Abstract
Currently, CTX-M β-lactamases are among the most prevalent and most heterogeneous extended-spectrum β-lactamases (ESBLs). In general, CTX-M enzymes are susceptible to inhibition by β-lactamase inhibitors. However, it is unknown if the pathway to inhibition by β-lactamase inhibitors for CTX-M ESBLs is similar to TEM and SHV β-lactamases and why bacteria possessing only CTX-M ESBLs are so susceptible to carbapenems. Here, we have performed a kinetic analysis and timed electrospray ionization mass spectrometry (ESI-MS) studies to reveal the intermediates of inhibition of CTX-M-9, an ESBL representative of this family of enzymes. CTX-M-9 β-lactamase was inactivated by sulbactam, tazobactam, clavulanate, meropenem, doripenem, ertapenem, and a 6-methylidene penem, penem 1. Ki values ranged from 1.6 ± 0.3 μM (mean ± standard error) for tazobactam to 0.02 ± 0.01 μM for penem 1. Before and after tryptic digestion of the CTX-M-9 β-lactamase apo-enzyme and CTX-M-9 inactivation by inhibitors (meropenem, clavulanate, sulbactam, tazobactam, and penem 1), ESI-MS and matrix-assisted laser desorption ionization–time of flight mass spectrometry (MALDI-TOF MS) identified different adducts attached to the peptide containing the active site Ser70 (+52, 70, 88, and 156 ± 3 atomic mass units). This study shows that a multistep inhibition pathway results from modification or fragmentation with clavulanate, sulbactam, and tazobactam, while a single acyl enzyme intermediate is detected when meropenem and penem 1 inactivate CTX-M-9 β-lactamase. More generally, we propose that Arg276 in CTX-M-9 plays an essential role in the recognition of the C3 carboxylate of inhibitors and that the localization of this positive charge to a “region of the active site” rather than a specific residue represents an important evolutionary strategy used by β-lactamases.
INTRODUCTION
CTX-M enzymes are becoming one of the most prevalent extended-spectrum β-lactamases (ESBLs) (3, 8, 9, 30–32) in the world. The widespread dissemination of CTX-M β-lactamases, especially Escherichia coli ST131 possessing CTX-M-15, has had a significant impact on the treatment of hospital- and community-acquired infections caused by E. coli and other enteric bacilli (6, 7, 13, 23, 36, 41, 44–49, 55, 59).
As class A family β-lactamases, CTX-Ms are the most genetically heterogeneous (5 major divisions, CTX-M-1, -2, -8, -25, and -9-like groups) (1, 24, 35, 44–46, 58, 60). Most CTX-M enzymes expressed in E. coli provide a high level of resistance to the oxyimino-cephalosporins, cefotaxime and ceftriaxone, and variable levels of resistance to cefepime and cefpirome (3, 43). Depending on the type of CTX-M expressed by the isolates, the MICs of ceftazidime are also increased (43). In addition, the MICs of combinations of clavulanate with amoxicillin or ticarcillin vary, and in some cases, low-level resistance has been observed (3).
As a consequence of their clinical importance, the reaction mechanism of CTX-M ESBLs has been the subject of intense study (10–12, 14, 16, 42). However, the molecular properties and characteristics of CTX-M that determine the level of susceptibility and resistance to β-lactam–β-lactamase inhibitor combinations and carbapenems are still unknown. Of the currently available inhibitors, tazobactam is the most active (50% inhibitory concentrations [IC50s] are 2 to 10 nM for tazobactam and 9 to 90 nM for clavulanate), and sulbactam is the least active (IC50s are 0.1 to 4.5 μM) (3).
In order to develop more effective and broader-spectrum β-lactamase inhibitors (18), detailed kinetic and biochemical measurements are needed to reveal the important intermediates in the inactivation of the CTX-M family. In TEM-1 and SHV-1, Arg244 is important in the mechanism of inactivation of carbapenems (imipenem and meropenem), clavulanic acid, sulbactam, and tazobactam (27, 28, 51, 53, 63). CTX-M-9 does not contain Arg244, and mutagenesis of a potential corresponding position, Arg276, does not firmly establish this amino acid as an “Arg244 equivalent” (42). Given the differences among class A enzymes, we wondered what the intermediates of inactivation by inhibitors would be for CTX-M-9. To answer this question, we examined the inactivation of CTX-M-9 β-lactamase with sulbactam, tazobactam, clavulanate, meropenem, doripenem, ertapenem, and a potent 6-methylidene penem (here referred to as penem 1) to gain deeper insight into the nature of β-lactamase inhibition in the CTX-M-9 β-lactamase. We chose penem 1, as it contains a bicyclic heterocycle which adopts the Z configuration at the C6 position and its chemistry has previously been shown to enhance affinity toward TEM-1, SHV-1, GC1, and class D OXA-1 β-lactamases (2, 37, 57). Penem 1 also contains certain chemical features that mimic carbapenems (a double bond between C2 and C3) (Fig. 1). Our analysis of the inactivation of CTX-M-9 indicates that a multistep inhibition mechanism is active for clavulanate, sulbactam, and tazobactam. In contrast, a single acyl enzyme intermediate was detected when meropenem and penem 1 were studied. In addition, we also discovered the key role Arg276 plays in the process of substrate/inhibitor recognition.
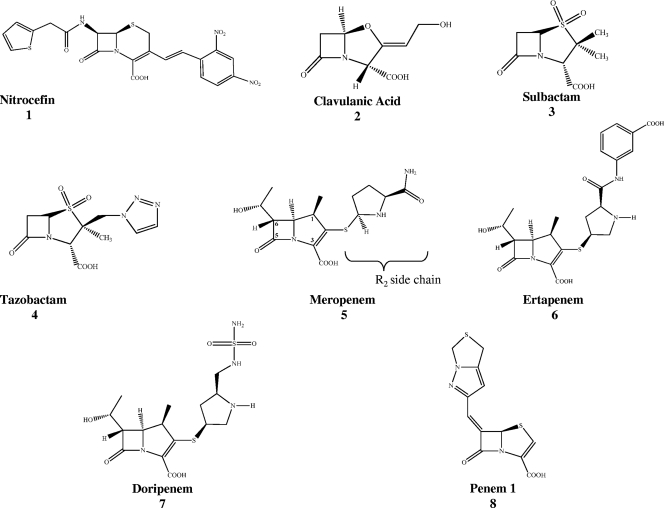
Fig. 1.
Chemical structures of indicator substrate (nitrocefin [1]) and β-lactamase inhibitors (clavulanate [2], sulbactam [3], tazobactam [4], meropenem [5; with the R2 side chain at the C2 position], ertapenem [6], doripenem [7], and penem 1 [8]) used in this study. The C atom numbering system is shown for meropenem.
MATERIALS AND METHODS
Cloning and β-lactamase purification.
The blaCTX-M-9 gene was cloned into the pET9a (+) plasmid vector (Novagen) as described previously and was maintained in E. coli BL21(DE3) cells (11, 16).
E. coli BL21(DE3) cells (Novagen) containing the pET 9a (+) expression vector were grown in superoptimal broth (SOB) containing 50 μg/ml kanamycin (11, 16). These cells were grown with agitation at 37°C until an optical density at 600 nm of 0.8 was attained. After addition of 0.2 mM isopropyl-β-d-thiogalactopyranoside (IPTG; Sigma), the cultures were grown for three additional hours. The cells were then centrifuged and the pellet was frozen. The next day, E. coli BL21(DE3) cells were lysed using lysozyme (Sigma); the β-lactamase was purified by performing preparative isoelectric focusing on a Multiphor II apparatus (GE Healthcare) according to a method described by Hujer et al. and Nukaga et al. (26, 39). Further purification was performed using size exclusion chromatography with a Pharmacia ÄKTA purifier system (GE Healthcare). We employed a HiLoad 16/60 Superdex 75 column (GE Healthcare) and eluted with 10 mM phosphate-buffered saline (PBS, pH 7.4). We determined the protein concentration by using Bio-Rad's protein assay with bovine serum albumin as a standard. β-Lactamase purity was assessed on a 5% stacking and 12% resolving sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) gel. After electrophoresis, SDS-PAGE gels were stained with Coomassie blue R250 (Fisher) to visualize the CTX-M-9 β-lactamase.
Kinetic experiments.
Antibiotics that were used as substrates and inhibitors for kinetic experiments were obtained from commercial suppliers, and their chemical structures are represented in Fig. 1. Nitrocefin (NCF) was purchased from Becton Dickinson, and clavulanic acid was a kind gift from GlaxoSmithKline. Tazobactam and penem 1 were obtained from Wyeth Pharmaceuticals, and sulbactam was donated by Pfizer. We obtained meropenem from AstraZeneca, doripenem from Johnson and Johnson, and ertapenem from Merck.
The kinetic parameters of hydrolysis and inhibition of CTX-M-9 β-lactamase were determined by continuous assays at room temperature (RT) using a diode array spectrophotometer (Agilent model 8453) in the manner described previously (22, 53). Each assay was performed in a 1-ml quartz cuvette with 10 mM phosphate-buffered saline at pH 7.4. Measurements were obtained using NCF (Δε482 of 17,400 M−1 cm−1).
The kinetic parameters Vmax and Km were obtained based on a nonlinear least squares fit of the data (Henri-Michaelis equation) using Enzfitter (Biosoft Corporation) according to equation 1:
| (1) |
In the experiments to determine Ki, each inhibitor (or “slow substrate”) was regarded as a “competitive inhibitor.” Therefore, Ki values were determined in a direct competition assay with NCF. In each case, initial velocities, v, were measured using a constant concentration of enzyme ([E]) and increasing concentrations of inhibitor ([I]) against the indicator substrate NCF [S]. After the Ki was obtained, the data were “corrected” based on equation 2 to account for the affinity of NCF for CTX-M-9, as follows:
| (2) |
The first-order rate constant for maximal inhibitor complex inactivation (kinact) was obtained directly by monitoring the reaction time course (t = 0 to 900 s) in the presence of inhibitors. A fixed concentration of enzyme, NCF, and increasing nanomolar concentrations of inactivator were used in each assay. The kobs for inactivation was determined using Origin 7.5 (OriginLab Co.) and equation 3. Each kobs was plotted as a function of inhibitor concentration and fit to equation 4 to determine kinact.
| (3) |
| (4) |
From the measurement of kinact, KI was determined (22, 53).
The turnover number (tn) or the partitioning of the initial enzyme-inhibitor complex between hydrolysis and enzyme inactivation, kcat/kinact (partition ratio), was determined as previously reported (2, 17, 53).
UV difference spectroscopy.
UV difference spectroscopy was performed to determine time-dependent absorption spectra for inhibitor that reacted with CTX-M-9 β-lactamase and was measured at wavelengths of 200 to 400 nm.
ESI-MS.
Electrospray ionization mass spectrometry (ESI-MS) of the intact CTX-M-9 β-lactamase inactivated by inhibitors was performed on an Applied Biosystems Q-STAR XL quadrupole time of flight mass spectrometer equipped with a nanospray source (Center for Proteomics and Bioinformatics, School of Medicine, Case Western Reserve University) as described previously (40, 53). Experiments were performed by desalting the reaction mixtures using C18 ZipTips (Millipore) according to the manufacturer's protocol. The protein sample was diluted with 50% acetonitrile–0.1% trifluoroacetic acid to a concentration of 10 μM. This protein solution was infused at a rate of 0.5 μl/min, and data were collected for 2 min. Spectra were deconvoluted using the Applied Biosystems Analyst program.
Tryptic digestion.
Proteolytic digestions were performed by adding trypsin (Sigma) to a solution of CTX-M-9 β-lactamase at a ratio of 1:25 (trypsin weight to protein weight). Tryptic digestions were carried out for 18 h at RT and terminated by the addition of a 1:10 volume of 1% trifluoroacetic acid. This solution was immediately desalted and concentrated by using a C18 ZipTip (Millipore), according to the manufacturer's protocol. The resulting digestion was analyzed by matrix-assisted laser desorption ionization–time of flight mass spectrometry (MALDI-TOF MS).
Molecular representations.
The crystal coordinates of CTX-M-9 (PDB accession code 2P74) were used to construct acyl enzyme models of meropenem and penem 1 in the active site of the wild-type (WT) β-lactamase using the Discovery Studio 2.1 (DS 2.1; Accelrys, Inc.) molecular modeling software.
The CTX-M-9 crystallographic structure was prepared for molecular docking by removing the crystallographic waters and minimizing the structure. The meropenem and penem 1 structures were constructed using Fragment Builder Tools and were minimized using a standard dynamics cascade protocol of DS 2.1. The minimized structures were used to generate a model for the acyl enzyme complexes. The models of meropenem and penem 1 in complex with the CTX-M-9 enzyme were developed using the flexible docking module of DS 2.1. This protocol was chosen because it allows for the docking of “flexible ligands” (i.e., meropenem and penem 1) in CTX-M-9, where the side chains of selected active site residues are allowed to move during docking. The low-energy ligand conformations were docked into the active site of CTX-M-9, and the structure was further refined using the ChiRotor algorithm. A final simulated annealing and energy minimization of each ligand pose was performed using the program CDOCKER (61).
The molecule was immersed in a water box, 7 Å from any face of the box, using the Solvation module of DS 2.1 with explicit periodic boundary conditions (PBC). All models were further minimized and equilibrated using a standard dynamic cascade protocol (minimization with steepest descent and minimization with conjugate gradient followed by a 6-ps dynamic simulation with heating, equilibration, and production steps).
All energy minimizations and molecular dynamics simulations (MDS) of the enzyme and enzyme complexes were carried out using force field parameters of CHARMm. The Particle Mesh Ewald (PME) method was used to take into account long-range electrostatics, and the bonds that involved hydrogen atoms were constrained with the SHAKE algorithm.
RESULTS AND DISCUSSION
Inhibition of CTX-M-9 β-lactamase.
In Table 1, we summarize our kinetic analysis of CTX-M-9 β-lactamase. CTX-M-9 hydrolyzes NCF with a catalytic efficiency (kcat/Km) of 8.0 ± 0.2 μM−1 s−1 (mean ± standard error); thus, NCF is used as a sensitive indicator substrate to characterize the inactivation of CTX-M-9 β-lactamase. As reported in Table 1, each commercially available inhibitor demonstrated comparable Ki values for CTX-M-9 β-lactamase (range, 0.8 to 1.6 μM). Moreover, the partition ratios (kcat/kinact or tn) were ≤50 for clavulanic acid, sulbactam, and tazobactam. These values are consistent with the increased susceptibility of ESBL-type enzymes to β-lactamase inhibitors (lower MICs in susceptibility testing and low Ki values when testing enzymes) and indicate a diminished ability to hydrolyze mechanism-based inactivators (lower kcat/kinact ratio and lower tn). This kinetic property (i.e., diminished turnover of inhibitors) is important in the inactivation pathway of CTX-M-9 (Fig. 2A to F).
Table 1.
Kinetic parameters of inhibition of CTX-M-9 β-lactamasea
| Compound | Ki (μM) | KI (μM) | kinact (s−1) | kinact/KI (μM−1 s−1) | tn |
|---|---|---|---|---|---|
| Clavulanate | 0.8 ± 0.1 | 0.60 ± 0.08 | 0.034 ± 0.001 | 0.06 ± 0.01 | 50 |
| Sulbactam | 1.2 ± 0.3 | 0.83 ± 0.07 | 0.09 ± 0.01 | 0.11 ± 0.02 | 40 |
| Tazobactam | 1.6 ± 0.3 | 1.6 ± 0.2 | 0.12 ± 0.01 | 0.08 ± 0.01 | 4 |
| Doripenem | 1.5 ± 0.1 | 1.3 ± 0.2 | 0.12 ± 0.01 | 0.09 ± 0.01 | 4 |
| Meropenem | 0.4 ± 0.1 | 0.50 ± 0.06 | 0.11 ± 0.01 | 0.22 ± 0.03 | 4 |
| Ertapenem | 0.4 ± 0.1 | 0.56 ± 0.10 | 0.21 ± 0.02 | 0.38 ± 0.08 | 4 |
| Penem 1 | 0.02 ± 0.01 | 0.031 ± 0.005 | 0.16 ± 0.01 | 5.16 ± 0.88 | 1 |
NCF was used as an indicator substrate. The Km of NCF was 19.4 ± 0.5 μM, and the kcat was 156.0 ± 2.0 s−1. The resulting catalytic efficiency, kcat/ Km, was 8.0 ± 0.2 μM−1 s−1.
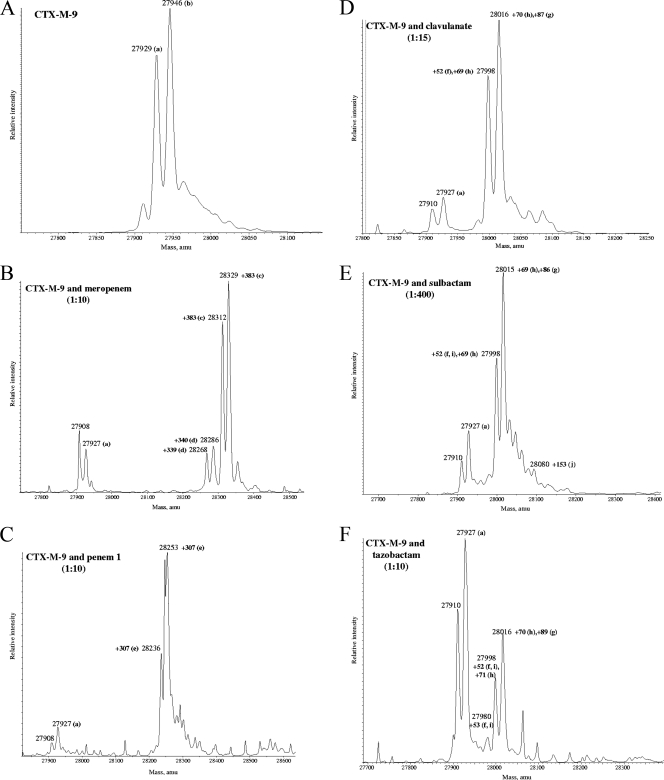
Fig. 2.
Deconvoluted mass spectra of CTX-M-9 β-lactamase (E, apo-enzyme) and CTX-M-9 β-lactamase with inhibitors (I) in a variable I/E ratio. Mass increases (products of inactivation) are shown. (A) CTX-M-9 β-lactamase alone; (B) CTX-M-9 with meropenem; (C) CTX-M-9 with penem 1; (D) CTX-M-9 β-lactamase with clavulanate; (E) CTX-M-9 β-lactamase with sulbactam; (F) CTX-M-9 with tazobactam. All measurements have an error of ±3 amu. For the commercially available inhibitors (clavulanate, sulbactam, and tazobactam), multiple products are shown.
In accordance with our investigations of meropenem with SHV-1 β-lactamase and with studies of carbapenems with class A β-lactamases (34, 38, 51, 63), we detected little hydrolysis of the carbapenems by CTX-M-9 (tn = 4). Among these compounds, meropenem and ertapenem displayed low Ki values (400 ± 100 nM). Compared to these two carbapenems, the doripenem Ki was 1.5 ± 0.1 μM. Penem 1 was the best inhibitor of CTX-M-9, with a Ki of 20 ± 1 nM and a partition ratio (kcat/kinact) of 1.
ESI-MS and MALDI-TOF.
To achieve insight as to why the inhibitors were potent inactivators of the CTX-M-9 β-lactamase, CTX-M-9 was reacted with clavulanate, sulbactam, tazobactam, penem 1, and meropenem (meropenem was chosen as a “representative” carbapenem), and the products of inactivation were determined using timed (15-min incubation) ESI-MS (Fig. 2 and 3 and Table 2). Analysis of the apo-enzyme by ESI-MS revealed two distinct species: 27,929 ± 3 atomic mass units (amu) and 27,946 ± 3 amu (Fig. 2A). As previously described using X-ray crystallography, these two isoforms represent the mature form of CTX-M-9 without the signal peptide and the same species with modification of the Glu at the N terminus (11). We note that the formation of pyroglutamate at the N-terminal glutamine stands as a unique observation among the class A β-lactamases (Fig. 3a).
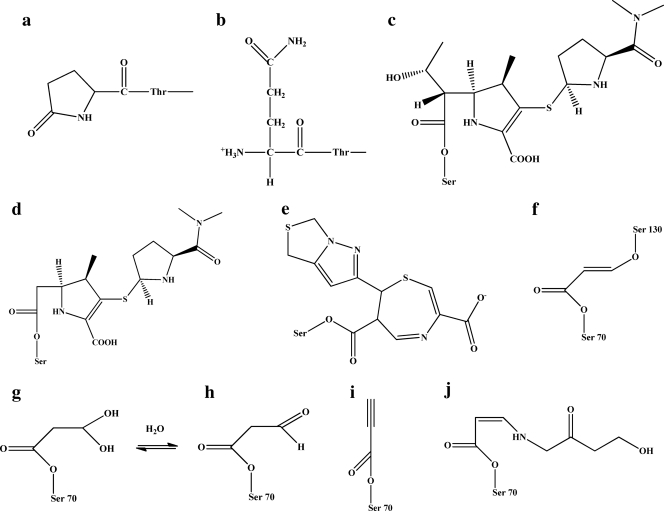
Fig. 3.
Postulated chemical structures of products of inactivation shown in Fig. 2. (a and b) The two forms of the N terminus of CTX-M-9. Panel a shows the pyroglutamate form. (c and d) The two forms of meropenem as the acyl enzyme (panel d shows the meropenem minus the ethoxy group from the C6 position). (e) The penem 1 acyl enzyme. In the case of the 52 ± 3 Da adduct shown in Fig. 2D, E, and F, we assigned the structure represented in panel i as the possible adduct for the structures shown in Fig. 2E and F (propynyl group). (f) It is also possible that the chemical moiety represented is present and represented in intermediates shown in Fig. 2D, E, and F. (g and h) The semialdlehye and aldehyde, respectively. (j) The 155-Da adduct.
Table 2.
Mass spectrometry analysis of CTX-M-9 β-lactamase alone and after incubation with inhibitors for 15 min
| Combination (inhibitor mass [Da]) | Mass of product (amu ± 3) |
||
|---|---|---|---|
| WT | Pyro-Glu-CTX-M-9 | Δa | |
| CTX-M-9 | 27,946 | 27,929 | |
| CTX-M-9 + clavulanate (199) | 27,998 | 52 | |
| 28,016 | 27,998 | 70b | |
| 28,034 | 28,016 | 88b | |
| 28,085 | 158 | ||
| CTX-M-9 + sulbactam (233) | 27,998 | 52 | |
| 28,015 | 27,998 | 70b | |
| 28,031 | 28,015 | 88b | |
| 28,080 | 156 | ||
| CTX-M-9 + tazobactam (300) | 27,998 | 27,980 | 52b |
| 28,016 | 27,998 | 70b | |
| 28,016 | 88 | ||
| CTX-M-9 + meropenem (383) | 28,329 | 28,312 | 383b |
| 28,286 | 28,268 | 340b | |
| CTX-M-9 + penem 1 (306) | 28,253 | 28,236 | 307 |
| 307 | |||
Mass of adducts formed during the inactivation of CTX-M-9 (amu).
The adduct can be attached to WT or pyro-Glu-CTX-M-9.
During the inactivation of CTX-M-9 by clavulanate, sulbactam, and tazobactam, we observed that many adducts were formed (Fig. 2D to F and 3f to j; Table 2). These adducts are indicative of a multistep pathway toward inactivation with several intermediates (possible formation of an imine or enamine [cis and/or trans], as well as inhibitor fragmentation [propynyl, +52 amu; aldehyde, +70 amu; semialdehyde, +88 amu; decarboxylated trans-enamine, +156] ± 3 amu) (22, 25, 50, 52, 53).
In contrast, meropenem (Fig. 2B) and penem 1 (Fig. 2C) formed a single species in inactivating CTX-M-9 β-lactamase. The major products observed were Δ +383 ± 3 amu and Δ +307 ± 3 amu, respectively (matching the molecular masses of the unfragmented meropenem and penem 1). In the case of meropenem, we also observed an adduct that was 43 ± 3 amu less than the main species (Δ +340 ± 3 Da). This secondary product is reminiscent of what has been seen in other ESI-MS analyses of inactivation of class A and class C enzymes by carbapenems, and we suspect it represents loss of the C6 ethoxy group (17, 20, 25).
Tryptic digestion.
A theoretical enzymatic digestion of the CTX-M-9 β-lactamase with trypsin (http://prospector.ucsf.edu/prospector/4.0.7/htmL/msdigest.htm) served as a guide for interpretation of the tryptic digestion of the CTX-M-9 β-lactamase before and after inactivation with meropenem, penem 1, clavulanate, sulbactam, and tazobactam. The unmatched fragments were compared to the matched fragments, and a series of candidate peptides were identified. Our results are summarized in the supplemental material.
Using MALDI-TOF MS, we detected peptides with amu increases (suggesting covalent attachment of the inhibitors) in the fragments containing the active site Ser70 (see the supplemental material). This finding supports the main reaction chemistry and that of the inhibitors involving the active site nucleophile Ser70, as identified with TEM-2 and clavulanate (4), TEM-1 with tazobactam (62), and PC1 with tazobactam (62) and as shown in the crystal structure and mass spectrometry analysis of SHV-1 with tazobactam and clavulanate (29, 50, 52, 53). We did not find definitive evidence that there was a resulting fragment that represented the “bridging species” connecting Ser70 to Ser130, but we did find a 52 ± 3 amu adduct (propynyl addition) when sulbactam and tazobactam inactivated CTX-M-9 via ESI-MS that may have served as such (Fig. 2 and 3 and Table 2).
UV difference spectroscopy.
UV difference spectroscopy is often able to provide insight into the natures or identities of reactive intermediates formed in the inactivation process (2, 50). Figure 4 reveals the spectral changes seen when CTX-M-9 was inactivated by meropenem and penem 1 as determined by UV difference spectroscopy. In the case of meropenem, the absorbance at 304 nm seen in the first 12 s of the reaction increased over the 60 min, indicating the breaking of the β-lactam bond. As we have seen in SHV-1 inactivated by meropenem, this long-lived intermediate suggests an acyl enzyme is directly formed with the CTX-M-9 β-lactamase.
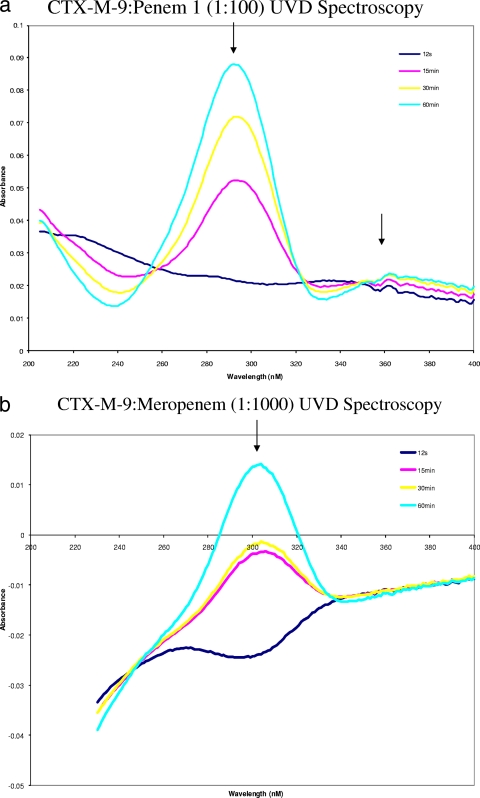
Fig. 4.
UV difference spectroscopy data were obtained by subtraction of free inhibitor from CTX-M-9 with added inhibitor at t = 0. (a) UV difference spectroscopy of CTX-M-9 reacted with penem 1 (I/E of 100/1). (b) Meropenem (I/E of 1,000/1). Note the formation of a chromophore at 304 nm and 360 nm. In accordance with previous data for these inhibitors with the class A β-lactamase SHV, we tentatively assigned the chromophore at 304 nm to the hydrolysis of the β-lactam bond and the chromophore at 360 to the formation of the bicyclic aromatic ring system (2).
For penem 1, chromophoric changes were evident in the near-UV spectrum, with the maximum absorbance at 294 nm and a change at 330 and 360 nm. In the case of penem 1, previous insights into the mechanism of inactivation coupled with the timed mass spectrometry and UV difference spectra suggest that penem 1 forms a linear imine species that undergoes 7-endo-trig cyclization to ultimately form a cyclic enamine, the 1,4-thiazepine derivative (appearance of a chromophore at 360 to 380 nm) (2, 5). We previously studied this penem inhibitor against OXA-1, a monomeric class D β-lactamase (2), SHV-1, and the R244S variant of SHV-1 β-lactamase, and we found similar results (2, 53). It is also possible that these UV difference spectra represent other chemical changes to the enzyme/inhibitor complex (e.g., closure of the C7 heterocycle ring of the penem).
Molecular modeling.
Why are meropenem and penem 1 more potent than clavulanate, sulbactam, and tazobactam? A major difference between penem 1 and meropenem compared to clavulanate, sulbactam, and tazobactam is the presence of a double bond between the C2 and C3 positions in the former compounds. As previously shown, this sp2 configuration plays a major role in the binding/catalysis of penems (clavulanate included) and carbapenems (2, 21, 37, 38). Sulbactam and tazobactam, which possess an sp3 hybridized R2 side chain, are positioned in a different manner in the active site. In addition, the presence of the leaving group at the C6 position also influences reaction chemistry (carbapenems versus penem 1 versus clavulanate) (21). To demonstrate the above findings as they pertain to the hybridization at the C2 position, we generated a model of meropenem and penem 1 in the active site.
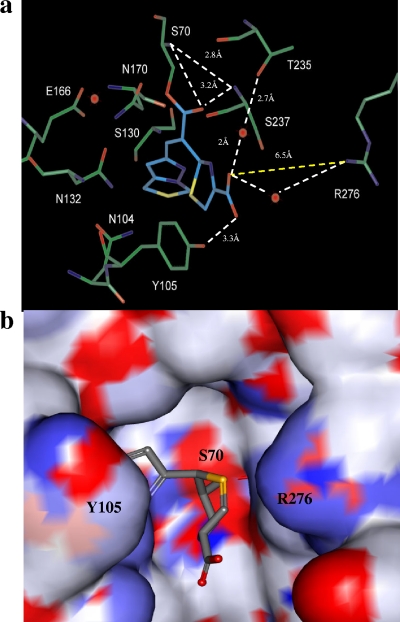
In the absence of an “Arg244 equivalent” (Thr is at Ambler position 244 in CTX-M-9), the necessary interaction with the C3 carboxylate for both compounds likely comes from long-range, but very important, electrostatic interactions from Arg276 (10–12, 14, 15) (Fig. 5 and 6). In the case of meropenem, we created a model for each tautomer, pyrroline Δ2 (Fig. 5a and b) and Δ1 (Fig. 5c and d). Our model for the Δ2 tautomer of meropenem, which has an sp2 hybridized R2 side chain at the C2 position, shows that the carbonyl oxygen of the β-lactam is positioned in the oxyanion hole, interacting with the backbone amides of Ser70 and Ser237. In addition, important hydrogen bonds with Asn132 and Asn104 are formed with the ethoxy side chain of meropenem. The C3 carboxylate also forms a hydrogen bond with a key water molecule and Thr235. These observations are interpreted as highly stabilizing interactions that contribute to the low turnover and high affinities (low Ki values) observed in our kinetic analysis. Lastly, electrostatic interactions are formed with the C3 carboxylate and guanidinium group of Arg 276 (approximate 3.1-Å distance), further stabilizing the acyl enzyme complex. The second model created for meropenem was with an sp3 hybridized R2 side chain at the C2 position (Δ1 tautomer), forming an acyl enzyme complex with CTX-M-9 (Fig. 5c and d). During the 6-ps standard dynamic minimization, the carbonyl oxygen of meropenem flips out of the oxyanion hole toward Lys73, making a hydrogen bond with its amine group. The carboxylate forms a complex hydrogen bond network with Arg276 and Thr235. In addition, the interaction mediated by a water molecule presented in the sp2 hybridized model is replaced by a hydrogen bond interaction. The movement of the water molecule has previously been observed in the crystal structure of BlaC with the Δ2 and Δ1tautomers of ertapenem (56).
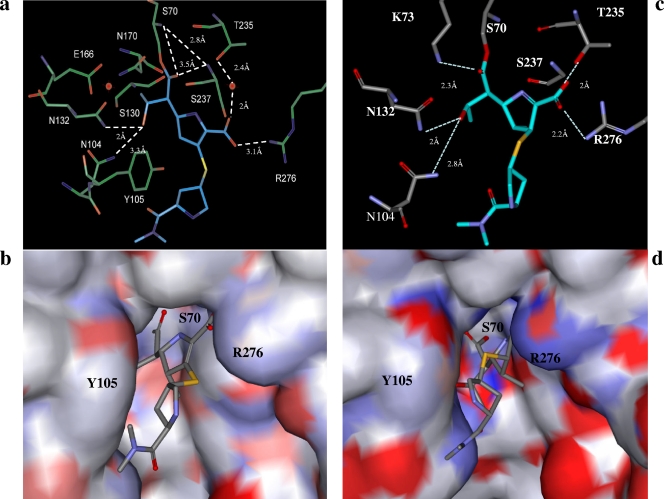
Fig. 5.
Molecular representations of CTX-M-9 and meropenem in the Δ2 tautomeric form (a and b) and Δ1 tautomeric form (c and d) docked in the active site of CTX-M-9. (a) The stable acyl enzyme species of meropenem and CTX-M-9 with the carbonyl oxygen of meropenem in the oxyanion hole formed by the amides of Ser70 and Ser237. In addition, the C3 carboxylate makes hydrogen bonds with T235 through a water molecule and the delocalized positively charged guanidinium group of Arg276. In this acyl enzyme model, meropenem's substituent at the C6 position interacts via hydrogen bonds to Asn132 and Asn104. (c) During the dynamic simulation, the carbonyl oxygen of meropenem flipped out of the oxyanion hole toward Lys73, making a hydrogen bond with its amine group. The carboxylate made a hydrogen bond network with Arg276 and Thr235 (the interaction mediated by a water molecule was replaced by a hydrogen bond interaction). The substituent at the C6 position group showed increased flexibility during MDS, maintaining the interactions with Asn132 and/or Asn104. (b and d) Connolly surface diagrams of meropenem in CTX-M-9, colored by atom charge. These representations show a change in the shape and electropositivity of the enzyme active site in the region of Arg276 upon binding with the two forms of meropenem. The larger positively charged region found near Arg276 in panel b is replaced by a more localized region closer to the oxyanion hole in panel d, suggesting that Arg276 possesses a flexible side chain and that conformational changes occur with the enzyme upon binding meropenem.
Fig. 6.
Molecular representation of penem 1 prior to endo-trig cyclization and formation of the 7-membered 1,4-thiazepine ring having R stereochemistry at the new C7 moiety, as seen with SHV-1 and penem 1, in the active site of CTX-M-9 (35). (a) The acyl enzyme with the carbonyl oxygen in the oxyanion hole formed by the amides of Ser70 and Ser237. Unlike the meropenem CTX-M-9 model presented in Fig. 5a, this model shows the C3 carboxylate making a hydrogen bond with the -OH of Tyr105 and losing the interactions with Thr235 (∼5 Å) and Arg276 (∼6 Å). In addition, two water molecules are positioned such that they may bridge the interaction between the two residues and the C3 carboxylate. (b) Connolly surface diagram of penem 1 in CTX-M-9.
We modeled penem 1 before secondary ring closure (Fig. 6a and b) (2, 37). Similar to the meropenem complex that was studied, we found the carbonyl oxygen of the acylated β-lactam positioned in the oxyanion hole. Here the carboxylate forms four important interactions. In marked contrast to meropenem, we saw a hydrogen bond that forms with the hydroxyl of Tyr105. This would suggest that there might be an accompanying conformational change at the opening of the enzyme active site as a result of inactivation by penem 1. We think this is likely, as the structure of SHV-1 with penems showed that the Tyr105 moves 20° compared to the apo-enzyme, resulting in resonance stabilization with the C7 heterocycle ring of the penem (37). From these constructions we postulate that a long-range interaction occurs between Arg276 and the carboxylate of penem 1, and it may be mediated by a water molecule. Another water molecule presented in the model also serves as a bridge to Thr235. However, computational analysis revealed that the interactions with the first and second water molecules are dynamic (during the 6-ps dynamic simulation, the observed water molecule and side chain of Arg276 were changing positions). We anticipate that crystallographic studies of this complex will yield deeper insights into the mechanism of inactivation by these compounds in these unique ESBLs.
In conclusion, we present the first analysis of the inactivation of CTX-M-9 ESBL by different β-lactamase inhibitors and by penems. Despite a unique three-dimensional structure and the absence of a direct “R244 equivalent,” inhibition proceeds efficiently (tn ≤ 50), especially as to the formation of intermediates that hinder deacylation (tn = 4). Our work also provides early insights as to why penems (especially carbapenems) are important “poor substrates” or inhibitors that are effective against the CTX-M-9 ESBLs and that these classes of compounds can be used as valuable agents against this emerging class A ESBL. We assert that β-lactam compounds that possess the sp2 hybridized R2 side chain at the C2 position offer interesting opportunities for lead compound development as the position of this side chain is strategic in the active site. Taken together, our results support the notion that intermediates that “occupy” the enzyme and prolonged time to recovery are important considerations in the future development of the next generation of inhibitors that target emerging β-lactamases (18). In addition, we also see the significance of a “region of positive charge” mediated by the Arg276 residue in the case of CTX-M-9 β-lactamases. The absence of an “Arg244 equivalent” is compensated by the positive charge at Arg276, a residue with a very flexible side chain that can be present in multiple conformations. These findings support the hypothesis advanced that a positive charge at Ambler position 244, 220, or 276 is important for class A enzyme evolution (33). We also hasten to add that this plasticity or delocalization of the positive charge to a region of the active site may be a more general phenomenon exhibited by class A and class C β-lactamases (19). These new insights are in line with previous notions developed by Todd and colleagues that (i) enzyme active sites are highly flexible; (ii) different functional groups may serve the same mechanistic role in catalysis; and (iii) this plasticity can arise by “functional residue hopping” (54). These perspectives could have profound implications for the future of β-lactamase inhibitor development, as it is possible that structural reorganization and its effects on protein stability maybe not be readily predicted.
Supplementary Material
ACKNOWLEDGMENTS
The Veterans Affairs Merit Review Program, VISN 10 GRECC, and the National Institutes of Health (RO1 AI063517-01) supported these studies.
We thank Tarek Mansour and Patricia Bradford, formerly of Wyeth Pharmaceuticals, for the kind gift of tazobactam and penem 1, and Tom Gootz, previously with Pfizer, for the kind gift of sulbactam. We also thank Sebastian Kurz of the Department of Medicine University Hospitals Case Medical Center and Paul Miller of Pfizer for their thoughtful reviews.
Footnotes
Supplemental material for this article may be found at http://aac.asm.org/.
Published ahead of print on 9 May 2011.
REFERENCES
- 1. Abdalhamid B., Pitout J. D., Moland E. S., Hanson N. D. 2004. Community-onset disease caused by Citrobacter freundii producing a novel CTX-M β-lactamase, CTX-M-30, in Canada. Antimicrob. Agents Chemother. 48:4435–4437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bethel C. R., et al. 2008. Inhibition of OXA-1 β-lactamase by penems. Antimicrob. Agents Chemother. 52:3135–3143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bonnet R. 2004. Growing group of extended-spectrum β-lactamases: the CTX-M enzymes. Antimicrob. Agents Chemother. 48:1–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Brown R. P., Aplin R. T., Schofield C. J. 1996. Inhibition of TEM-2 β-lactamase from Escherichia coli by clavulanic acid: observation of intermediates by electrospray ionization mass spectrometry. Biochemistry 35:12421–12432 [DOI] [PubMed] [Google Scholar]
- 5. Bulychev A., Massova I., Lerner S. A., Mobashery S. 1995. Penem BRL 42715: an effective inactivator for β-lactamases. J. Am. Chem. Soc. 117:4797–4801 [Google Scholar]
- 6. Bush K. 2010. Alarming β-lactamase-mediated resistance in multidrug-resistant Enterobacteriaceae. Curr. Opin. Microbiol. 13:558–564 [DOI] [PubMed] [Google Scholar]
- 7. Bush K. 2010. Bench-to-bedside review: the role of β-lactamases in antibiotic-resistant Gram-negative infections. Crit. Care 14:224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Canton R., Coque T. M. 2006. The CTX-M β-lactamase pandemic. Curr. Opin. Microbiol. 9:466–475 [DOI] [PubMed] [Google Scholar]
- 9. Canton R., et al. 2002. Epidemiology of extended-spectrum β-lactamase-producing Enterobacter isolates in a Spanish hospital during a 12-year period. J. Clin. Microbiol. 40:1237–1243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Chen Y., Bonnet R., Shoichet B. K. 2007. The acylation mechanism of CTX-M β-lactamase at 0.88Å resolution. J. Am. Chem. Soc. 129:5378–5380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Chen Y., Delmas J., Sirot J., Shoichet B., Bonnet R. 2005. Atomic resolution structures of CTX-M β-lactamases: extended spectrum activities from increased mobility and decreased stability. J. Mol. Biol. 348:349–362 [DOI] [PubMed] [Google Scholar]
- 12. Chen Y., Shoichet B., Bonnet R. 2005. Structure, function, and inhibition along the reaction coordinate of CTX-M β-lactamases. J. Am. Chem. Soc. 127:5423–5434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Cheng J., et al. 2010. Phenotypic and molecular characterization of two novel CTX-M enzymes carried by Klebsiella pneumoniae. Mol. Biol. Rep. 37:1261–1267 [DOI] [PubMed] [Google Scholar]
- 14. Delmas J., et al. 2008. Structure and dynamics of CTX-M enzymes reveal insights into substrate accommodation by extended-spectrum β-lactamases. J. Mol. Biol. 375:192–201 [DOI] [PubMed] [Google Scholar]
- 15. Delmas J., et al. Structural insights into substrate recognition and product expulsion in CTX-M enzymes. J. Mol. Biol. 400:108–120 [DOI] [PubMed] [Google Scholar]
- 16. Delmas J., Robin F., Carvalho F., Mongaret C., Bonnet R. 2006. Prediction of the evolution of ceftazidime resistance in extended-spectrum β-lactamase CTX-M-9. Antimicrob. Agents Chemother. 50:731–738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Drawz S. M., et al. 2010. Inhibition of the class C β-lactamase from Acinetobacter spp.: insights into effective inhibitor design. Biochemistry 49:329–340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Drawz S. M., Bonomo R. A. 2010. Three decades of β-lactamase inhibitors. Clin. Microbiol. Rev. 23:160–201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Drawz S. M., Taracila M., Caselli E., Prati F., Bonomo R. A. 14 March 2011. Exploring sequence requirements for the C3/C4 carboxylate recognition in the Pseudomonas aeruginosa cephalosporinase: insights into the plasticity of the AmpC β-lactamase. Protein Sci. doi:10.1002/pro.612. [Epub ahead of print.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Endimiani A., et al. 2010. Enhancing resistance to cephalosporins in class C β-lactamases: impact of Gly214Glu in CMY-2. Biochemistry 49:1014–1023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Fisher J. F., Meroueh S. O., Mobashery S. 2005. Bacterial resistance to β-lactam antibiotics: compelling opportunism, compelling opportunity. Chem. Rev. 105:395–424 [DOI] [PubMed] [Google Scholar]
- 22. Helfand M. S., et al. 2003. Understanding resistance to β-lactams and β-lactamase inhibitors in the SHV β-lactamase: lessons from the mutagenesis of SER-130. J. Biol. Chem. 278:52724–52729 [DOI] [PubMed] [Google Scholar]
- 23. Helfand M. S., Bonomo R. A. 2005. Current challenges in antimicrobial chemotherapy: the impact of extended-spectrum β-lactamases and metallo-β-lactamases on the treatment of resistant Gram-negative pathogens. Curr. Opin. Pharmacol. 5:452–458 [DOI] [PubMed] [Google Scholar]
- 24. Ho P. L., et al. 2007. Community emergence of CTX-M type extended-spectrum β-lactamases among urinary Escherichia coli from women. J. Antimicrob. Chemother. 60:140–144 [DOI] [PubMed] [Google Scholar]
- 25. Hugonnet J. E., Blanchard J. S. 2007. Irreversible inhibition of the Mycobacterium tuberculosis β-lactamase by clavulanate. Biochemistry 46:11998–12004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Hujer A. M., Hujer K. M., Bonomo R. A. 2001. Mutagenesis of amino acid residues in the SHV-1 β-lactamase: the premier role of Gly238Ser in penicillin and cephalosporin resistance. Biochim. Biophys. Acta 1547:37–50 [DOI] [PubMed] [Google Scholar]
- 27. Imtiaz U., Billings E. M., Knox J. R., Mobashery S. 1994. A structure-based analysis of the inhibition of class A β-lactamases by sulbactam. Biochemistry 33:5728–5738 [DOI] [PubMed] [Google Scholar]
- 28. Imtiaz U., Manavathu E. K., Mobashery S., Lerner S. A. 1994. Reversal of clavulanate resistance conferred by a Ser-244 mutant of TEM-1 β-lactamase as a result of a second mutation (Arg to Ser at position 164) that enhances activity against ceftazidime. Antimicrob. Agents Chemother. 38:1134–1139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kuzin A. P., et al. 2001. Inhibition of the SHV-1 β-lactamase by sulfones: crystallographic observation of two reaction intermediates with tazobactam. Biochemistry 40:1861–1866 [DOI] [PubMed] [Google Scholar]
- 30. Livermore D. M., et al. 2007. CTX-M: changing the face of ESBLs in Europe. J. Antimicrob. Chemother. 59:165–174 [DOI] [PubMed] [Google Scholar]
- 31. Livermore D. M., Hawkey P. M. 2005. CTX-M: changing the face of ESBLs in the UK. J. Antimicrob. Chemother. 56:451–454 [DOI] [PubMed] [Google Scholar]
- 32. Livermore D. M., Woodford N. 2006. The β-lactamase threat in Enterobacteriaceae, Pseudomonas and Acinetobacter. Trends Microbiol. 14:413–420 [DOI] [PubMed] [Google Scholar]
- 33. Marciano D. C., Brown N. G., Palzkill T. 2009. Analysis of the plasticity of location of the Arg244 positive charge within the active site of the TEM-1 β-lactamase. Protein Sci. 18:2080–2089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Maveyraud L., et al. 1998. Structural basis for clinical longevity of carbapenem antibiotics in the face of challenge by the common class A β-lactamases from the antibiotic-resistant bacteria. J. Am. Chem. Soc. 120:9748–9752 [Google Scholar]
- 35. Moubareck C., et al. 2005. Countrywide spread of community- and hospital-acquired extended-spectrum β-lactamase (CTX-M-15)-producing Enterobacteriaceae in Lebanon. J. Clin. Microbiol. 43:3309–3313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Nguyen N. T., et al. 2010. The sudden dominance of blaCTX-M harbouring plasmids in Shigella spp. circulating in southern Vietnam. PLoS Negl. Trop. Dis. 4:e702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Nukaga M., et al. 2003. Inhibition of class A and class C β-lactamases by penems: crystallographic structures of a novel 1,4-thiazepine intermediate. Biochemistry 42:13152–13159 [DOI] [PubMed] [Google Scholar]
- 38. Nukaga M., et al. 2008. Inhibition of class A β-lactamases by carbapenems: crystallographic observation of two conformations of meropenem in SHV-1. J. Am. Chem. Soc. 130:12656–12662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Nukaga M., Mayama K., Hujer A. M., Bonomo R. A., Knox J. R. 2003. Ultrahigh resolution structure of a class A β-lactamase: on the mechanism and specificity of the extended-spectrum SHV-2 enzyme. J. Mol. Biol. 328:289–301 [DOI] [PubMed] [Google Scholar]
- 40. Pattanaik P., et al. 2009. Strategic design of an effective β-lactamase inhibitor: LN-1-255, a 6-alkylidene-2′-substituted penicillin sulfone. J. Biol. Chem. 284:945–953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Peirano G., et al. 2010. High prevalence of ST131 isolates producing CTX-M-15 and CTX-M-14 among extended-spectrum-β-lactamase-producing Escherichia coli isolates from Canada. Antimicrob. Agents Chemother. 54:1327–1330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Perez-Llarena F. J., et al. 2008. Structure-function studies of arginine at position 276 in CTX-M β-lactamases. J. Antimicrob. Chemother. 61:792–797 [DOI] [PubMed] [Google Scholar]
- 43. Pitout J. D., Gregson D. B., Campbell L., Laupland K. B. 2009. Molecular characteristics of extended-spectrum-β-lactamase-producing Escherichia coli isolates causing bacteremia in the Calgary Health Region from 2000 to 2007: emergence of clone ST131 as a cause of community-acquired infections. Antimicrob. Agents Chemother. 53:2846–2851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Pitout J. D., Gregson D. B., Church D. L., Elsayed S., Laupland K. B. 2005. Community-wide outbreaks of clonally related CTX-M-14 β-lactamase-producing Escherichia coli strains in the Calgary health region. J. Clin. Microbiol. 43:2844–2849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Pitout J. D., Laupland K. B. 2008. Extended-spectrum β-lactamase-producing Enterobacteriaceae: an emerging public-health concern. Lancet Infect. Dis. 8:159–166 [DOI] [PubMed] [Google Scholar]
- 46. Rodriguez-Bano J., et al. 2006. Bacteremia due to extended-spectrum β-lactamase-producing Escherichia coli in the CTX-M era: a new clinical challenge. Clin. Infect. Dis. 43:1407–1414 [DOI] [PubMed] [Google Scholar]
- 47. Rodriguez-Bano J., et al. 2010. Risk factors and prognosis of nosocomial bloodstream infections caused by extended-spectrum-β-lactamase-producing Escherichia coli. J. Clin. Microbiol. 48:1726–1731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Rodriguez-Bano J., et al. 2010. Community-onset bacteremia due to extended-spectrum β-lactamase-producing Escherichia coli: risk factors and prognosis. Clin. Infect. Dis. 50:40–48 [DOI] [PubMed] [Google Scholar]
- 49. Sidjabat H. E., Derrington P., Nimmo G. R., Paterson D. L. 2010. Escherichia coli ST131 producing CTX-M-15 in Australia. J. Antimicrob. Chemother. 65:1301–1303 [DOI] [PubMed] [Google Scholar]
- 50. Sulton D., et al. 2005. Clavulanic acid inactivation of SHV-1 and the inhibitor-resistant S130G SHV-1 β-lactamase. Insights into the mechanism of inhibition. J. Biol. Chem. 280:35528–35536 [DOI] [PubMed] [Google Scholar]
- 51. Taibi P., Mobashery S. 1995. Mechanism of turnover of imipenem by the TEM β-lactamase revisited. J. Am. Chem. Soc. 117:7600–7605 [Google Scholar]
- 52. Thomson J. M., Distler A. M., Bonomo R. A. 2007. Overcoming resistance to β-lactamase inhibitors: comparing sulbactam to novel inhibitors against clavulanate resistant SHV enzymes with substitutions at Ambler position 244. Biochemistry 46:11361–11368 [DOI] [PubMed] [Google Scholar]
- 53. Thomson J. M., Distler A. M., Prati F., Bonomo R. A. 2006. Probing active site chemistry in SHV β-lactamase variants at Ambler position 244. Understanding unique properties of inhibitor resistance. J. Biol. Chem. 281:26734–26744 [DOI] [PubMed] [Google Scholar]
- 54. Todd A. E., Orengo C. A., Thornton J. M. 2002. Plasticity of enzyme active sites. Trends Biochem. Sci. 27:419–426 [DOI] [PubMed] [Google Scholar]
- 55. Tollentino F. M., et al. 2010. High prevalence of blaCTX-M extended spectrum β-lactamase genes in Klebsiella pneumoniae isolates from a tertiary care hospital: first report of blaSHV-12, blaSHV-31, blaSHV-38, and blaCTX-M-15 in Brazil. Microb. Drug Resist. 17:7–16 [DOI] [PubMed] [Google Scholar]
- 56. Tremblay L. W., Fan F., Blanchard J. S. Biochemical and structural characterization of Mycobacterium tuberculosis β-lactamase with the carbapenems ertapenem and doripenem. Biochemistry 49:3766–3773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Venkatesan A. M., et al. 2004. Structure-activity relationship of 6-methylidene penems bearing tricyclic heterocycles as broad-spectrum β-lactamase inhibitors: crystallographic structures show unexpected binding of 1,4-thiazepine intermediates. J. Med. Chem. 47:6556–6568 [DOI] [PubMed] [Google Scholar]
- 58. Villegas M. V., Kattan J. N., Quinteros M. G., Casellas J. M. 2008. Prevalence of extended-spectrum β-lactamases in South America. Clin. Microbiol. Infect. 14(Suppl. 1):154–158 [DOI] [PubMed] [Google Scholar]
- 59. Woerther P. L., et al. 2010. Emergence and dissemination of extended-spectrum β-lactamase-producing Escherichia coli in the community: lessons from the study of a remote and controlled population. J. Infect. Dis. 202:515–523 [DOI] [PubMed] [Google Scholar]
- 60. Woodford N., Kaufmann M. E., Karisik E., Hartley J. W. 2007. Molecular epidemiology of multiresistant Escherichia coli isolates from community-onset urinary tract infections in Cornwall, England. J. Antimicrob. Chemother. 59:106–109 [DOI] [PubMed] [Google Scholar]
- 61. Wu G., Robertson D. H., Brooks C. L., III, Vieth M. 2003. Detailed analysis of grid-based molecular docking: a case study of CDOCKER-A CHARMm-based MD docking algorithm. J. Comput. Chem. 24:1549–1562 [DOI] [PubMed] [Google Scholar]
- 62. Yang Y., et al. 2000. Mechanism of inhibition of the class A β-lactamases PC1 and TEM-1 by tazobactam. Observation of reaction products by electrospray ionization mass spectrometry. J. Biol. Chem. 275:26674–26682 [DOI] [PubMed] [Google Scholar]
- 63. Zafaralla G., Mobashery S. 1992. Facilitation of the Δ 2 to Δ 1 pyrroline tautomerization of carbapenem antibiotics by the highly conserved arginine-244 of class A β-lactamases during the course of turnover. J. Am. Chem. Soc. 114:1506–1507 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.