Abstract
The emergence of multidrug-resistant enterococci as a leading cause of hospital-acquired infection is an important public health concern. Little is known about the genetic mechanisms by which enterococci adapt to strong selective pressures, including the use of antibiotics. The lipopeptide antibiotic daptomycin is approved to treat Gram-positive bacterial infections, including those caused by enterococci. Since its introduction, resistance to daptomycin by strains of Enterococcus faecalis and Enterococcus faecium has been reported but is still rare. We evolved daptomycin-resistant strains of the multidrug-resistant E. faecalis strain V583. Based on the availability of a fully closed genome sequence for V583, we used whole-genome resequencing to identify the mutations that became fixed over short time scales (∼2 weeks) upon serial passage in the presence of daptomycin. By comparison of the genome sequences of the three adapted strains to that of parental V583, we identified seven candidate daptomycin resistance genes and three different mutational paths to daptomycin resistance in E. faecalis. Mutations in one of the seven candidate genes (EF0631), encoding a putative cardiolipin synthase, were found in each of the adapted E. faecalis V583 strains as well as in daptomycin-resistant E. faecalis and E. faecium clinical isolates. Alleles of EF0631 from daptomycin-resistant strains are dominant in trans and confer daptomycin resistance upon a susceptible host. These results demonstrate a mechanism of enterococcal daptomycin resistance that is genetically distinct from that occurring in staphylococci and indicate that enterococci possessing alternate EF0631 alleles are selected for during daptomycin therapy. However, our analysis of E. faecalis clinical isolates indicates that resistance pathways independent from mutant forms of EF0631 also exist.
INTRODUCTION
Enterococci are members of the gastrointestinal tract consortium in humans and most other organisms (1, 69). Enterococcus faecalis and Enterococcus faecium are also leading causes of hospital-acquired infection, including urinary tract infections, bacteremia, and postsurgical wound infections (24, 46). Several lineages of E. faecalis and E. faecium are repeatedly found among multidrug-resistant hospital-acquired infection isolates and appear to have become highly hospital adapted (34, 61). Much of the antibiotic resistance of hospital strains of enterococci is ascribable to the horizontal acquisition of mobile elements (35, 41, 51, 67, 72).
The lipopeptide antibiotic daptomycin is used to treat Gram-positive bacterial infections, including those caused by enterococci and staphylococci. Daptomycin is approved to treat complicated skin and skin structure infections (11) and has been used to treat vancomycin-resistant enterococcal bacteremia and endocarditis, among other infections (8). Daptomycin causes rapid depolarization of the bacterial cell membrane, resulting in cell death in the absence of lysis (10, 66). One model for daptomycin action proposes that daptomycin molecules insert into the cell membrane in a calcium-dependent manner and, once there, oligomerize, forming pores that allow ion leakage from the cell (45, 66). Since gaining FDA approval in 2003, daptomycin nonsusceptibility has been reported, although it remains rare (reviewed by Kelesidis et al. [29]). In this report, we refer to daptomycin nonsusceptibility (defined by the Clinical and Laboratory Standards Institute [CLSI] as an MIC of >4 μg/ml for enterococci [8]) as daptomycin resistance.
Daptomycin resistance has been extensively studied in Staphylococcus aureus, where it results from chromosomal mutation. Microarray-based comparative genome analysis of S. aureus strains subjected to in vitro daptomycin serial passage revealed that certain genes and intergenic regions (mprF, rpoB, yycG, and others) acquired mutations during the evolution of daptomycin resistance (18). Mutations in these regions in many, but not all, daptomycin-resistant S. aureus clinical isolates have also been detected (18, 28, 47, 53, 73, 75). The impact of these genetic changes has not been fully delineated.
Compared to S. aureus, little is known about enterococcal daptomycin resistance mechanisms or, more broadly, about how enterococci adapt to strong selective pressures in the absence of exogenous resistance element acquisition. However, there is evidence that the genome is prone to rapid change (38, 64). Because of the clinical significance of daptomycin, we examined underlying genetic changes in three independent, parallel experiments that involved passaging multidrug-resistant E. faecalis V583 in the presence of increasing levels of daptomycin in vitro. We applied next-generation whole-genome resequencing to identify genetic changes that occurred during the evolution of daptomycin resistance. Bioinformatic methods were adapted to permit analysis of strains possessing a large number of endogenous mobile elements, which occur in the V583 genome (>25% of genomic content [51]). We identified changes in seven genes associated with the development of daptomycin resistance in E. faecalis and three mutational pathways that led to elevated (≥256 μg/ml) daptomycin resistance. The importance of these polymorphisms was verified by identifying identical or similar polymorphisms in daptomycin-resistant E. faecalis and E. faecium clinical isolates and by expressing altered alleles of one candidate gene in trans in a susceptible host, converting it to daptomycin resistant.
MATERIALS AND METHODS
Bacterial strains and media.
Bacterial strains used in this study are shown in Table 1. Unless otherwise noted, E. faecalis and E. faecium were routinely cultured on brain heart infusion (BHI) agar or in BHI broth without agitation at 37°C. Spectinomycin at 750 μg/ml was included where appropriate for selection in E. faecalis.
Table 1.
Enterococcal strains used in this study
| E. faecalis straina | Descriptionb | DAP MIC (μg/ml) | Reference/source |
|---|---|---|---|
| Laboratory strains | |||
| V583 (CB807) | Vancomycin-resistant (vanB) clinical isolate | 2 | 62 |
| DAP-A (CB2502) | Day 12 isolate from V583 serial passage expt A | 256 | This study |
| DAP-B (CB2503) | Day 13 isolate from V583 serial passage expt B | 256 | This study |
| DAP-C (CB2504) | Day 14 isolate from V583 serial passage expt C | 512 | This study |
| OG1RF | Human oral cavity isolate | 4 | 19 |
| OG1RF pAT28 | OG1RF plus shuttle vector pAT28 | 4 | This study |
| KP101 | OG1RF plus pAT28 containing a 1,733-bp KpnI/XbaI fragment with V583 EF0631 and the putative EF0631 promoter region | 4 | This study |
| KP102 | OG1RF plus pAT28 containing a 1,724-bp KpnI/XbaI fragment with DAP-A EF0631 and the putative EF0631 promoter region | 8 | This study |
| KP103 | OG1RF plus pAT28 containing a 1,733-bp KpnI/XbaI fragment with DAP-C EF0631 and the putative EF0631 promoter region | 64 | This study |
| Clinical strains | |||
| ACL-1781ES | VSE, 12 March 2009 urine isolate; Indianac | 8 | LSId |
| ESJH-1310ES | VSE, 20 July 2008 coccyx wound isolate; Colorado | 8 | LSI |
| MCA-1970ES | VSE, 12 June 2009 body fluid isolate; Colorado | 32 | LSI |
| NYU-529ES | VSE, 19 January 2006 blood isolate; New York | 8 | LSI |
| OHMC-239ES | VSE, 27 February 2005 blood isolate; Washington | 8 | LSI |
| SFOK-980ES | VSE, 7 November 2007 blood isolate; Oklahoma | 8 | LSI |
| Sequential clinical strains | |||
| Patient 1 | |||
| CMC-041EM | VRE, 28 April 2004 blood isolate; New York | 4 | 56; LSI |
| CMC-087EM | VRE, 30 May 2004 blood isolate; New York | >32 | 56; LSI |
| CMC-088EM | VRE, 1 June 2004 blood isolate; New York | >32 | 56; LSI |
| Patient 2 | |||
| UTHS-218EM | VRE, 10 September 2004 urine isolate; Texas | 4 | 37; LSI |
| UTHS-219EM | VRE, 6 October 2004 blood isolate; Texas | 32 | 37; LSI |
Where appropriate, Cubist strain identifiers (CB numbers) are shown.
VSE, vancomycin-sensitive Enterococcus; VRE, vancomycin-resistant Enterococcus.
Location of submitting laboratory.
LSI = Daptomycin Reference Lab, Laboratory Specialists, Inc., Westlake, Ohio.
PCR and routine DNA sequencing.
PCR was performed using Taq polymerase (New England Biolabs) per the manufacturer's recommendations. Colony lysates were used as PCR templates as previously described (50). Primers utilized in this study are listed in Table S1 in the supplemental material. Routine DNA sequencing was performed by the Massachusetts General Hospital DNA Sequencing Core Facility.
Analysis of growth and antibiotic MIC.
E. faecalis growth was monitored by recording the optical density at 600 nm (OD600) using a BioTek Synergy 2 microplate reader. Late-exponential-phase bacteria were diluted to an OD600 of 0.0001 in fresh BHI broth, and 200 μl was transferred to the wells of 96-well plates. Growth was then measured at 20-min intervals over 12 h. Average generation times and growth yields were calculated based on 6 growth curves per strain. Statistical significance was assessed using a two-tailed Student's t test for samples with equal variance. Daptomycin MICs were determined by broth microdilution, using Mueller-Hinton broth supplemented with 50 mg/liter Ca2+ (MHBc).
In vitro evolution of daptomycin-resistant E. faecalis.
Daptomycin-resistant variants of E. faecalis V583 were generated by serial passage through increasing concentrations of daptomycin. E. faecalis V583 serial passage experiments were performed in triplicate, parallel experiments, designated A, B, and C. In each passage, E. faecalis V583 was exposed to daptomycin concentrations below, at, or above the MIC. The daptomycin MIC for E. faecalis V583 was determined to be 2 μg/ml. On day 1, E. faecalis V583 was adjusted to an OD600 of 0.1 and exposed to 1 μg/ml, 2 μg/ml, 4 μg/ml, and 8 μg/ml daptomycin in MHBc in triplicate experiments. Following 24 h of incubation at 37°C with shaking at 200 rpm, the highest drug concentration that contained visible growth in each experiment was used to determine the daptomycin concentrations to be tested on day 2. The culture in the well with the highest drug concentration containing growth was adjusted to an OD600 of 0.1 and used as the inoculum for the next round of daptomycin selection. Cultures were incubated for 24 h at 37°C with shaking as described above. This was repeated with increasing concentrations of daptomycin until MIC values plateaued, which occurred over a period of 12 to 14 days. Daily aliquots of serial passage cultures for each of the triplicate experiments were recovered and stored at −80°C. Finally, isolates obtained from the final serial passage of each experiment were passaged at least three times on daptomycin-free medium and confirmed to be stably resistant to daptomycin.
Resequencing and identification of polymorphisms.
The three daptomycin-resistant strains chosen for resequencing, DAP-A, DAP-B, and DAP-C, were isolated on day 12, day 13, and day 14, respectively, of independent serial passage experiments and had the highest daptomycin MIC values (MICs of 256 μg/ml, 256 μg/ml, and 512 μg/ml, respectively). The control strain used for resequencing was the E. faecalis parent strain V583. Genomic DNA was harvested from overnight control V583, DAP-A, DAP-B, and DAP-C E. faecalis broth cultures by phenol-chloroform extraction as described previously (39). Libraries for resequencing were prepared by the Tufts University DNA Core Facility. A modified Illumina protocol (D. Lazinski and A. Camilli, Tufts University School of Medicine, personal communication) was used. Briefly, genomic DNA was nebulized, and the resulting DNA fragments were blunted using the Quick Blunting kit (New England Biolabs). 3′ dATP overhangs were added to blunt-ended fragments using Klenow (New England BioLabs), and Illumina OLJ131/OLJ137 adapters were ligated to 3′ overhangs using the Quick Ligation kit (New England Biolabs). Ligations were analyzed by agarose gel electrophoresis, and ∼400- to 500-bp fragments were recovered by gel purification using the QIAquick gel extraction kit (Qiagen). Gel-purified DNA was finally used as a template in 15 cycles of PCR amplification with primers targeting the OLJ131/OLJ137 adapters. Libraries were sequenced using an Illumina genome analyzer II at the Tufts University Core Facility. Single-end sequencing reads were filtered to exclude reads with a quality score (Q score) of <25 at any position. Reads for the control assembly were 40 bp in size; reads for DAP genomes were 38 bp.
To generate consensus resequencing assemblies, filtered reads were assembled by comparison to the E. faecalis reference strain V583 chromosome (GenBank accession no. AE016830) and plasmids pTEF1 (GenBank accession no. AE016833), pTEF2 (GenBank accession no. AE016831), and pTEF3 (GenBank accession no. AE016832) and to the OLJ131 and OLJ137 Illumina adapters using the CLC Genomics Workbench (CLC Bio). Alignments were generated using the following settings: limit, 11; mismatch cost, 2; gapped alignment, yes; insertion and deletion costs, 3; global alignment, no; and random assignment of nonspecific matches, yes. The software was directed to randomly assign reads corresponding to repetitive sequences such as rRNA operons and insertion sequence (IS) elements. A summary of read assemblies is shown in Table S2 in the supplemental material. Fourteen zero-coverage regions were identified in the DAP-A, DAP-B, and DAP-C assemblies (see Table S3 in the supplemental material); no zero-coverage regions were detected in the control strain assembly. All zero-coverage regions were closed by PCR amplification and sequencing using appropriate primers.
Single nucleotide polymorphisms (SNPs) and deletion-insertion polymorphisms (DIPs) in resequencing assemblies were detected using CLC Genomics Workbench default settings, which analyze regions with at least 4-fold sequencing coverage and report polymorphisms with a minimum variant frequency of 35%. Polymorphisms unique to the daptomycin-resistant strains (see Table 3) were independently confirmed by PCR amplification and sequencing. PCR confirmation reactions were performed for all four strains, irrespective of the strain in which the polymorphism was detected. Polymorphisms between the control V583 sequence determined here and the published V583 genome sequence (51) were removed from further analysis.
Table 3.
Putative mobile element translocation sites detected by computational analysis and results of confirmation
| Strain | Approx reference positiona | Overlapping annotation | Confirmation resultb | Amino acid change |
|---|---|---|---|---|
| B | 1739313 | EF1797, conserved hypothetical protein | IS256insertion in EF1797 coding region | V16insIS256 |
| A | 671048 | EF0716, conserved hypothetical protein | 9-bp (TTCTGGGAA) duplication | W148_F150dup |
| C | 671036 | EF0716, conserved hypothetical protein | 9-bp (TTCTGGGAA) deletion | W148_F150del |
| C | 2391641 | EF2470, HD domain protein | 4-bp (ATTG) duplication | W82X |
Location of putative translocation sites in resequencing assemblies identified by computational analysis.
Confirmed mobile element translocation shown in boldface; other confirmed mutations shown in lightface.
To detect putative IS and other mobile element translocations, software settings were manipulated to generate assemblies based on global, rather than local, alignments. Resequencing assemblies were regenerated from filtered sequencing reads as described above, except that the global alignment setting was enabled. Utilization of this option forced alignment of unmatched read ends, which were masked in local alignments. Unmasking resulted in detection of multiple polymorphisms flanking putative transposition sites. Regions of interest were visually examined in resequencing assemblies, and those with alignment signatures similar to that observed for the IS256 insertion in DAP-B EF1797 (see Fig. S1 in the supplemental material for an illustration) were confirmed by PCR amplification and sequencing using the appropriate primers.
Bioinformatic analysis of daptomycin resistance candidates in other enterococcal strains.
Draft genome data for 16 E. faecalis and 8 E. faecium strains (49) were accessed at the Broad Institute Enterococcus database website (http://www.broadinstitute.org/annotation/genome/enterococcus_faecalis/MultiHome.html). The predicted frameshift mutation observed in EF0631 of E. faecalis T2 was confirmed by PCR and sequencing. Select Enterococcus and Listeria protein sequences were downloaded from GenBank. DNA and protein alignments were generated using ClustalW in MacVector. PSORTb (76; http://www.psort.org/psortb/index.html) and TMHMM 2.0 (33; http://www.cbs.dtu.dk/services/TMHMM/) were used to generate protein localization and structure predictions. BLASTP (2; http://blast.ncbi.nlm.nih.gov/Blast.cgi) and Pfam (16; http://pfam.janelia.org/) were used to identify homologues of candidate proteins. The NCBI Conserved Domains search tool (40; http://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi) and Pfam-A were used for functional classification of candidate proteins.
Assessment of phenotypic dominance of EF0631 alleles in E. faecalis OG1RF.
EF0631 alleles were amplified from wild-type V583 and daptomycin-resistant DAP-A and DAP-C derivatives using the primers EF0631 KpnI For and EF0631 XbaI Rev (see Table S1 in the supplemental material). Products were digested with the appropriate restriction enzymes (New England Biolabs) and ligated with KpnI/XbaI-digested pAT28 (71) using T4 DNA ligase (New England Biolabs). Ligations were directly transformed into E. faecalis OG1RF (19) by electroporation (65) after constructs were observed to be toxic to Escherichia coli (data not shown). Colonies obtained from OG1RF electroporation reactions were screened by PCR using a primer with identity to the pAT28 vector and a primer with identity to the EF0631 inserts (Table S1). Plasmid was isolated from PCR-positive strains, and sequences of EF0631 inserts were confirmed. Strains with wild-type EF0631 (KP101), DAP-A EF0631 (KP102), and DAP-C EF0631 (KP103) inserts were obtained.
Nucleotide sequence accession number.
DNA sequence data generated in this study have been submitted to the NCBI Sequence Read Archive under accession number SRA029563.1.
RESULTS
In vitro evolution of daptomycin-resistant E. faecalis V583.
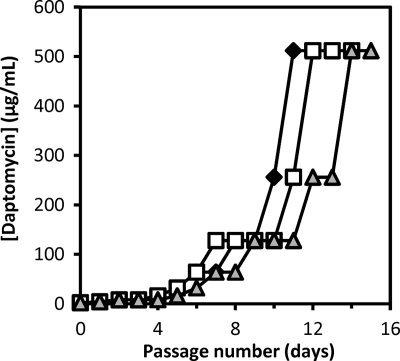
To identify the genetic changes that underlie the development of daptomycin resistance in enterococci, we generated daptomycin-resistant variants of multidrug-resistant E. faecalis V583. E. faecalis V583 is a member of an enterococcal clonal complex associated with hospital infection outbreaks and endemicity (61) and is vancomycin resistant (62) but daptomycin sensitive (MIC = 2 μg/ml). Daptomycin-resistant variants of E. faecalis V583 were generated by serial passage in increasing concentrations of daptomycin, using an approach similar to that described for S. aureus (18). A step pattern of resistance to daptomycin was observed in each of 3 independent experiments over the course of 15 days (Fig. 1). Daptomycin MICs for mutants obtained through serial passage plateaued at 512 μg/ml, representing a 256-fold increase in the MIC from the parent strain. Strains recovered from day 12 of experiment A (DAP-A), day 13 of experiment B (DAP-B), and day 14 of experiment C (DAP-C) were passaged on drug-free medium and were found to have stable daptomycin MICs of 256 μg/ml, 256 μg/ml, and 512 μg/ml, respectively. The vancomycin MICs of these strains were comparable to each other (32 to 64 μg/ml) and to that previously reported for E. faecalis V583 (64 μg/ml [62]).
Fig. 1.
Daptomycin serial passage experiments. The highest daptomycin concentration demonstrating growth (in μg/ml) is shown on the y axis for each day of passage. Diamonds, experiment A; squares, experiment B; triangles, experiment C.
In antibiotic-free BHI broth, DAP-A had an average doubling time similar to that of the parent V583 strain (38.3 ± 1.2 min and 38.5 ± 1.0 min, respectively), while the DAP-B and DAP-C doubling times were slightly, but significantly, longer (DAP-B, 44.6 ± 1.4 min, P = 0.001; DAP-C, 47.4 ± 2.8 min, P = 0.005). Growth yields were not significantly different among the four strains (data not shown). Thus, there does not appear to be a substantial fitness cost in vitro for daptomycin resistance. The small but reproducible differences in growth rates observed among the evolved strains suggest that DAP-B and DAP-C have altered physiologies compared to that of DAP-A.
Resequencing of daptomycin-resistant E. faecalis to identify candidate genes.
A closed genome sequence for E. faecalis V583 is available (51). For maximum accuracy, we used highly redundant, short-read Illumina whole-genome resequencing to identify genetic changes in DAP-A, DAP-B, and DAP-C. The E. faecalis V583 parent strain used in these experiments was also resequenced to control for changes that may have occurred since its isolation. The number of reads generated per strain after quality filtering of sequence data is shown in Table S2 in the supplemental material. Reads were aligned to the reference V583 chromosome (3.218-Mbp), pTEF1 (66.32-kb), pTEF2 (57.66-kb), and pTEF3 (17.96-kb) sequences to generate consensus genome assemblies for each strain. The range of fold coverage, average fold coverage, and additional assembly data are shown in Table S2. For >99.5% of nucleotide positions in each strain assembly, at least 20-fold coverage was attained.
While the control strain was sequenced to a minimum of 3-fold coverage (see Table S2 in the supplemental material), 14 zero-coverage regions were detected in the DAP strain assemblies (see Table S3 in the supplemental material). Most of these gaps occur in vanB or the genes surrounding vanB, a region of anomalously high GC content in the V583 genome (51). We further analyzed these gap regions by PCR and sequencing. A total of 13 of 14 gaps were the result of legitimate zero coverage during Illumina resequencing and were closed (Table S3). The remaining gap represents a bona fide 75-bp deletion in the strain DAP-C in EF1797, which encodes a conserved hypothetical protein. Because 38-bp reads were obtained, this 75-bp deletion created a gap in the DAP-C resequencing assembly. This deletion identified EF1797 as a candidate gene contributing to daptomycin resistance in DAP-C.
We compared reference and consensus assemblies to identify putative polymorphisms in control and DAP strains, using standard software settings (see Materials and Methods). The control genome sequence possesses 39 SNPs and DIPs relative to those of the V583 GenBank reference sequence, all occurring in the chromosome (see Table S4 in the supplemental material) and all shared with DAP-A, DAP-B, and DAP-C. These polymorphisms may result from the different sequencing methodologies used to generate the V583 GenBank reference sequence (Sanger sequencing) and that used here (Illumina). Alternatively, they may be the result of laboratory passage, as has recently been documented for Pseudomonas aeruginosa PAO1 strains (30). As these polymorphisms were shared by both DAP strains and the control, we removed them from further analysis.
Ten polymorphisms were detected in the DAP-A, DAP-B, and DAP-C genome assemblies that did not occur in the control strain (Table 2). All of these polymorphisms occur within coding regions on the chromosome and lead to either nonsynonymous or frameshift mutations in seven candidate daptomycin resistance genes. No synonymous mutations were found. Importantly, all three DAP strains possess polymorphisms in EF0631, encoding a putative cardiolipin (CL) synthase (Cls).
Table 2.
Polymorphisms detected by SNP/DIP analysis
| Strain | Reference position(s)a | Variation type | Referencef | Variation(s) | Fold coverageb | Count | Variation frequency(ies) (%) | Overlapping annotationc | Position within gened | Amino acid change |
|---|---|---|---|---|---|---|---|---|---|---|
| A | 206948 | Complex SNP | C | C/A | 40 | 23/17 | 57.5/42.5 | EF0224, rpsE | 283/501 | L95I |
| C | 219603 | Complex SNP | A | A/C | 148 | 86/62 | 58.1/41.9 | EF0243, brnQ | 1101/1356 | S367R |
| A | 585518–585519 | DIP | A | 26 | 20 | 76.9 | EF0631, putative cardiolipin synthetase | 226-7/1446 | N77fse | |
| B | 585945 | SNP | G | A | 113 | 113 | 100 | EF0631, putative cardiolipin synthetase | 653/1446 | R218Q |
| C | 585945 | SNP | G | A | 128 | 127 | 99.2 | EF0631, putative cardiolipin synthetase | 653/1446 | R218Q |
| A | 740197 | SNP | C | G | 78 | 78 | 100 | EF0782, rpoN | 296/1314 | T99R |
| B | 1699646 | DIP | T | 98 | 82 | 83.7 | EF1753, conserved hypothetical protein | 1168/1602 | I390fs | |
| C | 1699646 | DIP | T | 73 | 55 | 75.3 | EF1753, conserved hypothetical protein | 1168/1602 | I390fs | |
| A | 1739262 | DIP | C | 62 | 62 | 100 | EF1797, conserved hypothetical protein | 88/645 | D30fs | |
| B | 2613619 | SNP | A | T | 83 | 83 | 100 | EF2698, putative tellurite resistance protein TelA | 829/1188 | I277F |
Position in the E. faecalis V583 reference sequence.
Fold coverage at the specified nucleotide in the resequencing assembly.
Annotated open reading frames (ORFs) overlapping computationally predicted polymorphisms.
Location of polymorphism relative to the gene sequence; shown as polymorphism position/total nucleotide positions within the gene.
N77fs predicted by computational analysis prior to confirmation.
Nucleotide in the E. faecalis V583 reference sequence.
Confirmation of polymorphisms identified by resequencing.
To confirm polymorphisms, we amplified and sequenced regions of the 7 genes of interest (EF0224 rpsE, EF0243 brnQ, EF0631, EF0782 rpoN, EF1753, EF1797, EF2698) using each of the strains (control, DAP-A, DAP-B, DAP-C) as templates. Mutations in EF0224 rpsE and EF0243 brnQ were not confirmed by this analysis. A region of the Illumina OLJ131 adapter has identity to rpsE, and examination of the DAP-A resequencing alignment indicates that OLJ131-rpsE hybrids aligned to rpsE, generating a false-complex SNP. Indeed, this erroneous alignment occurred in all four genome assemblies, although it exceeded the SNP detection threshold only in DAP-A (data not shown). Conversely, there is no evidence for adapter interference in the DAP-C brnQ complex SNP call. Variation at this nucleotide position does not occur in the control, DAP-A, and DAP-B resequencing assemblies, nor does examination of DAP-C sequencing chromatograms (n = 3) support variation at this position. Therefore, the initial polymorphism identified in brnQ in DAP-C was deemed a computational artifact and not considered further.
The remaining polymorphisms, occurring in EF0631, EF0782 rpoN, EF1753, EF1797, and EF2698, were confirmed by PCR and sequencing. Note that the frameshift mutation computationally predicted in DAP-A EF0631 is actually a 9-bp deletion. Two 9-bp repeats (GAATTTCCA) occur in tandem in EF0631, and one of these repeats is deleted in DAP-A.
Since some mutations were predicted to occur in one strain and not in others, each of these genes was resequenced in each strain to confirm the absence of polymorphism. Each was verified with one exception. Amplification of EF1797 from DAP-B yielded an amplicon of anomalously large size (data not shown). Sequencing of this product revealed a novel IS256 insertion in DAP-B EF1797. This insertion was not detected in the initial computational analysis because of masking in the local alignment used for genome assembly (see Fig. S1 in the supplemental material).
Identification of putative mobile element translocation in resequencing assemblies.
The E. faecalis V583 genome possesses 38 IS elements (51). It is possible that other novel IS insertions, in addition to that observed for DAP-B EF1797, occurred in the DAP strains or that movements of larger elements such as phage, plasmids, or the pathogenicity island (64) occurred. We modified methods to generate resequencing assemblies such that the known DAP-B EF1797 IS256 insertion could be detected by polymorphism analysis (see Materials and Methods). Using this approach, we detected four additional putative mobile element translocations, two shared by the control and daptomycin-resistant strains (data not shown) and two unique to daptomycin-resistant strains (Table 3). These unique sites occur within DAP-A and DAP-C EF0716 and DAP-C EF2470.
We examined EF0716 and EF2470 in control and DAP strains by PCR and sequencing (Table 3). This analysis revealed that microdeletions and duplications, not mobile element translocations, occur in these genes, resulting in anomalous alignments similar to that observed for the DAP-B EF1797 IS256 insertion. An internal 9-bp sequence (TTCTGGGAA) of EF0716 is duplicated in DAP-A, while the same sequence is deleted in DAP-C. EF0716 is identical to the reference in the control and DAP-B strains. For DAP-C EF2470, a 4-bp internal sequence (ATTG) is duplicated. EF2470 is identical to the reference in the control and DAP-A strains. In repeated sequencing attempts, we failed to resolve one nucleotide position of DAP-B EF2470 (position 350 of 516). Sequencing chromatograms reveal a T/G variation at this position; T is the ancestral nucleotide. Examination of the DAP-B resequencing assembly confirmed that the T/G variation occurs at this position, although below the threshold used for SNP detection, as follows: 83 counts for T (81.4% frequency) and 19 counts for G (18.6% frequency), with 102-fold coverage. No variation at this position was observed in the control, DAP-A, and DAP-C resequencing assemblies.
In summary, closure of zero-coverage regions in resequencing assemblies and confirmation of point mutations, microdeletions, and duplications identified by computational analysis yielded 7 candidate daptomycin resistance genes, 2 of which (EF0631 and EF1797) are common to all three daptomycin-resistant strains. The predicted amino acid changes associated with these mutations are shown in Table 4.
Table 4.
Summary of amino acid changes in E. faecalis V583 DAP strains
| ORF | Gene | Annotated protein function | Amino acid changea |
||
|---|---|---|---|---|---|
| DAP-A | DAP-B | DAP-C | |||
| EF0631 | Putative cardiolipin synthase | N77_Q79del | R218Q | R218Q | |
| EF1797 | Hypothetical protein | D30fs | V16insIS256 | G130_F154del | |
| EF0716 | Hypothetical protein | W148_F150dup | W148_F150del | ||
| EF0782 | rpoN | Alternative σ factor σ54 | T99R | ||
| EF1753 | Hypothetical protein | I390fs | I390fs | ||
| EF2470 | HD domain-containing protein | (I117S) | W82X | ||
| EF2698 | Putative tellurite resistance protein TelA | I277F | |||
Daptomycin MICs for DAP-A, DAP-B, and DAP-C were 256, 256, and 512 μg/ml, respectively.
EF0631 putative cardiolipin synthase.
Cardiolipin (bisphosphatidylglycerol) is an acidic, negatively charged phospholipid component of prokaryotic and mitochondrial membranes (63). Cardiolipin synthase (Cls) catalyzes the reversible transphosphatidylation of two phosphatidylglycerol (PG) molecules, generating cardiolipin (63). Cls belongs to the phospholipase D (PLD) protein superfamily, characterized by two conserved PLD signature motifs [HxK(x)4D(x)6G(G/S), referred to as an HKD motif] (54, 68). Cls exhibits PLD activity, converting phosphatidylglycerol to a phosphatidyl-enzyme intermediate that can subsequently transfer the phosphatidyl group to a phosphatidylglycerol acceptor (63). EF0631 encodes a putative cardiolipin synthase (PRK01642 Cls; expectation value of 1.92 × 10−99) with two PLD active-site signature motifs and two N-terminal transmembrane (TM) helices predicted by TMHMM (Fig. 2). The R218Q substitution shared by DAP-B and DAP-C occurs between conserved H and K residues required for PLD catalytic activity in other organisms (36, 60). Replacements between the H and K residues in each HKD domain of a plant PLD significantly altered enzyme activities, with effects ranging from a complete loss of activity to an enhancement of hydrolysis and transphosphatidylation activities (36). This suggests that the R218Q substitution in DAP-B and DAP-C could significantly alter the activity of EF0631 Cls. N77_Q79del in DAP-A does not occur within a predicted catalytic domain.
Fig. 2.
EF0631 predicted cardiolipin synthase. The EF0631 protein sequence with predicted transmembrane (TM) helices and phospholipase D (PLD) domains is shown. The location of DAP-A N77_Q79del is boxed in green. The location of the DAP-B and DAP-C R218Q substitution is indicated by an asterisk. Transmembrane helices were predicted by TMHMM, and PLD domains were predicted by Pfam-A.
EF0631 homologues from E. faecalis V583 and 17 other E. faecalis strains were aligned and are shown in Fig. S2 in the supplemental material. With the exception of that in E. faecalis T2, EF0631 is highly conserved. E. faecalis T2 possesses a single base pair deletion in an A-rich region, resulting in a frameshift. This mutation was confirmed by PCR and sequencing (data not shown). The frameshift in E. faecalis T2 EF0631 occurs upstream of the coding region for the first EF0631 HKD signature motif. The presence of this mutation in E. faecalis T2 suggests that EF0631 is a nonessential gene. E. faecalis T2 was found to be daptomycin sensitive (MIC of 1 μg/ml).
E. faecalis V583 possesses a second predicted cardiolipin synthase encoded by EF1608. The EF0631 and EF1608 proteins share two predicted HKD domains and 45% overall amino acid sequence identity; the EF0631 N77 to Q79 amino acid residues are not conserved (see Fig. S3 in the supplemental material). EF1608 homologues were identified in 17 additional E. faecalis strains (data not shown), suggesting that, like EF0631, EF1608 is core to the E. faecalis genome. The significance of this potential redundancy is unknown.
EF1797 hypothetical protein.
EF1797 is a predicted membrane protein (PSORTb) with 6 TM helices predicted by TMHMM (Fig. 3) (but posterior probability values for TM helix 6 are weak). The frameshift in DAP-A and the IS256 disruption in DAP-B occur in regions coding for predicted TM helix 1 and the extracellular loop between TM helices 1 and 2, respectively (Fig. 3). The EF1797 protein is likely to be nonfunctional in DAP-A and DAP-B. The DAP-C G130_F154 deletion encompasses a portion of predicted TM helix 4 and the intracellular loop between TM helices 4 and 5 (Fig. 3), suggesting that residues within this region are important for EF1797 activity. Alternatively, it is possible that this region interacts with daptomycin at the cell membrane or that the deletion alters the accessibility of other regions of the protein.
Fig. 3.
EF1797 hypothetical protein. The EF1797 protein sequence with predicted transmembrane (TM) helices is shown. Transmembrane helices were predicted by TMHMM. The location of the DAP-A frameshift is denoted by a blue asterisk. The location of the DAP-B IS256 insertion is denoted by a red asterisk. The location of the DAP-C G130_F154 deletion is shown.
BLASTP and Pfam-B analysis of EF1797 reveals a limited phylogenetic distribution. Pfam-B analysis assigns EF1797 to Pfam-B family 12657 (PB012657; 210/214 amino acids aligned, expectation value of 8.2 × 10−111), which has a species distribution limited to E. faecalis, E. faecium, and Listeria. This limited phylogenetic distribution is supported by BLASTP analysis of EF1797 against the NCBI nonredundant protein and Broad Institute Enterococcus databases. An alignment of proteins from 18 E. faecalis strains, 9 E. faecium strains, and selected Listeria spp., each assigned to Pfam-B family 12657 with expectation values of <1.9 × 10−103 and with ≥41% protein sequence identity to EF1797, is shown in Fig. S4 in the supplemental material. All 9 E. faecium strains carry what appears to be a core EF1797 protein but vary in content of an additional EF1797 protein.
A potential lead for EF1797 function was identified. Pfam-A analysis assigns EF1797 amino acids 114 to 207 to the serine incorporator (Serinc) family with a weak expectation value (0.0093). Serinc is a recently discovered family of eukaryotic transmembrane proteins that facilitate phosphatidylserine and sphingolipid biosynthesis in eukaryotic cells and facilitate phosphatidylserine biosynthesis when heterologously expressed in Escherichia coli (26). In a model for Serinc function, Serinc acts a scaffold for serine and phospholipid biosynthesis machinery and facilitates transport of the polar substrate serine into the hydrophobic membrane (26). EF1797 could play a similar role in E. faecalis.
Early mutations in daptomycin resistance.
To determine whether a hierarchy in mutations occurred during the evolution of daptomycin resistance, culture aliquots obtained directly from days 4 and 5 of serial passage were streaked on BHI agar, and 10 to 15 colonies were mixed and used as PCR templates to identify changes in candidate gene sequences. These serial passage days were chosen because the levels of daptomycin resistance (8 to 32 μg/ml) (Table 5) resemble those observed for clinical daptomycin-resistant enterococcal isolates (Table 1) (29).
Table 5.
Early mutations in serial passage experiments
| Day | Passage MIC (μg/ml) | Mutation ina: |
||||||
|---|---|---|---|---|---|---|---|---|
| EF0631 | EF0716 | RpoN | EF1753 | EF1797 | EF2470 | EF2698 | ||
| Expt A | ||||||||
| 4 | 16 | N77_Q79del | D30fs | |||||
| 5 | 32 | N77_Q79del | wt/T99Rb | D30fs | ||||
| 13 | N77_Q79del | W148_F150dup | T99R | D30fs | ||||
| Expt B | ||||||||
| 4 | 16 | R218Q | I390fs | I277F | ||||
| 5 | 32 | R218Q | I390fs | I277F | ||||
| 14 | R218Q | I390fs | V16insIS256 | I117S | I277F | |||
| Expt C | ||||||||
| 4 | 8 | wt/R218Qb | wt/I390fsb | |||||
| 5 | 16 | R218Q | I390fs | |||||
| 15 | R218Q | W148_F150del | I390fs | G130_F154del | W82X | |||
Amino acid changes are shown.
A mixture of wild-type (wt) and mutant alleles were detected.
By examining sequencing chromatograms, we detected mixed populations of wild-type and mutant alleles of a subset of candidate daptomycin resistance genes at days 4 and 5 of passage (Table 5). The pattern of mutation among the three experiments differs: EF0631, EF1797, and rpoN mutations occurred early in experiment A, EF0631, EF1753, and EF2698 mutations occurred early in experiment B, and EF0631 and EF1753 mutations occurred early in experiment C. Interestingly, EF0631 mutations occurred early in all three serial passage experiments, at the latest by day 4 of serial passage, and this was the only commonality among the three experiments.
EF0631 alleles in daptomycin-resistant clinical isolates.
With the exception of that in E. faecalis T2, the EF0631 predicted amino acid sequence is highly conserved among a collection of 18 E. faecalis strains for which genome sequence information is available (see Fig. S2 in the supplemental material). Because EF0631 mutations occurred early in the evolution of daptomycin resistance (Table 5), we surmised that alternate EF0631 alleles, presumably encoding Cls proteins with altered activities, could be present in daptomycin-resistant enterococcal clinical isolates. We used PCR and sequencing to profile EF0631 alleles in 6 daptomycin-resistant clinical isolates of E. faecalis. A summary of the amino acid changes observed in these strains, using V583 EF0631 as a reference, is shown in Table 6; an alignment of these sequences is shown in Fig. S2 in the supplemental material. The clinical isolate with the highest daptomycin MIC (MCA-1970ES; 32 μg/ml) lacks amino acids N77 to Q79, identical to DAP-A EF0631. An R267S substitution was observed in a second strain having a lower daptomycin MIC (SFOK-980ES; 8 μg/ml). The remaining 4 daptomycin-resistant E. faecalis strains (MICs of 8 μg/ml) encode predicted EF0631 proteins that are identical to E. faecalis V583 EF0631. This result indicates that alternate paths to daptomycin resistance, not involving early EF0631 mutation, exist.
Table 6.
Predicted Cls amino acid changes in daptomycin-resistant clinical isolates
| E. faecalis strain | Description | Daptomycin MIC (μg/ml) | EF0631/EFTG_00614 amino acid change |
|---|---|---|---|
| Clinical strainsa | |||
| OHMC-239ES | 27 February 2005 blood isolate | 8 | |
| NYU-529ES | 19 January 2006 blood isolate | 8 | |
| ESJH-1310ES | 10 July 2008 coccyx wound isolate | 8 | |
| ACL-1781ES | 12 March 2009 urine isolate | 8 | |
| SFOK-980ES | 7 November 2007 blood isolate | 8 | R267S |
| MCA-1970ES | 12 June 2009 body fluid isolate | 32 | N77_Q79del |
| Sequential strains from patient 1b | |||
| CMC-041EM | 28 April 2004 blood isolate | 4 | |
| CMC-087EM | 30 May 2004 blood isolate | >32 | V37A |
| CMC-088EM | 1 June 2004 blood isolate | >32 | V37A |
| Sequential isolates from patient 2c | |||
| UTHS-218EM | 10 September 2004 urine isolate | 4 | |
| UTHS-219EM | 6 October 2004 blood isolate | 32 | R218Q |
Two sets of sequential E. faecium isolates, from cases in which strains had converted to resistance during treatment (37, 56), were also examined. The clonality of each isolate set was previously demonstrated (37, 56). We first identified EF0631 homologues in E. faecium using BLASTP analysis. An alignment of EF0631 proteins from E. faecalis V583 and from 9 E. faecium strains is shown in Fig. S5 in the supplemental material. We then designed primers to amplify interior fragments of E. faecium 1,231,410 EFTG_00614, which is annotated as a phospholipase D/transphosphatidylase and shares 69% amino acid identity with EF0631. E. faecium 1,231,410 also possesses a predicted cardiolipin synthase, EFTG_01168, with lower identity to EF0631 (40% amino acid identity). Highly conserved homologues of EFTG_01168 were found in an additional 8 E. faecium strains (data not shown). Thus, similar to E. faecalis, E. faecium encodes at least two putative cardiolipin synthases, and both of these are core components of the E. faecium genome. We used EFTG_00614-specific primers to profile EF0631 homologues in 5 E. faecium isolates. The results of this analysis are shown in Table 6 and in Fig. S5. Strikingly, in strains from patient 2 (37), an R218Q substitution occurred—the same HKD motif substitution observed for DAP-B and DAP-C EF0631. In strains from patient 1 (56), a V37A substitution occurred.
DAP-A and DAP-C EF0631 alleles confer trans-dominant daptomycin resistance to E. faecalis OG1RF.
Data obtained from our V583 evolution experiments and from EF0631 sequencing of daptomycin-resistant E. faecalis and E. faecium clinical isolates strongly support a role for altered EF0631 alleles in enterococcal daptomycin resistance. To demonstrate a functional link between EF0631 mutations and daptomycin resistance, we amplified EF0631 alleles from the wild-type V583, DAP-A, and DAP-C strains and cloned these fragments into the multicopy shuttle vector pAT28 (71). Cloned fragments included the EF0631 coding region and 243 bp of upstream sequence to include a putative EF0631 promoter. These constructs were electroporated into the natively daptomycin-sensitive E. faecalis strain OG1RF (MIC of 4 μg/ml), confirmed for correct EF0631 construct sequence, and then tested for daptomycin MICs. As expected, OG1RF transformed with either pAT28 or pAT28 harboring a wild-type EF0631 insert (KP101) (Table 1) had daptomycin MICs comparable to that of wild-type OG1RF (4 μg/ml). The daptomycin MIC increased to 8 μg/ml in OG1RF transformed with pAT28 with a DAP-A EF0631 insert (KP102) (Table 1). Strikingly, the daptomycin MIC increased to 64 μg/ml in OG1RF transformed with pAT28 with a DAP-C EF0631 insert (KP103) (Table 1). These results demonstrate that EF0631 mutations confer daptomycin resistance to E. faecalis.
DISCUSSION
In this report, we describe genetic mutations accompanying increasing daptomycin resistance in E. faecalis V583 and in daptomycin-resistant clinical isolates of E. faecalis and E. faecium. Whole-genome resequencing was used to identify changes in the E. faecalis V583 genome following adaptation to increasing concentrations of daptomycin. Overall genome coverage was high, and the few gaps occurring in resequencing assemblies were closed. Each putative polymorphism in the evolved strains was confirmed by independent amplification and sequencing. Additionally, comparative analysis methods were modified to permit detection of potential movements of mobile elements.
Our analysis demonstrates that diverse genetic changes—among them, transitions, transversions, deletions ranging from 1 bp to 75 bp in size, duplications, and IS translocations—occur readily in E. faecalis populations (Table 7) and can become fixed in a short time (≤2 weeks) under antibiotic selection. Although mutation to daptomycin resistance was demonstrable in vitro, as of yet, it is rare among clinical isolates. Strain background may influence the frequency at which mutations arise. Evidence supporting this includes the observation that spontaneous lipiarmycin resistance rates are ∼100-fold higher for E. faecalis V583 than for E. faecalis OG1RF (22), a natively drug-sensitive human oral isolate lacking much of the mobile element content of V583 (4). Further, daptomycin serial passage of vancomycin-sensitive E. faecalis and E. faecium strains by an independent group obtained lower levels of daptomycin resistance (stable MICs of 16 μg/ml; a 2 to 3-fold increase over parent MICs) over a longer period of serial passage (21 to 24 days) (32). We selected E. faecalis V583 as a starting point for these experiments for several reasons: V583 is vancomycin resistant, and daptomycin resistance has been documented primarily in vancomycin-resistant enterococci (29); V583 is representative of leading hospital-associated E. faecalis lineages in sequence type and in mobile element content (51, 61); and a closed genome sequence is available for V583 (51). In vitro analysis of evolved V583 mutants identified EF0631 polymorphisms in all daptomycin-resistant strains, and identical or similar polymorphisms were found in daptomycin-resistant clinical isolates but not in susceptible strains for which genome sequence data exists.
Table 7.
Genetic changes observed in enterococci in this studya
| Mutation type | Mutationb | ORF | Strain(s) | Commentb |
|---|---|---|---|---|
| Transition | G→A | EF0631 cls | DAP-B, DAP-C, E. faecium UTHS-219EM | |
| EFTG_00614 cls | ||||
| T→C | EFTG_00614 cls | E. faecium CMC-087EM, E. faecium CMC-088EM | ||
| Transversion | C→A | EF0631 cls | E. faecalis SFOK-980ES | |
| C→G | EF0782 rpoN | DAP-A | ||
| T→G | EF2470 | DAP-B | ||
| A→T | EF2698 | DAP-B | ||
| Deletion (del) | 1-bp del | EF0631 | E. faecalis T2 | 1 A residue in homopolymeric tract (A8) deleted |
| 1-bp del | EF1753 | DAP-B, DAP-C | 1 A residue in homopolymeric tract (A7) deleted | |
| 1-bp del | EF1797 | DAP-C | G residue is deleted in sequence CAAAGAT | |
| 9-bp del | EF0631 cls | DAP-A, E. faecalis MCA-1970ES | 1 of 2 tandem 9-bp repeats deleted | |
| 9-bp del | EF0716 | DAP-C | ||
| 75-bp del | EF1797 | DAP-B | ||
| Duplication (dup) | 9-bp dup | EF0716 | DAP-A | Same 9-bp sequence is deleted in DAP-C |
| 4-bp dup | EF2470 | DAP-C | ||
| Mobile element translocation | IS insertion | EF1797 | DAP-B | IS256 insertion |
Only nonsynonymous mutations are shown for E. faecalis clinical isolates.
Nucleotide residues shown relative to gene sequence.
To our knowledge, this is the first published report associating enterococcal daptomycin resistance with specific genetic lesions. Daptomycin resistance has been most extensively studied in S. aureus, in which the mprF gene contributes to resistance. Mutations in mprF have been detected in both in vitro-evolved and clinical daptomycin-resistant S. aureus isolates (15, 18, 28, 32, 47, 55, 73, 75). S. aureus mprF mutations associated with daptomycin resistance appear to be gain-of-function mutations that enhance MprF expression and/or activity (27, 43, 59, 74). MprF catalyzes the lysinylation of phosphatidylglycerol (PG), generating lysylphosphatidylglycerol (Lys-PG) (52). In Bacillus subtilis, daptomycin preferentially interacts with PG-rich regions of the membrane (23). Because lysine modification converts negatively charged PG to positively charged Lys-PG (52), it is thought that overall increased membrane Lys-PG or increased flipping of Lys-PG to the outer leaflet interferes with daptomycin-membrane interactions (27, 43, 74). mprF sequences were previously profiled in daptomycin-resistant E. faecium strains isolated from a daptomycin-treated patient, and as for our current study of E. faecalis, no mprF mutations were detected (44). However, a role for mprF in enterococcal resistance cannot be excluded, as discussed further below.
By cloning and expressing DAP EF0631 cls alleles in trans, we could confer daptomycin resistance to the natively daptomycin-sensitive E. faecalis strain OG1RF, demonstrating a functional link between these alleles and daptomycin resistance. Because of trans-dominance, EF0631 mutations in the DAP strains are unlikely to be loss-of-function or reduced activity mutations. Cardiolipin (CL; bis-PG) is a negatively charged phospholipid associated with septal and polar membrane protein-lipid microdomains in B. subtilis and other bacteria (42). CL has the potential to significantly impact local membrane structure and charge-charge interactions at the membrane. EF0631 mutations observed in the DAP strains theoretically could result in decreased CL synthesis or increased CL degradation (as Cls catalyzes reversible transphosphatidylation), as either scenario could lead to overall decreased levels of CL in membranes of daptomycin-resistant enterococci. Again, the observed trans-dominance reduces the likelihood of the former possibility but does not exclude the latter one. Alternatively, it is possible that EF0631 mutations alter the subcellular localization of Cls. Cls is localized to septa in B. subtilis, among other proteins associated with phospholipid biosynthesis (48). Finally, Cls can exhibit relaxed substrate specificity (63), and the mutations observed in this study could modulate that specificity, perhaps allowing for the incorporation of modified PG substrates into CL. E. faecium MprF can synthesize arginine-modified PG (Arg-PG), alanine-modified PG (Ala-PG), and Lys-PG, distinguishing E. faecium MprF from S. aureus MprF (58). It is possible that any of these modified PGs could serve as substrates for enterococcal Cls. Of note, Listeria spp., which are closely related to enterococci, synthesize both Lys-PG and lysine-modified cardiolipin (Lys-CL) (17, 70). Synthesis of Lys-CL by Listeria spp. is mprF dependent; however, it is unclear whether MprF directly catalyzes lysine transfer to CL or if MprF-generated Lys-PG is utilized as a substrate for CL biosynthesis by Cls (70). If substrate specificity of Cls has been altered to better accommodate modified PG substrates, we would expect to detect aminoacylated CL in membranes of daptomycin-resistant enterococci.
Enterococcal membrane phospholipid composition and biosynthetic pathways are not well characterized. Enterococcus hirae ATCC 9790 (formerly Streptococcus faecalis 9790) was extensively studied in the 1960s and 1970s and was found to produce a diverse array of modified PGs, identified as Ala-PG, Lys-PG, diLys-PG, Arg-PG, and diglucosyl-PG (14), and CL (14, 25). Ala-PG, Lys-PG, and CL were detected in a strain described as Streptococcus faecalis 10C1 (31); however, it is unknown if this strain would be classified as E. faecalis today. A greater understanding of the membrane phospholipid compositions of E. faecalis and E. faecium, and how their membrane compositions change in response to stress and other stimuli, will be critical to understanding the precise mechanism of daptomycin resistance. These analyses are the focus of ongoing work.
Interestingly, EF0631 cls mutations were not detected in four daptomycin-resistant E. faecalis clinical isolates (Table 6); however, a role for cls mutations in the daptomycin resistance observed for these strains cannot be fully excluded. While mutations in the related copy of the cls gene at EF1608 were not detected in our E. faecalis V583 evolution experiments, EF1608 mutations could occur in these clinical strains. Alternatively, other pathways to daptomycin resistance may be involved, analogous to S. aureus daptomycin resistance, where paths to resistance that do not involve mprF mutation exist (7, 12, 28, 53).
Mutations in six additional E. faecalis genes were detected by our resequencing analysis, four encoding predicted cytoplasmic proteins (rpoN, EF1753, EF2470, EF2698) and two (EF1797 and EF0716) encoding predicted membrane-associated proteins. Three of these genes have previously been implicated in resistance to membrane-active antimicrobials in E. faecalis and Listeria monocytogenes. rpoN is important for resistance to subclass IIa bacteriocins in E. faecalis and L. monocytogenes (6, 13, 57). Differential expression of L. monocytogenes lmo2487, whose predicted protein shares 43% similarity with EF1753, is associated with spontaneous nisin resistance in that species (20, 21). Further, lmo1967, encoding TelA, was recently shown to be important for nisin and gallidermin resistance in L. monocytogenes (9), and TelA levels are increased in cold-adapted L. monocytogenes compared to those in cells grown at 37°C (5). Collectively, these results suggest that the RpoN, EF1753, and EF2698 proteins help E. faecalis cope with stresses imposed by certain membrane perturbations. It is as yet unclear what roles EF1797, EF2470, and EF0716 could play in daptomycin resistance. We found a weak link between EF1797 and membrane phospholipid biosynthesis using Pfam-A analysis; whether EF1797 plays a role in these pathways remains to be experimentally confirmed. Regardless, it appears that inactivation of EF1797 is important for daptomycin resistance in E. faecalis. HD domain-containing proteins such as EF2470 are predicted metal-dependent phosphohydrolases, with diverse roles in nucleotide metabolism, signaling, and other cellular functions (3). We were unable to elucidate any functional predictions for EF0716.
The genomic data presented here implicate seven proteins, among them, EF0631 Cls and other proteins with putative roles in membrane stability and biogenesis, in E. faecalis daptomycin resistance. We have demonstrated a functional link between specific EF0631 cls mutations and daptomycin resistance. Our work indicates that daptomycin treatment can select for alternate EF0631/EFTG_00614 (cls) alleles in clinical populations of E. faecalis and E. faecium. While genetically distinct from S. aureus, a role for altered membrane phospholipid composition in both staphylococcal and enterococcal daptomycin resistance is likely.
Supplementary Material
ACKNOWLEDGMENTS
We gratefully thank Aileen Rubio of Cubist Pharmaceuticals, Inc., for manuscript feedback and Kip Bodi of the Tufts University Core Facility and Veronica Kos for helpful discussions.
Portions of this work were supported by NIH grant AI072360 (to M.S.G.), the Harvard-wide antibiotic resistance project AI083214, NIH fellowship support grant EY020734 to K.L.P., and Cubist Pharmaceuticals, Inc.
Footnotes
Supplemental material for this article may be found at http://aac.asm.org/.
Published ahead of print on 18 April 2011.
The authors have paid a fee to allow immediate free access to this article.
REFERENCES
- 1. Aarestrup F. M., Butaye P., Witte W. 2002. Nonhuman reservoirs of enterococci, p. 55–99 In Gilmore M. S. (ed.), The enterococci: pathogenesis, molecular biology, and antibiotic resistance. ASM Press, Washington, DC [Google Scholar]
- 2. Altschul S. F., et al. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Aravind L., Koonin E. V. 1998. The HD domain defines a new superfamily of metal-dependent phosphohydrolases. Trends Biochem. Sci. 23:469–472 [DOI] [PubMed] [Google Scholar]
- 4. Bourgogne A., et al. 2008. Large scale variation in Enterococcus faecalis illustrated by the genome analysis of strain OG1RF. Genome Biol. 9:R110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Cacace G., et al. 2010. Proteomics for the elucidation of cold adaptation mechanisms in Listeria monocytogenes. J. Proteomics 73:2021–2030 [DOI] [PubMed] [Google Scholar]
- 6. Calvez S., Rince A., Auffray Y., Prevost H., Drider D. 2007. Identification of new genes associated with intermediate resistance of Enterococcus faecalis to divercin V41, a pediocin-like bacteriocin. Microbiology 153:1609–1618 [DOI] [PubMed] [Google Scholar]
- 7. Camargo I. L., Neoh H. M., Cui L., Hiramatsu K. 2008. Serial daptomycin selection generates daptomycin-nonsusceptible Staphylococcus aureus strains with a heterogeneous vancomycin-intermediate phenotype. Antimicrob. Agents Chemother. 52:4289–4299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Canton R., Ruiz-Garbajosa P., Chaves R. L., Johnson A. P. 2010. A potential role for daptomycin in enterococcal infections: what is the evidence? J. Antimicrob. Chemother. 65:1126–1136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Collins B., Joyce S., Hill C., Cotter P. D., Ross R. P. 2010. TelA contributes to the innate resistance of Listeria monocytogenes to nisin and other cell wall-acting antibiotics. Antimicrob. Agents Chemother. 54:4658–4663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Cotroneo N., Harris R., Perlmutter N., Beveridge T., Silverman J. A. 2008. Daptomycin exerts bactericidal activity without lysis of Staphylococcus aureus. Antimicrob. Agents Chemother. 52:2223–2225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Cubist Pharmaceuticals, Inc 2010. Cubicin (daptomycin for injection) package insert. Cubist Pharmaceuticals, Inc., Lexington, MA [Google Scholar]
- 12. Cui L., et al. 2010. An RpoB mutation confers dual heteroresistance to daptomycin and vancomycin in Staphylococcus aureus. Antimicrob. Agents Chemother. 54:5222–5233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Dalet K., Briand C., Cenatiempo Y., Hechard Y. 2000. The rpoN gene of Enterococcus faecalis directs sensitivity to subclass IIa bacteriocins. Curr. Microbiol. 41:441–443 [DOI] [PubMed] [Google Scholar]
- 14. dos Santos Mota J. M., den Kamp J. A., Verheij H. M., van Deenen L. L. 1970. Phospholipids of Streptococcus faecalis. J. Bacteriol. 104:611–619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Farrell D. J., Robbins M., Rhys-Williams W., Love W. G. 2011. Investigation of the potential for mutational resistance to XF-73, retapamulin, mupirocin, fusidic acid, daptomycin, and vancomycin in methicillin-resistant Staphylococcus aureus isolates during a 55-passage study. Antimicrob. Agents Chemother. 55:1177–1181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Finn R. D., et al. 2010. The Pfam protein families database. Nucleic Acids Res. 38:D211–D222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Fischer W., Leopold K. 1999. Polar lipids of four Listeria species containing L-lysylcardiolipin, a novel lipid structure, and other unique phospholipids. Int. J. Syst. Bacteriol. 49(Pt. 2):653–662 [DOI] [PubMed] [Google Scholar]
- 18. Friedman L., Alder J. D., Silverman J. A. 2006. Genetic changes that correlate with reduced susceptibility to daptomycin in Staphylococcus aureus. Antimicrob. Agents Chemother. 50:2137–2145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Gold O. G., Jordan H. V., van Houte J. 1975. The prevalence of enterococci in the human mouth and their pathogenicity in animal models. Arch. Oral Biol. 20:473–477 [DOI] [PubMed] [Google Scholar]
- 20. Gravesen A., et al. 2004. pbp2229-mediated nisin resistance mechanism in Listeria monocytogenes confers cross-protection to class IIa bacteriocins and affects virulence gene expression. Appl. Environ. Microbiol. 70:1669–1679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Gravesen A., Sorensen K., Aarestrup F. M., Knochel S. 2001. Spontaneous nisin-resistant Listeria monocytogenes mutants with increased expression of a putative penicillin-binding protein and their sensitivity to various antibiotics. Microb. Drug Resist. 7:127–135 [DOI] [PubMed] [Google Scholar]
- 22. Gualtieri M., Tupin A., Brodolin K., Leonetti J. P. 2009. Frequency and characterisation of spontaneous lipiarmycin-resistant Enterococcus faecalis mutants selected in vitro. Int. J. Antimicrob. Agents 34:605–606 [DOI] [PubMed] [Google Scholar]
- 23. Hachmann A. B., Angert E. R., Helmann J. D. 2009. Genetic analysis of factors affecting susceptibility of Bacillus subtilis to daptomycin. Antimicrob. Agents Chemother. 53:1598–1609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hidron A. I., et al. 2008. NHSN annual update: antimicrobial-resistant pathogens associated with healthcare-associated infections: annual summary of data reported to the National Healthcare Safety Network at the Centers for Dis. Control and Prevention, 2006–2007. Infect. Control Hosp. Epidemiol. 29:996–1011 [DOI] [PubMed] [Google Scholar]
- 25. Ibbott F. A., Abrams A. 1964. The phospholipids in membrane ghosts from Streptococcus faecalis protoplasts. Biochemistry 3:2008–2012 [DOI] [PubMed] [Google Scholar]
- 26. Inuzuka M., Hayakawa M., Ingi T. 2005. Serinc, an activity-regulated protein family, incorporates serine into membrane lipid synthesis. J. Biol. Chem. 280:35776–35783 [DOI] [PubMed] [Google Scholar]
- 27. Jones T., et al. 2008. Failures in clinical treatment of Staphylococcus aureus infection with daptomycin are associated with alterations in surface charge, membrane phospholipid asymmetry, and drug binding. Antimicrob. Agents Chemother. 52:269–278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Julian K., et al. 2007. Characterization of a daptomycin-nonsusceptible vancomycin-intermediate Staphylococcus aureus strain in a patient with endocarditis. Antimicrob. Agents Chemother. 51:3445–3448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kelesidis T., Humphries R., Uslan D. Z., Pegues D. A. 2011. Daptomycin nonsusceptible enterococci: an emerging challenge for clinicians. Clin. Infect. Dis. 52:228–234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Klockgether J., et al. 2010. Genome diversity of Pseudomonas aeruginosa PAO1 laboratory strains. J. Bacteriol. 192:1113–1121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kocun F. J. 1970. Amino acid containing phospholipids as major components of the phospholipids of Streptococcus faecalis 10C1. Biochim. Biophys. Acta 202:277–282 [DOI] [PubMed] [Google Scholar]
- 32. Kosowska-Shick K., et al. 2009. Activity of telavancin against staphylococci and enterococci determined by MIC and resistance selection studies. Antimicrob. Agents Chemother. 53:4217–4224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Krogh A., Larsson B., von Heijne G., Sonnhammer E. L. 2001. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J. Mol. Biol. 305:567–580 [DOI] [PubMed] [Google Scholar]
- 34. Leavis H. L., Bonten M. J., Willems R. J. 2006. Identification of high-risk enterococcal clonal complexes: global dispersion and antibiotic resistance. Curr. Opin. Microbiol. 9:454–460 [DOI] [PubMed] [Google Scholar]
- 35. Leavis H. L., et al. 2007. Insertion sequence-driven diversification creates a globally dispersed emerging multiresistant subspecies of E. faecium. PLoS Pathog. 3:e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Lerchner A., Mansfeld J., Kuppe K., Ulbrich-Hofmann R. 2006. Probing conserved amino acids in phospholipase D (Brassica oleracea var. capitata) for their importance in hydrolysis and transphosphatidylation activity. Protein Eng. Des. Sel. 19:443–452 [DOI] [PubMed] [Google Scholar]
- 37. Lewis J. S., II, et al. 2005. Emergence of daptomycin resistance in Enterococcus faecium during daptomycin therapy. Antimicrob. Agents Chemother. 49:1664–1665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Manson J. M., Hancock L. E., Gilmore M. S. 2010. Mechanism of chromosomal transfer of Enterococcus faecalis pathogenicity island, capsule, antimicrobial resistance, and other traits. Proc. Natl. Acad. Sci. U. S. A. 107:12269–12274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Manson J. M., Keis S., Smith J. M., Cook G. M. 2003. A clonal lineage of VanA-type Enterococcus faecalis predominates in vancomycin-resistant enterococci isolated in New Zealand. Antimicrob. Agents Chemother. 47:204–210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Marchler-Bauer A., et al. 2009. CDD: specific functional annotation with the Conserved Domain Database. Nucleic Acids Res. 37:D205–D210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. McBride S. M., Fischetti V. A., Leblanc D. J., Moellering R. C., Jr., Gilmore M. S. 2007. Genetic diversity among Enterococcus faecalis. PLoS One 2:e582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Mileykovskaya E., Dowhan W. 2009. Cardiolipin membrane domains in prokaryotes and eukaryotes. Biochim. Biophys. Acta 1788:2084–2091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Mishra N. N., et al. 2009. Analysis of cell membrane characteristics of in vitro-selected daptomycin-resistant strains of methicillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 53:2312–2318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Montero C. I., Stock F., Murray P. R. 2008. Mechanisms of resistance to daptomycin in Enterococcus faecium. Antimicrob. Agents Chemother. 52:1167–1170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Muraih J. K., Pearson A., Silverman J., Palmer M. 2011. Oligomerization of daptomycin on membranes. Biochim. Biophys. Acta [DOI] [PubMed] [Google Scholar]
- 46. Murray B. E. 1990. The life and times of the Enterococcus. Clin. Microbiol. Rev. 3:46–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Murthy M. H., Olson M. E., Wickert R. W., Fey P. D., Jalali Z. 2008. Daptomycin non-susceptible meticillin-resistant Staphylococcus aureus USA 300 isolate. J. Med. Microbiol. 57:1036–1038 [DOI] [PubMed] [Google Scholar]
- 48. Nishibori A., Kusaka J., Hara H., Umeda M., Matsumoto K. 2005. Phosphatidylethanolamine domains and localization of phospholipid synthases in Bacillus subtilis membranes. J. Bacteriol. 187:2163–2174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Palmer K. L., et al. 2010. High-quality draft genome sequences of 28 Enterococcus sp. isolates. J. Bacteriol. 192:2469–2470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Palmer K. L., Gilmore M. S. 2010. Multidrug-resistant enterococci lack CRISPR-cas. mBio 1(4):e00227–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Paulsen I. T., et al. 2003. Role of mobile DNA in the evolution of vancomycin-resistant Enterococcus faecalis. Science 299:2071–2074 [DOI] [PubMed] [Google Scholar]
- 52. Peschel A., et al. 2001. Staphylococcus aureus resistance to human defensins and evasion of neutrophil killing via the novel virulence factor MprF is based on modification of membrane lipids with l-lysine. J. Exp. Med. 193:1067–1076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Pillai S. K., et al. 2007. Daptomycin nonsusceptibility in Staphylococcus aureus with reduced vancomycin susceptibility is independent of alterations in MprF. Antimicrob. Agents Chemother. 51:2223–2225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Ponting C. P., Kerr I. D. 1996. A novel family of phospholipase D homologues that includes phospholipid synthases and putative endonucleases: identification of duplicated repeats and potential active site residues. Protein Sci. 5:914–922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Quinn B., Hussain S., Malik M., Drlica K., Zhao X. 2007. Daptomycin inoculum effects and mutant prevention concentration with Staphylococcus aureus. J. Antimicrob. Chemother. 60:1380–1383 [DOI] [PubMed] [Google Scholar]
- 56. Ramos L. G., Strasfeld L. M., Soave R., Larone D. H. 2005. Development of daptomycin-resistant vancomycin-resistant Enterococcus faecium (VREF) bacteremia in an allogenic peripheral blood stem cell transplant (PBSCT) patient, abstr. L-2141. Abstr. 45th Intersci. Conf. Antimicrob. Agents Chemother. [Google Scholar]
- 57. Robichon D., et al. 1997. The rpoN (sigma54) gene from Listeria monocytogenes is involved in resistance to mesentericin Y105, an antibacterial peptide from Leuconostoc mesenteroides. J. Bacteriol. 179:7591–7594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Roy H., Ibba M. 2009. Broad range amino acid specificity of RNA-dependent lipid remodeling by multiple peptide resistance factors. J. Biol. Chem. 284:29677–29683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Rubio A., et al. 2011. Regulation of mprF by antisense RNA restores daptomycin susceptibility to daptomycin-resistant isolates of Staphylococcus aureus. Antimicrob. Agents Chemother. 55:364–367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Rudolph A. E., et al. 1999. Expression, characterization, and mutagenesis of the Yersinia pestis murine toxin, a phospholipase D superfamily member. J. Biol. Chem. 274:11824–11831 [DOI] [PubMed] [Google Scholar]
- 61. Ruiz-Garbajosa P., et al. 2006. Multilocus sequence typing scheme for Enterococcus faecalis reveals hospital-adapted genetic complexes in a background of high rates of recombination. J. Clin. Microbiol. 44:2220–2228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Sahm D. F., et al. 1989. In vitro susceptibility studies of vancomycin-resistant Enterococcus faecalis. Antimicrob. Agents Chemother. 33:1588–1591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Schlame M. 2008. Cardiolipin synthesis for the assembly of bacterial and mitochondrial membranes. J. Lipid Res. 49:1607–1620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Shankar N., Baghdayan A. S., Gilmore M. S. 2002. Modulation of virulence within a pathogenicity island in vancomycin-resistant Enterococcus faecalis. Nature 417:746–750 [DOI] [PubMed] [Google Scholar]
- 65. Shepard B. D., Gilmore M. S. 1995. Electroporation and efficient transformation of Enterococcus faecalis grown in high concentrations of glycine. Methods Mol. Biol. 47:217–226 [DOI] [PubMed] [Google Scholar]
- 66. Silverman J. A., Perlmutter N. G., Shapiro H. M. 2003. Correlation of daptomycin bactericidal activity and membrane depolarization in Staphylococcus aureus. Antimicrob. Agents Chemother. 47:2538–2544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Solheim M., et al. 2011. Comparative genomic analysis reveals significant enrichment of mobile genetic elements and genes encoding surface structure-proteins in hospital-associated clonal complex 2 Enterococcus faecalis. BMC Microbiol. 11:3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Stuckey J. A., Dixon J. E. 1999. Crystal structure of a phospholipase D family member. Nat. Struct. Biol. 6:278–284 [DOI] [PubMed] [Google Scholar]
- 69. Tannock G. W., Cook G. 2002. Enterococci as members of the intestinal microflora of humans, p. 101–132 In Gilmore M. S. (ed.), The enterococci: pathogenesis, molecular biology, and antibiotic resistance. ASM Press, Washington, DC [Google Scholar]
- 70. Thedieck K., et al. 2006. The MprF protein is required for lysinylation of phospholipids in listerial membranes and confers resistance to cationic antimicrobial peptides (CAMPs) on Listeria monocytogenes. Mol. Microbiol. 62:1325–1339 [DOI] [PubMed] [Google Scholar]
- 71. Trieu-Cuot P., Carlier C., Poyart-Salmeron C., Courvalin P. 1990. A pair of mobilizable shuttle vectors conferring resistance to spectinomycin for molecular cloning in Escherichia coli and in gram-positive bacteria. Nucleic Acids Res. 18:4296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. van Schaik W., et al. 2010. Pyrosequencing-based comparative genome analysis of the nosocomial pathogen Enterococcus faecium and identification of a large transferable pathogenicity island. BMC Genomics 11:239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Yang S. J., et al. 2009. Enhanced expression of dltABCD is associated with the development of daptomycin nonsusceptibility in a clinical endocarditis isolate of Staphylococcus aureus. J. Infect. Dis. 200:1916–1920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Yang S. J., et al. 2010. Cell wall thickening is not a universal accompaniment of the daptomycin nonsusceptibility phenotype in Staphylococcus aureus: evidence for multiple resistance mechanisms. Antimicrob. Agents Chemother. 54:3079–3085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Yang S. J., et al. 2009. Regulation of mprF in daptomycin-nonsusceptible Staphylococcus aureus strains. Antimicrob. Agents Chemother. 53:2636–2637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Yu N. Y., et al. 2010. PSORTb 3.0: improved protein subcellular localization prediction with refined localization subcategories and predictive capabilities for all prokaryotes. Bioinformatics 26:1608–1615 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.