acetate.
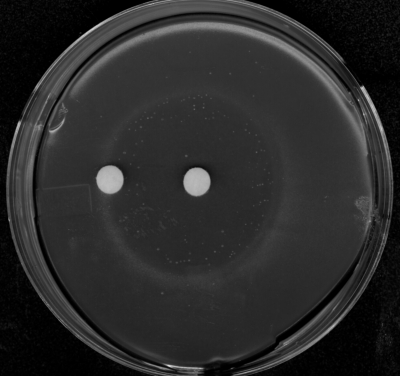
Because MAP is a known inhibitor of pyruvate dehydrogenase and pyruvate oxidase, we examined whether the bioactivity of dehydrophos could be relieved by supplementation with acetate, which is required for growth of pdh pox double mutants (4). To this end, we performed a bioassay in the presence of dehydrophos and acetate. The zone of inhibition remained unaltered, a finding consistent with the idea that dehydrophos has multiple, essential cellular targets (Fig. 6).
Fig. 6.
Acetate fails to relieve antibiotic inhibition generated by dehydrophos. The central filter disc was spotted with synthetic dehydrophos, while the outer disc contained 10 μl of a 40 mM acetate solution.
1-HEP O-Me is a product of dehydrophos breakdown.
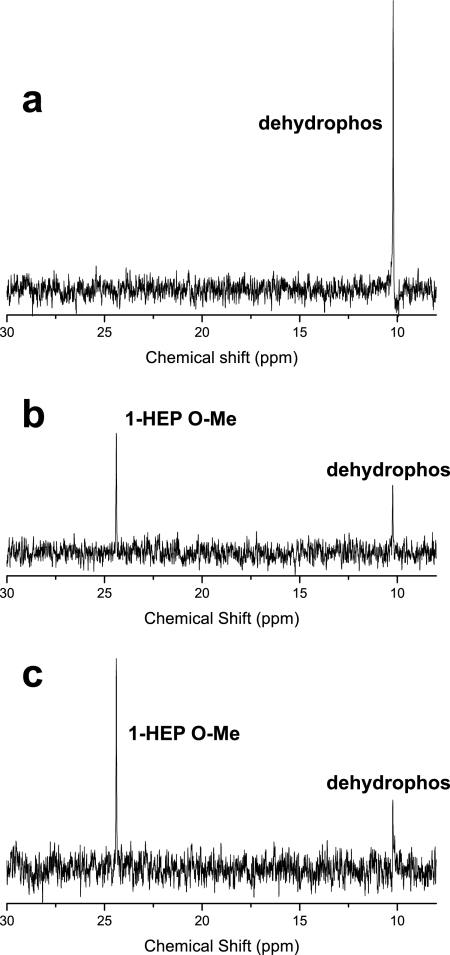
Early investigations into dehydrophos biosynthesis were confounded by the presence of an unidentified phosphonate observed by 31P NMR spectroscopy in the culture supernatants of native and heterologous dehydrophos producers (5). Our observation of MAP as the product of in vivo peptidase processing of dehydrophos suggested a potential source and identity of this unknown phosphonate. Spiking of samples containing the unidentified phosphonate with authentic standards showed that the unknown compound was not MAP (data not shown), leading to the hypothesis that cells might detoxify MAP via reduction to form 1-HEP O-Me, analogous to the thermodynamically favorable reduction of pyruvate to lactate by lactate dehydrogenase. To test this idea, we fed dehydrophos to Streptomyces lividans 66 and monitored the culture supernatant with phosphorus NMR spectroscopy. Over time, the phosphorus peak associated with dehydrophos diminishes in intensity corresponding with the appearance and growth of a second peak, which was confirmed to be 1-HEP O-Me by the addition of a synthetic standard (Fig. 7).
Fig. 7.
S. lividans 66 converts exogenous dehydrophos to 1-HEP O-Me. 31P NMR spectra demonstrate the fate of dehydrophos fed to S. lividans 66 over time. (a) Prior to inoculation, dehydrophos can be observed at a chemical shift of 10.2 ppm. (b) After the inoculation of S. lividans 66 and 48 h of growth, the peak corresponding to dehydrophos has decreased in intensity, and a second peak has appeared at 24.5 ppm. (c) The addition of a synthetic standard increased the intensity of the 24.5-ppm peak and confirmed its identity as 1-hydroxyethylphophonate O-methyl ester (1-HEP O-Me).
DISCUSSION
Many known phosphonate antibiotics utilize short peptides as a “Trojan horse” to obtain access into the cellular environment. The data presented here clearly indicate that dehydrophos can be added to this list. The in vivo requirement for both peptide transport and hydrolysis, coupled with the results of in vitro peptidase digestion, strongly suggest that the bioactive agent derived from dehydrophos is MAP, a structural analog of pyruvate. MAP is a known inhibitor of both pyruvate dehydrogenase and pyruvate oxidase. With a Ki of 5 × 10−8 M, MAP binds to pyruvate dehydrogenase 10,000 times better than the natural substrate, pyruvate (24). MAP has relatively modest Ki for pyruvate oxidase (1 mM, pH 7.0), which is a 10-fold lower affinity than the enzyme has for its natural substrate pyruvate (8, 24). Mutants lacking both pyruvate dehydrogenase and pyruvate oxidase are incapable of growth unless the media are supplemented with acetate, potentially explaining the mechanisms of growth inhibition by dehydrophos. Nevertheless, these two enzymes cannot be the sole targets of dehydrophos because supplementation of the media with acetate fails to relieve growth inhibition. This finding is not altogether surprising given the number of essential cellular processes that involve pyruvate. Indeed, while MAP is best known as a potent pyruvate dehydrogenase inhibitor, it has also been shown to inhibit other pyruvate-utilizing enzymes (24).
Our model for in vivo production of MAP involves enzymatic cleavage to produce an unstable compound that undergoes spontaneous chemical conversion to the pyruvate analog. Interestingly, our 31P NMR data show the transient accumulation of a phosphonate intermediate at 6.2 ppm that could correspond to the unstable intermediate, which could be either 1-aminovinylphosphonate O-methyl ester or 1-iminovinylphosphonate O-methyl ester (Fig. 1). Although we could not rigorously establish the identity of this compound, we believe the intermediate is probably the latter compound because known examples of enamine/imine tautomerization occur rapidly and favor the imine form (18). Regardless of which intermediate accumulates, the observation of this transient peak in the 31P NMR spectrum raises interesting questions regarding the mode of action of dehydrophos. It is possible that one of the transient reactive intermediates produced during dehydrophos catabolism might contribute to the toxicity of dehydrophos. Importantly, these data suggest that the peptide linkages have a dual role in the antibiotic activity of dehydrophos. Not only do they promote uptake of the inactive tripeptide, they also stabilize an unstable phosphonate moiety by preventing the enamine/imine tautomerization of the vinyl amine functional group.
Finally, it should be noted that our results suggest that there is significant substrate specificity in both the uptake and processing of dehydrophos in S. enterica. These findings are reminiscent of the specificity seen in other phosphonate tripeptide antibiotics. For example, rhizocticin and plumbemycin contain the same bioactive component but differ in the amino acid side chains. Interestingly, rhizocticin is an antifungal compound with little activity toward bacteria, whereas plumbemycin is antibacterial, with little activity toward fungi. It has been proposed that the differences in bioactivity are a reflection of substrate specificity in either uptake or processing (3, 6). This observation has significant implications in the development and design of new, more specific antibiotics. Given the varied nature of oligopeptide uptake and cleavage, an intriguing idea would be the attachment of small molecule inhibitors to multiple peptide scaffolds. Applied appropriately, this could open the door for the treatment of specific infections with specific antibiotics, while avoiding the deleterious effects on the natural host-associated microbial community.
ACKNOWLEDGMENTS
This study was supported by the National Institutes of Health (GM PO1 GM077596). B.T.C. was supported by a National Institutes of Health Chemistry-Biology Interface Training Program (GM070421).
The contents of this study are solely the responsibility of the authors and do not necessarily represent the official views of the NIGMS or NIH.
Footnotes
Published ahead of print on 2 May 2011.
REFERENCES
- 1. Abouhamad W. N., Manson M., Gibson M. M., Higgins C. F. 1991. Peptide transport and chemotaxis in Escherichia coli and Salmonella typhimurium: characterization of the dipeptide permease (Dpp) and the dipeptide-binding protein. Mol. Microbiol. 5:1035–1047 [DOI] [PubMed] [Google Scholar]
- 2. Akif M., et al. 2010. Crystal structure of a phosphonotripeptide K-26 in complex with angiotensin converting enzyme homologue (AnCE) from Drosophila melanogaster. Biochem. Biophys. Res. Commun. 398:532–536 [DOI] [PubMed] [Google Scholar]
- 3. Borisova S. A., Circello B. T., Zhang J. K., A. van der Donk W., Metcalf W. W. 2010. Biosynthesis of rhizocticins, antifungal phosphonate oligopeptides produced by Bacillus subtilis ATCC 6633. Chem. Biol. 17:28–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Chang Y. Y., Cronan J. E., Jr. 1983. Genetic and biochemical analyses of Escherichia coli strains having a mutation in the structural gene (poxB) for pyruvate oxidase. J. Bacteriol. 154:756–762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Circello B. T., Eliot A. C., Lee J. H., A. van der Donk W., Metcalf W. W. 2010. Molecular cloning and heterologous expression of the dehydrophos biosynthetic gene cluster. Chem. Biol. 17:402–411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Diddens H., Zahner H., Kraas E., Gohring W., Jung G. 1976. On the transport of tripeptide antibiotics in bacteria. Eur. J. Biochem. 66:11–23 [DOI] [PubMed] [Google Scholar]
- 7. Gibson M. M., Price M., Higgins C. F. 1984. Genetic characterization and molecular cloning of the tripeptide permease (tpp) genes of Salmonella typhimurium. J. Bacteriol. 160:122–130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Hager L. P. 1957. Trypsin activation of a ferricyanide-linked pyruvic acid oxidation. J. Biol. Chem. 229:251–263 [PubMed] [Google Scholar]
- 9. Hamilton S., Miller C. G. 1992. Cloning and nucleotide sequence of the Salmonella typhimurium dcp gene encoding dipeptidyl carboxypeptidase. J. Bacteriol. 174:1626–1630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Higgins C. F., Gibson M. M. 1986. Peptide transport in bacteria. Methods Enzymol. 125:365–377 [DOI] [PubMed] [Google Scholar]
- 11. Hiles I. D., Gallagher M. P., Jamieson D. J., Higgins C. F. 1987. Molecular characterization of the oligopeptide permease of Salmonella typhimurium. J. Mol. Biol. 195:125–142 [DOI] [PubMed] [Google Scholar]
- 12. Hiles I. D., Powell L. M., Higgins C. F. 1987. Peptide transport in Salmonella typhimurium: molecular cloning and characterization of the oligopeptide permease genes. Mol. Gen. Genet. 206:101–109 [DOI] [PubMed] [Google Scholar]
- 13. Hogarth B. G., Higgins C. F. 1983. Genetic organization of the oligopeptide permease (opp) locus of Salmonella typhimurium and Escherichia coli. J. Bacteriol. 153:1548–1551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Jamieson D. J., Higgins C. F. 1984. Anaerobic and leucine-dependent expression of a peptide transport gene in Salmonella typhimurium. J. Bacteriol. 160:131–136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Johnson R. D., Kastner R. M., Larsen S. H., Ose E. E. 5 April 1984. Antibiotic A53868 and process for production thereof. U.S. patent 4,482,488
- 16. Kuemin M., van der Donk W. A. 2010. Structure-activity relationships of the phosphonate antibiotic dehydrophos. Chem. Commun. (Camb.) 46:7694–7696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Laber B., Lindell S. D., Pohlenz H. D. 1994. Inactivation of Escherichia coli threonine synthase by dl-Z-2-amino-5-phosphono-3-pentenoic acid. Arch. Microbiol. 161:400–403 [DOI] [PubMed] [Google Scholar]
- 18. Lu S. P., Lewin A. H. 1998. Enamine/imine tautomerism in α,β-unsaturated-α-amino acids. Tetrahedron 54:15097–15104 [Google Scholar]
- 19. Mathew Z., Knox T. M., Miller C. G. 2000. Salmonella enterica serovar Typhimurium peptidase B is a leucyl aminopeptidase with specificity for acidic amino acids. J. Bacteriol. 182:3383–3393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. McHugh G. L., Miller C. G. 1974. Isolation and characterization of proline peptidase mutants of Salmonella typhimurium. J. Bacteriol. 120:364–371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Miller C. G. 1975. Peptidases and proteases of Escherichia coli and Salmonella typhimurium. Annu. Rev. Microbiol. 29:485–504 [DOI] [PubMed] [Google Scholar]
- 22. Miller C. G., Mackinnon K. 1974. Peptidase mutants of Salmonella typhimurium. J. Bacteriol. 120:355–363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ntai I., Manier M. L., Hachey D. L., Bachmann B. O. 2005. Biosynthetic origins of C-P bond containing tripeptide K-26. Organic Lett. 7:2763–2765 [DOI] [PubMed] [Google Scholar]
- 24. O'Brien T. A., Kluger R., Pike D. C., Gennis R. B. 1980. Phosphonate analogues of pyruvate: probes of substrate binding to pyruvate oxidase and other thiamin pyrophosphate-dependent decarboxylases. Biochim. Biophys. Acta 613:10–17 [DOI] [PubMed] [Google Scholar]
- 25. Rosenthal P. J., et al. 1996. Antimalarial effects of vinyl sulfone cysteine proteinase inhibitors. Antimicrob. Agents Chemother. 40:1600–1603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Vogel H. J., Bonner D. M. 1956. Acetylornithinase of Escherichia coli: partial purification and some properties. J. Biol. Chem. 218:97–106 [PubMed] [Google Scholar]
- 27. Vogt V. M. 1970. Purification and properties of an aminopeptidase from Escherichia coli. J. Biol. Chem. 245:4760–4769 [PubMed] [Google Scholar]
- 28. Whitteck J. T., et al. 2007. Reassignment of the structure of the antibiotic A53868 reveals an unusual amino dehydrophosphonic acid. Angew Chem. Int. Ed. Engl. 46:9089–9092 [DOI] [PubMed] [Google Scholar]