Abstract
The antimicrobial susceptibility of clinical isolates of Brachyspira hyodysenteriae in Spain was monitored, and the underlying molecular mechanisms of resistance were investigated. MICs of tylosin, tiamulin, valnemulin, lincomycin, and tylvalosin were determined for 87 B. hyodysenteriae isolates recovered from 2008 to 2009 by broth dilution. Domain V of the 23S rRNA gene and the ribosomal protein L3 gene were sequenced in 20 isolates for which the tiamulin MIC was ≥4 μg/ml, presenting decreased susceptibility, and in 18 tiamulin-susceptible isolates (MIC ≤ 0.125 μg/ml), and all isolates were typed by multiple-locus variable-number tandem repeats analysis. A comparison with antimicrobial susceptibility data from 2000 to 2007 showed an increase in pleuromutilin resistance over time, doubling the number of isolates with decreased susceptibility to tiamulin. No alteration in susceptibility was detected for lincomycin, and the MIC of tylosin remained high (MIC50 > 128 μg/ml). The decreased susceptibility to tylosin and lincomycin can be explained by mutations at position A2058 of the 23S rRNA gene (Escherichia coli numbering). A2058T was the predominant mutation, but A2058G also was found together with a change of the neighboring base pair at positions 2057 to 2611. The role of additional point mutations in the vicinity of the peptidyl transferase center and mutations in the L3 at amino acids 148 and 149 and their possible involvement in antimicrobial susceptibility are considered. An association between G2032A and high levels of tiamulin and lincomycin MICs was found, suggesting an increasing importance of this mutation in antimicrobial resistance of clinical isolates of B. hyodysenteriae.
INTRODUCTION
Brachyspira hyodysenteriae is the etiological agent of swine dysentery, a severe mucohemorrhagic colitis that affects pigs primarily during the grow-finish period, and it has a significant economic impact (15). The treatment and control of swine dysentery are based mainly on the use of antimicrobials, as no commercial vaccine against B. hyodysenteriae is available.
In Spain, swine dysentery is involved in more than 30% of diarrhea outbreaks in commercial pig farms (7). A trend toward antimicrobial resistance has been detected in B. hyodysenteriae isolates from 2000 to 2007 (17), and Spanish isolates with reduced susceptibility to several antimicrobial products registered against swine dysentery have been reported recently (18). Such isolates have been detected in many pig-producing countries (25, 27, 37), and they represent a serious threat to the pig industry (15). Accordingly, the antimicrobial susceptibility testing of clinical isolates of B. hyodysenteriae has become essential to assist practitioners in selecting swine dysentery treatment strategies. Moreover, a monitoring program may help to detect new resistance trends and to evaluate the usefulness of the few available drugs on a national level.
The genetic basis of resistance in clinical isolates of B. hyodysenteriae to macrolides and lincosamides has been explained by an A→T transversion mutation at position 2058 of the 23S rRNA gene (Escherichia coli numbering) (22). Moreover, Pringle et al. (34) related resistance to tiamulin in laboratory-selected mutants of B. hyodysenteriae to point mutations in domain V of the 23S rRNA gene and/or the ribosomal protein L3 gene. In that study, two or more point mutations were frequently detected after in vitro selection with tiamulin, although they were not proved by genetic evidence to be the cause of resistance. The corresponding domain V 23S rRNA mutations were investigated individually later in Mycobacterium smegmatis, and the genetic basis of resistance to pleuromutilins was confirmed (28, 29). Three clinical isolates of B. hyodysenteriae with reduced susceptibility to tiamulin, probably linked to an Asn148Ser change in ribosomal protein L3 (Brachyspira pilosicoli numbering), have been described (34). Mutations in the L3 gene after in vitro selection with tiamulin also have been detected in Staphylococcus aureus (13, 30) and in Escherichia coli (4).
This study was performed to monitor resistance to tylosin, tiamulin, valnemulin, and lincomycin in clinical isolates of B. hyodysenteriae recovered in 2008 and 2009 and to report on antimicrobial susceptibility to tylvalosin, which recently has been registered for the treatment of swine dysentery in Spain. In addition, the mechanisms of resistance in B. hyodysenteriae field isolates to tylosin, tiamulin, valnemulin, lincomycin, and tylvalosin were investigated by relating mutational changes in the domain V part of the 23S rRNA gene and the L3 gene to changes in MICs.
MATERIALS AND METHODS
Bacterial strains.
A set of 87 Spanish isolates of B. hyodysenteriae from the bacterial collection held at the Animal Health Department at the University of León was used in this study. Isolates had been recovered from fecal samples of pigs suffering from diarrhea submitted for diagnostic examination between January 2008 and December 2009 by following the methodology described previously (17). Subsequently, pure cultures of strongly beta-hemolytic intestinal spirochetes were confirmed as B. hyodysenteriae using a species-specific PCR based on the tlyA gene (36). Isolates were selected to represent the main Spanish pig-producing regions of the country, and a single B. hyodysenteriae isolate was included per farm (n = 87).
Antimicrobial agents and broth dilution procedure at the monitoring stage.
Susceptibility testing was performed by broth dilution (24) using VetMIC Brachy antibiotic panels (SVA, Sweden) according to the manufacturer's recommendations. The antibiotic panels consisted of 48-well tissue culture trays with dried antimicrobial agents, including one well without drug as a positive growth control. Two-fold serial dilutions of the following antimicrobial agents were tested: tiamulin (0.063 to 8 μg/ml), valnemulin (0.031 to 4 μg/ml), tylosin (2 to 128 μg/ml), lincomycin (0.5 to 64 μg/ml), and tylvalosin (0.25 to 32 μg/ml). The MIC was determined as the lowest concentration of antimicrobial agent that prevented visible growth. The absence of contamination was confirmed by phase-contrast microscopy. The B. hyodysenteriae type strain B78T (ATCC 27164) was used as a quality control strain as previously proposed (35).
Detecting changes in antimicrobial susceptibility patterns over time.
Trends in the antimicrobial susceptibility of Spanish B. hyodysenteriae field isolates to tiamulin, valnemulin, tylosin, and lincomycin were studied using a survival analysis. This approach relates the proportion of the isolates that are not inhibited to the concentration of antibiotic present, resulting in a survival curve for a particular drug. Changes in bacterial growth inhibition for a given antimicrobial agent can be tested across the entire range of concentrations, and the use of cutoff values for resistance is avoided (38). Survival curves were plotted using the nonparametric Kaplan-Meier method. Curves were right-censored when there was no growth inhibition at the highest concentration of antimicrobial tested. A detailed description of this methodology has been reported previously (17). Moreover, a log-rank test (α = 0.05) was performed to compare survival curves from 2008 to 2009 (this study) to those from 2000 to 2004 and 2006 to 2007 (17), which had been obtained from MICs against 50 and 58 Spanish clinical isolates of B. hyodysenteriae, respectively. All estimations were done using the statistical package SPSS for Windows, version 17.0 (SPSS, Chicago, IL).
MICs determined in other studies (17, 23, 25, 37), which have been used for comparison, were obtained by the broth dilution method used in this study.
Study of in vivo-acquired resistance mechanisms.
Two groups of B. hyodysenteriae field isolates were defined according to their particularly high or low tiamulin MICs. One group, subset A (Table 1), comprised 20 isolates with reduced susceptibility to tiamulin (MIC ≥ 4 μg/ml), and a second group, subset B (Table 2), included 18 field isolates for which the tiamulin MIC was ≤0.125 μg/ml. DNA was extracted from all isolates after a boiling step and was used for the PCR amplification of part of the genes for 23S rRNA (domain V) and ribosomal protein L3. PCR mixtures contained 1× PCR buffer (20 mM Tris HCl [pH 8.4], 50 mM KCl), 3 mM MgCl2, 1 U of platinum Taq DNA polymerase (Invitrogen, Carlsbad, CA), 200 μM deoxynucleoside triphosphate mix (Invitrogen), 0.25 μM each forward and reverse primers, 2 μl of extracted DNA (5 to 20 ng/μl), and sterile distilled water to a final volume of 50 μl. The primer pairs described by Pringle et al. (34) were used for PCR in a Mastercycler apparatus (Eppendorf Scientific Inc., Westbury, NY) with an initial step of 95°C for 5 min, followed by 30 cycles of a three-step cycle protocol consisting of 95°C for 20 s, 68 or 58°C (domain V or L3 amplification, respectively) for 20 s, and 72°C for 1 min, with a final extension step of 72°C for 5 min. The resulting fragments were sequenced in both directions. Nucleotide positions of the 23S rRNA gene were numbered according to the numbering used for E. coli, whereas translated sequences of ribosomal protein L3 were numbered according to B. pilosicoli numbering, enabling comparison to results from other studies. The E. coli 23S rRNA gene (J01695) and B. pilosicoli ribosomal protein L3 gene (AF114845) sequences were retrieved from GenBank and aligned with the homologous B. hyodysenteriae sequences. Ribosomal protein L3 gene sequences were translated using CLC Sequence Viewer (CLC Bio). 23S rRNA analyses were performed at the nucleotide level, and protein L3 analyses were performed at the amino acid level.
Table 1.
MICs, MLVA types, and point mutations of partial sequences of 23S rRNA and ribosomal protein L3 genes recovered in Spain between 2008 and 2009, isolate subset Aa
| Isolate | MLVA type | MICd (μg/ml) |
Point mutation in 23S rRNA gene atb: |
L3 mutationc | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| TIA | VAL | LINC | TYLO | TYLV | A2031 | G2032 | G2057 | A2058 | A2059 | G2087 | C2146 | C2362 | G2365 | G2535 | C2611 | |||
| RSp1 | 3 | 32 | 32 | 32 | 1,024 | 8 | T | A | ||||||||||
| RSp2 | 3 | 32 | 32 | 64 | 2,048 | 4 | T | T | C | N148S | ||||||||
| RSp3 | 3 | 32 | 8 | 128 | 512 | 4 | A | T | A | |||||||||
| RSp7 | 3 | 8 | 4 | >64 | >128 | >32 | T | G | ||||||||||
| RSp9 | 3 | 4 | 2 | 16 | >128 | 4 | T | |||||||||||
| RSp19 | 3 | 8 | 4 | 64 | >128 | 4 | T | |||||||||||
| RSp12 | 9 | 64 | >32 | 256 | 1,024 | 16 | A | T | T | |||||||||
| RSp13 | 9 | 16 | 8 | 32 | 1,024 | 32 | T | T | ||||||||||
| RSp4 | 12 | 16 | 4 | 128 | 1,024 | 4 | A | G | T | |||||||||
| RSp10 | 12 | 4 | 2 | >64 | >128 | 2 | A | G | T | |||||||||
| RSp20 | 12 | 8 | >4 | 64 | >128 | 8 | A | G | T | |||||||||
| RSp18 | 13 | 8 | 4 | 16 | >128 | 16 | T | |||||||||||
| RSp5 | 14 | 32 | 2 | 32 | 512 | 4 | T | T | ||||||||||
| RSp15 | 22 | 4 | 2 | 32 | >128 | 4 | T | |||||||||||
| RSp6 | 24 | 4 | 2 | >64 | >128 | 4 | A | T | ||||||||||
| RSp8 | 24 | 8 | 2 | >64 | >128 | 2 | A | T | ||||||||||
| RSp11 | 24 | 8 | 2 | >64 | >128 | 16 | T | A | T | S149T | ||||||||
| RSp14 | 24 | 4 | 1 | >64 | >128 | 2 | T | A | T | S149T | ||||||||
| RSp17 | 24 | 8 | 2 | >64 | >128 | 4 | A | T | ||||||||||
| RSp16 | 45 | 4 | >4 | 64 | >128 | 8 | A | T | ||||||||||
Subset A includes 20 B. hyodysenteriae isolates for which the tiamulin MICs were ≥4 μg/ml.
23S rRNA positions numbered according to Escherichia coli (J01695) numbering.
Ribosomal protein L3 amino acids numbered according to B. pilosicoli (AF114845) numbering.
TIA, tiamulin; VAL, valnemulin; LINC, lincomycin; TYLO, tylosin; TYLV, tylvalosin.
Table 2.
MICs, MLVA types, and point mutations of partial sequences of 23S rRNA and ribosomal protein L3 genes recovered in Spain between 2008 and 2009, isolate subset Ba
| Isolate | MLVA type | MICd (μg/ml) |
23S rRNA gene point mutation atb: |
L3 mutationsc | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| TIA | VAL | LINC | TYLO | TYLV | A2058 | G2087 | C2146 | G2201 | C2362 | G2365 | G2535 | |||
| RSp37 | 1 | 0.125 | ≤0.031 | 16 | >128 | 4 | T | T | C | |||||
| RSp21 | 3 | 0.125 | ≤0.031 | 16 | >128 | 1 | T | T | C | |||||
| RSp38 | 3 | 0.125 | 0.25 | 4 | >128 | 2 | T | A | ||||||
| RSp22 | 9 | ≤0.063 | ≤0.031 | 4 | 128 | 1 | T | |||||||
| RSp23 | 9 | ≤0.063 | ≤0.031 | 16 | >128 | 4 | T | T | ||||||
| RSp24 | 9 | ≤0.063 | ≤0.031 | 16 | >128 | 2 | T | T | ||||||
| RSp25 | 11 | ≤0.063 | ≤0.031 | 16 | >128 | 16 | T | T | ||||||
| RSp26 | 14 | ≤0.063 | ≤0.031 | 8 | >128 | 2 | T | T | ||||||
| RSp27 | 14 | ≤0.063 | ≤0.031 | 8 | >128 | 2 | T | T | ||||||
| RSp28 | 14 | 0.125 | ≤0.031 | 16 | >128 | 4 | T | T | ||||||
| RSp29 | 14 | 0.125 | ≤0.031 | 16 | >128 | 32 | T | T | ||||||
| RSp30 | 14 | 0.125 | ≤0.031 | 16 | >128 | 4 | T | T | ||||||
| RSp34 | 14 | ≤0.063 | ≤0.031 | 8 | >128 | 2 | T | T | ||||||
| RSp35 | 14 | 0.125 | ≤0.031 | 8 | >128 | 1 | T | T | ||||||
| RSp36 | 14 | 0.125 | ≤0.031 | 16 | >128 | 2 | T | T | ||||||
| RSp31 | 18 | 0.125 | ≤0.031 | 8 | >128 | 8 | T | C | A | N148S | ||||
| RSp32 | 18 | 0.125 | ≤0.031 | 16 | >128 | 4 | T | C | A | N148S | ||||
| RSp33 | 37 | 0.125 | ≤0.031 | 8 | >128 | 2 | T | A | N148S | |||||
Subset B includes 18 B. hyodysenteriae isolates for which the tiamulin MIC was ≤0.125 μg/ml.
23S rRNA positions numbered according to Escherichia coli (J01695) numbering.
Ribosomal protein L3 amino acids numbered according to B. pilosicoli (AF114845) numbering.
TIA, tiamulin; VAL, valnemulin; LINC, lincomycin; TYLO, tylosin; TYLV, tylvalosin.
Those isolates that had not been inhibited by the highest concentration of tiamulin at the monitoring stage (8 μg/ml) were tested for higher antimicrobial concentrations using VetMIC Brachy QCR High panels (SVA, Sweden) as described above. Antimicrobial ranges were 1 to 128 μg/ml for tiamulin, 0.25 to 32 μg/ml for valnemulin, 16 to 2,048 μg/ml for tylosin, and 2 to 256 μg/ml for lincomycin.
Finally, the 38 selected B. hyodysenteriae field isolates were typed by multiple-locus variable-number tandem-repeat analysis (MLVA) as previously described (19). Reference and type strains of B. hyodysenteriae, B204R (ATCC 31212) and B78T (ATCC 27164), were included as typing controls. The Hunter-Gaston diversity index (HGDI) (21) was used to measure the degree of discrimination of the MLVA typing method in the 38 selected isolates of B. hyodysenteriae.
The VMD program (20) and Protein Data Bank file 3OFZ were used to visualize E. coli 23S RNA and L3 to obtain distances between mutated positions and L3.
Nucleotide sequence accession numbers.
The nucleotide sequences of partial 23S rRNA (domain V) and protein L3 genes have been deposited in GenBank under accession numbers JF412548 to JF412585 and JF412586 to JF412623, respectively.
RESULTS AND DISCUSSION
Antimicrobial susceptibility of pathogenic B. hyodysenteriae isolates.
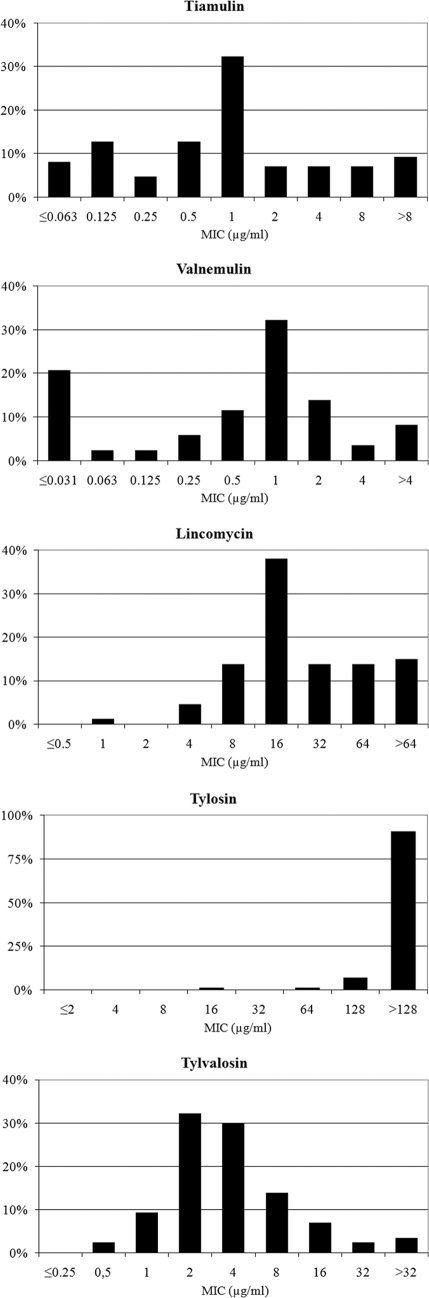
An initial study on MICs of tylosin, tiamulin, valnemulin, lincomycin, and tylvalosin was performed for 87 B. hyodysenteriae isolates recovered between 2008 and 2009 from fecal samples of pigs suffering from diarrhea. The values of the lowest concentrations of tiamulin, valnemulin, tylosin, lincomycin, and tylvalosin that completely inhibited the growth of 50% and 90% of the B. hyodysenteriae isolates, MIC50 and MIC90, respectively, are shown in Table 3. The corresponding MIC distributions of the antimicrobial agents are presented in Fig. 1.
Table 3.
MIC50, MIC90, and ranges for five antimicrobial agents against 87 Spanish field isolates of B. hyodysenteriae recovered between 2008 and 2009
| Drug | MIC (μg/ml) |
||
|---|---|---|---|
| 50% | 90% | Range | |
| Tiamulin | 1 | 8 | ≤0.063–>8 |
| Valnemulin | 1 | 4 | ≤0.031–>4 |
| Tylosin | >128 | >128 | 16–>128 |
| Lincomycin | 16 | >64 | 1–>64 |
| Tylvalosin | 4 | 16 | 0.5–>32 |
Fig. 1.
Distribution of MICs of five antimicrobials for 87 Spanish field isolates of B. hyodysenteriae recovered between 2008 and 2009.
The pleuromutilins, tiamulin and valnemulin, demonstrated similar distributions, in which the MIC was below 0.5 μg/ml for approximately 30% of the isolates tested, while the MIC was 1 μg/ml for another 30%. The 2008-2009 data showed a marked decrease in susceptibility to valnemulin and tiamulin compared to that of previous years. The MIC90 reached 8 μg/ml for tiamulin and 4 μg/ml for valnemulin, while the MIC50 for both drugs was 1 μg/ml, showing an increase of at least 4-fold compared to their respective MIC50s from 2000 to 2007 (17). Similar results were reported in Germany for 71 and 40 B. hyodysenteriae field isolates recovered in 2000 and 2001, respectively (37). However, several of the Spanish field isolates had higher tiamulin and valnemulin MICs than those reported by Rohde et al. (37), being similar to German and British isolates for which the tiamulin MICs were particularly high, as reported by Karlsson et al. (25).
The MIC of tylosin was ≥128 μg/ml for 98% of the isolates, showing that the decreased susceptibility of Spanish B. hyodysenteriae isolates to this macrolide is widespread. Although tylvalosin is a derivative of tylosin, it showed a much more susceptible distribution than tylosin, which was unimodal with a peak at 2 μg/ml. In accordance with suggested clinical breakpoints (6), tylvalosin can be useful in the treatment of swine dysentery in Spain, while the use of tylosin clearly is not advisable. The use of tylosin might even worsen the situation by providing pressure to keep the high resistance level. As shown in the present study, tylosin MICs for Spanish B. hyodysenteriae isolates have been consistently high during the last decade, with the MIC50 exceeding 128 μg/ml.
The distribution for lincomycin showed one large peak at a concentration of 16 μg/ml, accounting for 38% of the isolates, while the MIC was lower for 20%. There were no remarkable changes in lincomycin MIC distribution from 2008 to 2009 compared to that of 2000 to 2007 (17). However, a comparison with the lincomycin MIC distribution of 76 Australian isolates (23) revealed that Spanish isolates were less susceptible to lincomycin, lacking a significant subpopulation below 4 μg/ml.
Monitoring antimicrobial susceptibility of pathogenic B. hyodysenteriae isolates over time by survival analysis.
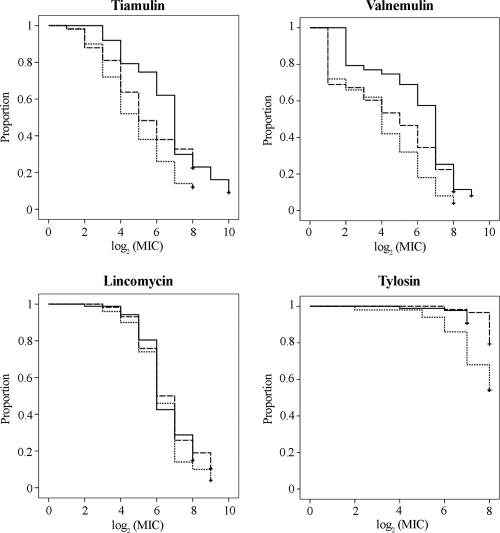
Changes in antimicrobial susceptibility were monitored by a survival analysis approach that tests changes in bacterial growth inhibition for a given antimicrobial agent across the entire range of tested concentrations. The survival curves for each antimicrobial agent in different periods of time (Fig. 2) were compared using the log-rank test at α = 0.05. As a result, no statistically significant differences for lincomycin were detected during the last decade when any of the studied periods were compared. On the other hand, statistically significant differences for MIC distributions of tiamulin (P < 0.001), valnemulin (P < 0.001), and tylosin (P = 0.001) were found between isolates collected in 2008 to 2009 relative to those collected in 2000 to 2004. When survival curves from 2008 to 2009 were compared to survival curves from 2006 to 2007, only valnemulin showed a statistically significant difference (P = 0.038). In all cases, the survival curves from 2008 to 2009 were above survival curves from previous periods of time. Tylosin differences have been reported previously between isolates from 2000 to 2004 and those from 2006 to 2007 (17). Hence, this study confirms that tylosin resistance has persisted since 2006. The survival analysis for tiamulin and valnemulin MICs showed an increase in resistance to the pleuromutilins in Spanish isolates in recent years, as suggested previously (17). It is remarkable that the tiamulin MICs for more than 60% of the isolates from 2008 to 2009 exceeded the microbiological breakpoint of 0.5 μg/ml proposed for monitoring decreased susceptibility to tiamulin by Karlsson et al. (24), thus doubling this percentage compared to that of preceding years (17). In agreement with Lobová et al. (27), cross-resistance between the two pleuromutilins was encountered for most of the isolates, with valnemulin MICs being one to two dilutions lower than tiamulin MICs for 85% (74 out of 87) of the isolates.
Fig. 2.
Survival curves of the log2 (MIC) values of tiamulin, valnemulin, lincomycin, and tylosin for 87 Spanish field isolates of B. hyodysenteriae recovered between 2008 and 2009 (solid lines). Survival curves for 50 isolates from 2000 to 2004 (dotted lines) and 58 isolates from 2006 to 2007 (dashed lines), obtained in a previous investigation (17), have been included for comparison.
Investigation of the molecular basis of the in vivo-acquired resistances.
Two groups of B. hyodysenteriae field isolates were defined according to their high or low tiamulin MICs. Subset A comprised 20 isolates with reduced susceptibility to tiamulin (MIC ≥ 4 μg/ml), while subset B comprised 18 field isolates for which the MIC was ≤0.125 μg/ml. Isolates that were not inhibited by tiamulin at the monitoring stage (8 μg/ml) were tested for concentrations up to 128 μg/ml tiamulin, 32 μg/ml valnemulin, 2,048 μg/ml tylosin, and 256 μg/ml lincomycin. MICs for all isolates in the two groups are presented in Table 1 (subset A) and Table 2 (subset B).
The MLVA typing of the two groups demonstrated that the 20 isolates included in subset A comprised eight different types, including the new MLVA type 45 (numerical profile [arranged according to Hidalgo et al. (19)]: 1, 1, 4, 2, 5, 2, 99, 1) and that subset B comprised seven different MLVA types. Three of the MLVA types (3, 9, and 14) were shared by isolates of both subsets (Tables 1 and 2). MLVA was chosen because it is a low-cost, portable, and highly discriminatory method for the strain typing of B. hyodysenteriae that retains a high phylogenetic value (19). A diversity index (HGDI) of 0.847 was obtained for the 38 selected isolates using MLVA. The HGDI calculated from data obtained in a previous diversity study of Spanish B. hyodysenteriae isolates using multilocus sequence typing was 0.749 (31). This suggests that MLVA is a more discriminatory technique than multilocus sequence typing when applied to the Spanish B. hyodysenteriae population.
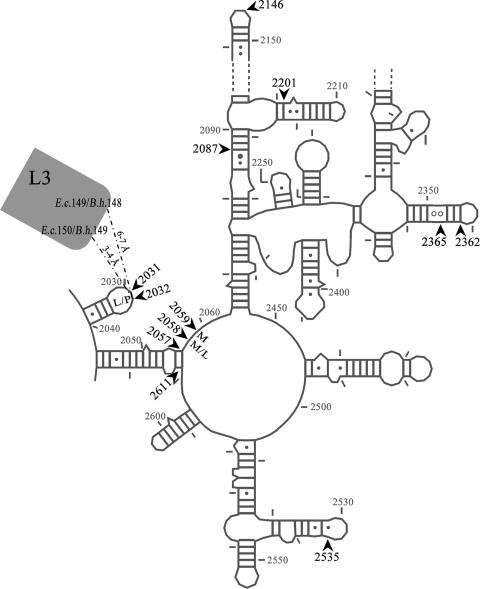
All of the tested antimicrobial agents bind to the large ribosomal subunit at or close to the so-called peptidyl transferase center (PTC) and thereby inhibit protein synthesis. Numerous studies have shown that ribosomal mutations can exhibit resistance to the tested drugs, although other resistance mechanisms also can play a role. Especially in organisms with few (one or two) rrn operons, 23S rRNA mutations often are found as antimicrobial resistance determinants (41). As previous studies of Brachyspira have shown or strongly indicated, the involvement of mutations in domain V of 23S rRNA and possibly L3 mutations in resistance to macrolides, lincosamides, and pleuromutilins (22, 34), the relevant regions of the 23S rRNA and L3 genes were sequenced from all 38 isolates in subsets A and B. The mutations are shown in Tables 1 and 2 together with the MICs and MLVA typing and are depicted on a secondary-structure model of domain V 23S rRNA in Fig. 3.
Fig. 3.
Secondary-structure model of domain V 23S rRNA showing locations of point mutations detected (arrows) in the present study. Macrolide (M), lincosamide (L), and pleuromutilin (P) resistance associated with a particular position has been indicated. Distances from position 2032 to the nearest ribosomal protein L3 amino acids are included. E.c., Escherichia coli; B.h., Brachyspira hyodysenteriae.
Mutations observed in 23S rRNA and their role in resistance to tiamulin and other antimicrobial agents.
We report MICs from five antibiotics authorized for the treatment of swine dysentery in Spain, which are members of three different groups: pleuromutilins, macrolides, and lincosamides. The exact binding of candidates from each group of antimicrobial agents to the 50S bacterial ribosomal subunit has been determined by X-ray structures (5, 9, 12, 14, 40). Pleuromutilins and lincosamides have essential overlapping sites at the PTC, and the macrolides bind at an adjacent site, with the larger macrolides (such as tylosin and tylvalosin) reaching slightly into the PTC. It thus is not surprising that single mutations in the PTC area can confer resistance to more than one group of antibiotics. Dealing with the presence of several mutations makes the cross-resistance pattern complex, especially as these mutations also can act synergistically (29).
Nucleotide A2058 of the 23S rRNA gene was mutated in all isolates tested. In 35 out of 38 isolates an A→T mutation was found in this position, while three isolates showed an A→G mutation. Thus, in agreement with a previous report on Swedish isolates (22), Spanish B. hyodysenteriae isolates presented mainly an A2058T transversion. Our detection of A2058G mutations is the first observation of this mutation in clinical isolates of B. hyodysenteriae, although it has been induced in vitro (22). It is well known that base substitutions at position 2058 of the 23S rRNA gene gives resistance to some or all of the macrolide-lincosamide-streptogramin B antibiotics (41). Therefore, the decreased susceptibility to tylosin in all of our clinical isolates of B. hyodysenteriae is likely to be explained by the presence of the A2058 point mutations. In five isolates, four in subset A (RSp9, RSp15, RSp18, and RSp19) and one in subset B (RSp22), the 2058 mutation was the only point mutation detected, so the resistance to tiamulin and valnemulin in RSp9, RSp15, RSp18, and RSp19 must be caused by mechanisms of resistance not detectable in this survey. These could be mutations in other regions of the ribosome, methylations of rRNA, or effects on efflux or influx.
It has been suggested that the binding of tylvalosin is affected by point mutations at position 2058 of the 23S rRNA gene (22, 25). However, our data do not fully support such cross-resistance between tylosin and tylvalosin in B. hyodysenteriae, as MICs as low as 1 μg/ml for tylvalosin were found in isolates for which the tylosin MICs were high and in which the A2058T mutation was present (Tables 1 and 2).
Isolate RSp7, belonging to MLVA type 3 (subset A), contained both the A2058T and an A2059G mutation in the 23S rRNA gene. Both 2058 and 2059 mutations previously have been associated with both macrolide and lincosamide resistance (33, 41). Therefore, we inferred that the increased MICs for lincomycin and tylvalosin in isolate RSp7 compared to those for isolates with only the 2058 mutation were due to the 2059 mutation. In addition, a decrease in pleuromutilin susceptibility has been associated with an A2059G-plus-A2503T change and not the single A2503T change in Mycoplasma gallisepticum (26).
Isolates RSp4, RSp10, and RSp20, belonging to MLVA type 12 (subset A), presented the A2058G mutation together with G2057A-plus-C2611T mutations at the adjacent base pair (Fig. 3). In Mycobacterium smegmatis, such a change of the bp 2057 and 2611 is thought to alleviate the fitness cost for A2058G (32). The fitness cost for A2058G alone has been considered for Helicobacter pylori (3, 10). In the present study, the low rate of A2058G (3 out of 38 isolates) compared to A2058T (35 out of 38) could reflect an associated biological cost of A2058G in B. hyodysenteriae that could be ameliorated by the 2057 and 2611 point mutations. In general, there were no clear indications that 2057 and 2611 mutations influenced pleuromutilin or macrolide susceptibility, since the resistance pattern of these isolates was not considerably different from the patterns of the isolates that only have A2058 mutations as described above. However, lincomycin MICs against RSp4, RSp10, and RSp20 were at the higher end. Interestingly, mutations at position 2611 previously have been related to moderate resistance to lincomycin and clindamycin in chloroplasts of Chlamydomonas reinhardtii, a green alga (16).
Point mutations detected in field isolates belonging to the same MLVA type but classified in different groups according to their susceptibility to tiamulin might be good indicators of the mechanism of in vivo-acquired resistance. On the other hand, single-nucleotide point mutations occurring in a given MLVA type in both susceptible and resistant clones likely are not involved in the resistance mechanism, although some synergistic effect on resistance cannot completely be ruled out. On this basis, G2087T for type 14, C2146T, G2365C, and G2535A for type 3, and C2362T for type 9 are considered irrelevant for the observed difference in resistance. All of these nucleotides are positioned 50 to 120 Å away from the tiamulin binding site in the ribosomal 50S subunit. The methylation of G2535 (39) and the G2535A mutation (1, 2) previously have been shown to confer resistance to the orthosomycin antibiotics avilamycin and evernimicin. Therefore, we infer that the G2535A mutation has been selected by a previous exposure to avilamycin, which has been used extensively for animal growth promotion in Europe.
Five 23S rRNA mutations were found in subset A but not in subset B. The base pair changes A2057G plus C2611T and A2059G were discussed above. There were two isolates, RSp11 and RSP14, in MLVA type 24 that contained A2031T mutations. To our knowledge, this mutation has not been related to any resistance phenotype in any bacteria. A comparison to other isolates from the same MLVA type did not point to any specific effects from this mutation and it might just be a random mutation, but it should be noted that it was present together with an L3 mutation (discussed below). The fifth mutation was G2032A.
G2032A mutation occurs frequently in tiamulin-resistant isolates.
Eight out of the 20 isolates in subset A (40%) presented a G→A transition mutation at position 2032, whereas no isolate in subset B contained this mutation. Besides the ever-present A2058 mutation, the additional 23S rRNA mutations found in these eight isolates were G2535A, C2362T, and A2031G, which were concluded not to have any major effect on the resistance investigated. By examining Table 1, it is very clear that the lincomycin MIC for all of the isolates with the G2032A mutation is high (≥64 μg/ml). This is in agreement with other studies linking 2032 mutations to lincosamide resistance (8, 11). Moreover, the tiamulin MICs for two of the eight isolates with G2032A mutations (RSp3 and RSp12) were markedly high (32 and 64 μg/ml, respectively), and the 2032 mutation could be a candidate for the MIC differences between RSp12 and RSp13 (both MLVA type 9) involving tiamulin, valnemulin, and lincomycin. On the other hand, the tiamulin MICs for MLVA types 24 and 45, with the 2032 mutation, were not increased to that level, thus it must be more complicated than this single change. The G2032A mutation initially was reported to appear together with other mutations after in vitro selection for tiamulin resistance in B. hyodysenteriae (34). The individual contribution to resistance from each of these 23S rRNA mutations (as well as other mutations) was later investigated in Mycobacterium smegmatis, and some of the mutant combinations have been investigated (28, 29). In M. smegmatis, both G2032A and G2032C showed a 4-fold-reduced susceptibility to valnemulin, but the G2032C mutation resulted in a considerably higher clindamycin MIC (a lincosamide) than that after G2032A mutation (29). These studies also showed that combinations of single mutations can cause synergistic effects on antibiotic susceptibility, and that positions distant from an antibiotic binding site can perturb local flexibility and the structure of the drug binding pocket.
Are L3 mutations in Brachyspira spp. resistance determinants?
Bacterial resistance to pleuromutilins in laboratory-induced resistant isolates has been associated with mutations in ribosomal protein L3 genes (4, 13, 30, 34). Therefore, the L3 gene was sequenced for all isolates from subsets A and B to search for further correlation, and six isolates showed changes. An Asn→Ser substitution in amino acid position 148 in protein L3 was detected in three isolates from subset B and one isolate from subset A (Tables 1 and 2). As this mutation was in subset B, it does not appear to provide any reduced susceptibility to tiamulin on its own. The two additional 23S rRNA mutations at 2146 and 2365 in isolate RSp2 (subset A) are not likely to act synergistically with the L3 mutation, as these 23S rRNA positions are far away from the antibiotic binding site. Thus, our data do not support a direct involvement of this mutation in the development of tiamulin resistance. The same mutation also was found previously in three isolates of B. hyodysenteriae with reduced tiamulin susceptibility and in laboratory strains selected for tiamulin resistance (34), although without genetic proof of correlation. The only genetic proof of the involvement of L3 mutations in reduced tiamulin resistance is from an N149D mutation (equivalent to position 148 in Brachyspira) on a plasmid-borne L3 gene in E. coli (4) and a triple L3 mutation in Staphylococcus aureus (13).
A Ser→Thr substitution was found at position 149 in two isolates from subset A (RSp11 and RSp14). Ser149 is close to 23S RNA position G3032 (3 to 4 Å). However, these isolates did not show higher pleuromutilin resistance than the other isolates from the same MLVA type, so again there was no clear indication of relevance for resistance. An S149I change also was observed in B. hyodysenteriae laboratory strains selected for tiamulin resistance (34) but with no genetic proof of correlation. More than 10 different L3 mutations and various combinations of these mutations have been reported in Staphylococcus and have been associated with tiamulin resistance (13, 30). These mutations were at positions equivalent to or relatively close to amino acids 148 and 149 in L3 from Brachyspira spp. It remains to be established whether there is a clear link between all of these L3 mutations and pleuromutilin resistance or if the L3 mutations appear for some other reason. They might be related to some compensatory adaptations or work in concert with unidentified mutations elsewhere. In some isolates they may have been selected by pressure from other antibiotics binding in the peptidyl transferase center.
Conclusion.
In summary, antimicrobial resistance to the main drugs used against B. hyodysenteriae is widespread in Spain. Moreover, the existence of several multiresistant isolates, which are genetically diverse, is reported herein. While mutations at nucleotide position 2058 are involved in tylosin resistance and decreased lincomycin susceptibility, nucleotide 2032 seems to be a key position in the advance toward pleuromutilin resistance and higher lincomycin MICs. A2058G and G2032A mutations had been observed previously by in vitro selection approaches and now also occur in clinical isolates of B. hyodysenteriae, underlining the importance of in vitro selection studies.
ACKNOWLEDGMENTS
We thank Gloria Fernández Bayón and Idoia Portillo Arias for excellent technical assistance.
Álvaro Hidalgo is supported by a grant from Consejería de Educación of the Junta de Castilla y León and the European Social Fund. This work was funded by the Ministerio de Educación y Ciencia (Spanish Ministry of Education and Science) and cofinanced by the European Regional Development Funds (ERDF) as projects AGL2005-01976/GAN (January 2006), AGL2010-18804, and PET 2006-0008.
Footnotes
Published ahead of print on 9 May 2011.
REFERENCES
- 1. Adrian P. V., et al. 2000. Evernimicin (SCH27899) inhibits a novel ribosome target site: analysis of 23S ribosomal DNA mutants. Antimicrob. Agents Chemother. 44:3101–3106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Belova L., Tenson T., Xiong L., McNicholas P. M., Mankin A. S. 2001. A novel site of antibiotic action in the ribosome: interaction of evernimicin with the large ribosomal subunit. Proc. Natl. Acad. Sci. U. S. A. 98:3726–3731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Björkholm B., et al. 2001. Mutation frequency and biological cost of antibiotic resistance in Helicobacter pylori. Proc. Natl. Acad. Sci. U. S. A. 98:14607–14612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bøsling J., Poulsen S. M., Vester B., Long K. S. 2003. Resistance to the peptidyl transferase inhibitor tiamulin caused by mutation of ribosomal protein L3. Antimicrob. Agents Chemother. 47:2892–2896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bulkley D. P., Innis C. A., Blaha G., Steitz T. A. 2010. Revisiting the structures of several antibiotics bound to the bacterial ribosome. Proc. Natl. Acad. Sci. U. S. A. 107:17158–17163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Burch D. G. S. 2005. Pharmacokinetic, pharmacodynamic and clinical correlations relating to the therapy of colonic infections in the pig and breakpoint determinations. Pig J. 56:8–24 [Google Scholar]
- 7. Carvajal A., et al. 2006. Prevalence of Brachyspira species in pigs with diarrhoea in Spain. Vet. Rec. 158:700–701 [DOI] [PubMed] [Google Scholar]
- 8. Cseplö A., Etzold T., Schell J., Schreier P. H. 1988. Point mutations in the 23S rRNA genes of four lincomycin resistant Nicotiana plumbaginifolia mutants could provide new selectable markers for chloroplast transformation. Mol. Gen. Genet. 214:295–299 [DOI] [PubMed] [Google Scholar]
- 9. Davidovich C., Bashan A., Auerbach-Nevo T., Yonath A. 2007. Induced-fit tightens pleuromutilins binding to ribosomes and remote interactions enable their selectivity. Proc. Natl. Acad. Sci. U. S. A. 104:4291–4296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Debets-Ossenkopp Y. J., Brinkman A. B., Kuipers E. J., Vandenbroucke-Grauls C. M., Kusters J. G. 1998. Explaining the bias in the 23S rRNA gene mutations associated with clarithromycin resistance in clinical isolates of Helicobacter pylori. Antimicrob. Agents Chemother. 42:2749–2751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Douthwaite S. 1992. Functional interactions within 23S rRNA involving the peptidyltransferase center. J. Bacteriol. 174:1333–1338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Dunkle J. A., Xiong L., Mankin A. S., Cate J. H. D. 2010. Structures of the Escherichia coli ribosome with antibiotics bound near the peptidyl transferase center explain spectra of drug action. Proc. Natl. Acad. Sci. U. S. A. 107:17152–17157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Gentry D. R., Rittenhouse S. F., McCloskey L., Holmes D. J. 2007. Stepwise exposure of Staphylococcus aureus to pleuromutilins is associated with stepwise acquisition of mutations in rplC and minimally affects susceptibility to retapamulin. Antimicrob. Agents Chemother. 51:2048–2052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Gürel G., Blaha G., Moore P. B., Steitz T. A. 2009. U2504 determines the species specificity of the A-site cleft antibiotics: the structures of tiamulin, homoharringtonine, and bruceantin bound to the ribosome. J. Mol. Biol. 389:146–156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hampson D. J., Fellström C., Thomson J. R. 2006. Swine dysentery, p. 785–805 In Straw B. E., Zimmerman J. J., D'Allaire S., Taylor D. J. (ed.), Diseases of swine, 9th ed. Blackwell Publishing, Ames, IA [Google Scholar]
- 16. Harris E. H., Burkhart B. D., Gillham N. W., Boynton J. E. 1989. Antibiotic resistance mutations in the chloroplast 16S and 23S rRNA genes of Chlamydomonas reinhardtii: correlation of genetic and physical maps of the chloroplast genome. Genetics 123:281–292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hidalgo Á., Carvajal A., García-Feliz C., Osorio J., Rubio P. 2009. Antimicrobial susceptibility testing of Spanish field isolates of Brachyspira hyodysenteriae. Res. Vet. Sci. 87:7–12 [DOI] [PubMed] [Google Scholar]
- 18. Hidalgo Á, Carvajal A., Pringle M., Rubio P., Fellström C. 2010. Characterization and epidemiological relationships of Spanish Brachyspira hyodysenteriae field isolates. Epidemiol. Infect. 138:76–85 [DOI] [PubMed] [Google Scholar]
- 19. Hidalgo Á., et al. 2010. Multiple-locus variable-number tandem repeats analysis of the swine dysentery pathogen, Brachyspira hyodysenteriae. J. Clin. Microbiol. 48:2859–2865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Humphrey W., Dalke A., Schulten K. 1996. VMD-visual molecular dynamics. J. Mol. Graphics 14:33–38 [DOI] [PubMed] [Google Scholar]
- 21. Hunter P. R., Gaston M. A. 1988. Numerical index of the discriminatory ability of typing systems: an application of Simpson's index of diversity. J. Clin. Microbiol. 26:2465–2466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Karlsson M., Fellström C., Heldtander M. U., Johansson K. E., Franklin A. 1999. Genetic basis of macrolide and lincosamide resistance in Brachyspira (Serpulina) hyodysenteriae. FEMS Microbiol. Lett. 172:255–260 [DOI] [PubMed] [Google Scholar]
- 23. Karlsson M., Oxberry S. L., Hampson D. J. 2002. Antimicrobial susceptibility testing of Australian isolates of Brachyspira hyodysenteriae using a new broth dilution method. Vet. Microbiol. 84:123–133 [DOI] [PubMed] [Google Scholar]
- 24. Karlsson M., Fellström C., Gunnarsson A., Landén A., Franklin A. 2003. Antimicrobial susceptibility testing of porcine Brachyspira (Serpulina) species isolates. J. Clin. Microbiol. 41:2596–2604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Karlsson M., Aspán A., Landén A., Franklin A. 2004. Further characterization of porcine Brachyspira hyodysenteriae isolates with decreased susceptibility to tiamulin. J. Med. Microbiol. 53:281–285 [DOI] [PubMed] [Google Scholar]
- 26. Li B. B., et al. 2010. Mutations in 23S rRNA gene associated with decreased susceptibility to tiamulin and valnemulin in Mycoplasma gallisepticum. FEMS Microbiol. Lett. 308:144–149 [DOI] [PubMed] [Google Scholar]
- 27. Lobová D., Smola J., Cizek A. 2004. Decreased susceptibility to tiamulin and valnemulin among Czech isolates of Brachyspira hyodysenteriae. J. Med. Microbiol. 53:287–291 [DOI] [PubMed] [Google Scholar]
- 28. Long K. S., et al. 2009. Single 23S rRNA mutations at the ribosomal peptidyl transferase centre confer resistance to valnemulin and other antibiotics in Mycobacterium smegmatis by perturbation of the drug binding pocket. Mol. Microbiol. 71:1218–1227 [DOI] [PubMed] [Google Scholar]
- 29. Long K. S., et al. 2010. Mutations in 23S rRNA at the peptidyl transferase center and their relationship to linezolid binding and cross resistance. Antimicrob. Agent Chemother. 54:4705–4713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Miller K., Dunsmore C. J., Fishwick C. W., Chopra I. 2008. Linezolid and tiamulin cross-resistance in Staphylococcus aureus mediated by point mutations in the peptidyl transferase center. Antimicrob. Agents Chemother. 52:1737–1742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Osorio J., et al. 2010. Genetic diversity and population structure of B. hyodysenteriae in Spain, p. 125 In grosse Beilage E., Blaha T. (ed.), Proceedings of the 2nd European Symposium on Porcine Health Management. European College of Porcine Health Management, Hannover, Germany [Google Scholar]
- 32. Pfister P., et al. 2005. 23S rRNA base pair 2057-2611 determines ketolide susceptibility and fitness cost of the macrolide resistance mutation 2058A→G. Proc. Natl. Acad. Sci. U. S. A. 102:5180–5185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Poehlsgaard J., Pfister P., Böttger E. C., Douthwaite S. 2005. Molecular mechanisms by which rRNA mutations confer resistance to clindamycin. Antimicrob. Agents Chemother. 49:1553–1555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Pringle M., Poehlsgaard J., Vester B., Long K. S. 2004. Mutations in ribosomal protein L3 and 23S ribosomal RNA at the peptidyl transferase centre are associated with reduced susceptibility to tiamulin in Brachyspira spp. isolates. Mol. Microbiol. 54:1295–1306 [DOI] [PubMed] [Google Scholar]
- 35. Pringle M., et al. 2006. Quality-control ranges for antimicrobial susceptibility testing by broth dilution of the Brachyspira hyodysenteriae type strain (ATCC 27164T). Microb. Drug Resist. 12:219–221 [DOI] [PubMed] [Google Scholar]
- 36. Råsbäck T., Fellström C., Gunnarsson A., Aspán A. 2006. Comparison of culture and biochemical tests with PCR for detection of Brachyspira hyodysenteriae and Brachyspira pilosicoli. J. Microbiol. Methods 66:347–353 [DOI] [PubMed] [Google Scholar]
- 37. Rohde J., Kessler M., Baums C. G., Amtsberg G. 2004. Comparison of methods for antimicrobial susceptibility testing and MIC values for pleuromutilin drugs for Brachyspira hyodysenteriae isolated in Germany. Vet. Microbiol. 102:25–32 [DOI] [PubMed] [Google Scholar]
- 38. Stegeman J. A., Vernooij J. C., Khalifa O. A., Van den Broek J., Mevius D. J. 2006. Establishing the change in antibiotic resistance of Enterococcus faecium strains isolated from Dutch broilers by logistic regression and survival analysis. Prev. Vet. Med. 74:56–66 [DOI] [PubMed] [Google Scholar]
- 39. Treede I., et al. 2003. The avilamycin resistance determinants AviRa and AviRb methylate 23S rRNA at the guanosine 2535 base and the uridine 2479 ribose. Mol. Microbiol. 49:309–318 [DOI] [PubMed] [Google Scholar]
- 40. Tu D., Blaha G., Moore P. B., Steitz T. A. 2005. Structures of MLSBK antibiotics bound to mutated large ribosomal subunits provide a structural explanation for resistance. Cell 121:257–270 [DOI] [PubMed] [Google Scholar]
- 41. Vester B., Douthwaite S. 2001. Macrolide resistance conferred by base substitutions in 23S rRNA. Antimicrob. Agents Chemother. 45:1–12 [DOI] [PMC free article] [PubMed] [Google Scholar]