Abstract
The clinical utility of the echinocandins is potentially compromised by the emergence of drug resistance. We investigated whether Candida albicans with amino acid substitutions at position Ser645 in Fks1 can be treated with either a conventional or an elevated dosage of micafungin. We studied Candida albicans (wild-type SC5314; MIC, 0.06 mg/liter) and four fks1 mutants (one FKS1/fks1 heterozygote mutant [MIC, 0.5 mg/liter] and three fks1/fks1 homozygous mutants [MICs for all, 2 mg/liter]) with a variety of amino acid substitutions at Ser645. The pharmacokinetic and pharmacodynamic relationships were characterized in a persistently neutropenic murine model of disseminated candidiasis. A mathematical model was fitted to all pharmacokinetic and pharmacodynamic data. This mathematical model was then used to “humanize” the murine pharmacokinetics, and the predicted antifungal effect was determined. The estimated maximal rate of growth and ultimate fungal densities in the kidney for each of the strains were similar. The administration of micafungin at 1 mg/kg of body weight to the wild type resulted in moderate antifungal activity, whereas the administration of 5 and 20 mg/kg resulted in rapid fungicidal activity. In contrast, the FKS1/fks heterozygote was killed only with 20 mg/kg, and the homozygous fks1 mutants failed to respond to any dosage. The bridging study revealed that human dosages of 100 and 400 mg/day were active only against the wild type, with no activity against either the heterozygote or the homozygote mutants. Ser645 Fks1 Candida albicans mutants cannot be treated with either conventional or elevated dosages of micafungin and should be deemed resistant.
INTRODUCTION
Disseminated candidiasis is a syndrome that is associated with significant morbidity and mortality (10). Micafungin is an echinocandin agent that exhibits rapid candidacidal activity and is effective for the treatment of patients with disseminated candidiasis (12, 16). In common with other members of this class, micafungin has a wide therapeutic index, with little evidence of clinically significant toxicity (7). The currently licensed dosage is 100 to 150 mg/day in adults, but dosages as high as 8 mg/kg of body weight/day have been safely administered to adults (21). Furthermore, dosages as high as 15 mg/kg have been administered to neonates without untoward effects (22).
Resistance to the echinocandins is increasingly described. This usually follows prolonged administration and clinically manifests as breakthrough infection (20). The majority of breakthrough isolates have elevated echinocandin MICs and amino acid substitutions in well-defined portions of the Fks subunits of glucan synthase (Fks1) (1, 3, 17, 20). Given the wide therapeutic index of all the three echinocandin agents and the increased frequency of breakthrough isolates, a clinically relevant question is whether higher human drug exposures can be used to successfully treat these Fks1mutants.
We investigated whether Fks1 mutants can be treated with micafungin by using a well-validated murine model of disseminated candidiasis. We studied the micafungin exposure-response relationships of four Ser645 Fks1 Candida albicans mutants. The experimental data were described using mathematical models, and the results were bridged from mice to humans. Our results suggest that amino acid substitutions at position 645 in glucan synthase cannot be successfully treated with micafungin.
MATERIALS AND METHODS
Strains of Candida albicans, MICs, and sequencing of FKS1.
The wild-type strain of Candida albicans was the well-characterized isolate SC5314. The Fks1 mutants of SC5314 used in this study and the conditions used to construct them have been described elsewhere (4). Briefly, the heterozygous mutant A15 (T1933C heterozygous; S645/S645P) was selected by serial passage on medium containing anidulafungin (0.5 mg/liter). C. albicans 20S (C1934T homozygous; Ser645F), 22S (C1934A homozygous Ser645Y), and 36S (T1933C homozygous Ser645P) were all selected using caspofungin containing yeast extract-peptone-dextrose (YPD) medium (8 mg/liter). The MICs were determined using both Clinical Laboratory Sciences Institute (CLSI) (5) and European Committee for Antimicrobial Susceptibility Testing (EUCAST) (8) methodologies in three separate independent experiments.
For all in vivo experiments, organisms were subcultured from beads stored at minus 80°C on Sabouraud dextrose agar (Oxoid, Basingstoke, United Kingdom) and incubated at 37°C for 48 h. Subsequently, strains were subcultured in Sabouraud dextrose broth (Oxoid, Basingstoke, United Kingdom) and incubated at 37°C for 16 h on an orbital shaker. Organisms were washed twice in phosphate-buffered saline (PBS). The inoculum was prepared using a hemocytometer with progressive dilution in PBS. The final inoculum was verified by quantitative culture on Sabouraud agar (Oxoid, Basingstoke, United Kingdom).
The Candida FKS1 gene was sequenced in the hot-spot regions by the Sanger methodology using a CEQ 8000 Beckman Coulter genetic analysis system, as previously described (15). Briefly, fungal genomic DNA was extracted from overnight cultures grown in YPD medium using a Q-Biogene FastDNA kit (Irvine, CA).
The FKS1 gene was amplified and sequenced using Candida albicans Fks1 primers flanking the Fks1 hot-spot regions with primers F2426 (CATTGCTGTGGCCACTTTAG), R2919 (GATTTCCATTTCCGTGGTAGC), F4590 (TACTATGGTCATCCAGGTTTCC), and R4954 (GGTCAAATCAGTGAAAACCG). The sequences obtained were edited with the BioEdit program (Ibis Therapeutics, Carlsbad, CA) and aligned with the FKS1 gene sequence of Candida albicans (GenBank accession number XM_716336) using ClustalW software (http://www.clustal.org/).
Drug.
The clinical formulation of micafungin was a gift from Astellas Pharma. The drug was reconstituted in 0.9% sodium chloride to produce a stock solution of 5 mg/ml. This stock was further diluted in 0.9% sodium chloride to produce the final desired concentrations for treatment. All dosages were administered intraperitoneally (i.p.) in a 0.25-ml volume.
Murine models of disseminated candidiasis.
All experiments were performed under United Kingdom Home Office project license 40/3101 and approved by the University of Manchester Ethics Committee. Male CD1 mice (Charles River Ltd., Kent, United Kingdom) weighing 22 to 24 g were used. Both pharmacokinetic-pharmacodynamic (PK-PD) and survival models were performed. Mice were housed in vented HEPA-filtered cages, and food and water were provided ad libitum. All mice received cyclophosphamide (Sigma, Poole, United Kingdom) at 200 mg/kg of body weight i.p. on day minus 3 relative to the day of infection. Previous studies in our laboratory have shown that this results in neutropenia for 5 to 6 days (data not shown). The target fungal inoculum for all strains was 1.6 × 104 organisms/mouse, which was administered intravenously in 0.2 ml of PBS.
Pharmacokinetic and pharmacodynamic experiments.
A serial sacrifice design was used to characterize the pharmacokinetics and pharmacodynamics of micafungin. Treatment was initiated at 5 h postinoculation and was given every subsequent 24 h thereafter. Cohorts of mice received micafungin at 0.1, 1, 5, and 20 mg/kg i.p. at 5, 29, and 53 h postinoculation. For the pharmacokinetic experiments, blood was obtained by terminal cardiac puncture using heparinized syringes. Samples were drawn throughout the second and third dosing intervals. Samples were centrifuged at 15,000 × g for 3 min, and the plasma was stored at minus 80°C prior to analysis. The tissue pharmacokinetics were simultaneously determined at the primary infection site in the kidney. Kidneys were harvested at the time of killing, placed in 1 ml PBS, homogenized, and then stored at minus 80°C prior to analysis. Groups of 4 mice were used for each dose-time point combination. For the pharmacodynamic experiments, the kidneys were harvested, homogenized in PBS, and plated to Sabouraud dextrose agar. Plates were incubated for 48 h at 37°C, after which quantitative counts were enumerated.
The same immunosuppression, fungal inoculum, and treatment regimen were used for the survival study. Groups of mice (n = 10 per group) were infected with the wild type and each of the four mutants. All mice received micafungin at 20 mg/kg/day i.p. for 3 doses and then observed. Survival over the experimental period was modeled using a Kaplan-Meier method.
Measurement of micafungin in plasma and kidneys.
Micafungin concentrations in both plasma and kidney tissue were measured using high-performance liquid chromatography (HPLC; Shimadzu Prominence, Shimadzu, Milton Keynes, United Kingdom). Forty microliters of extracted sample was injected onto a Hypersil BDS C18 5-μm column (250 by 4.6 mm; Thermo Fisher Scientific, Loughborough, United Kingdom). A standard curve encompassing 0.05 to 25 mg/liter was constructed in either plasma or kidney homogenate from stock solutions of pure micafungin powder at 1,000 mg/liter in ethanol (Fisher Scientific, Loughborough, United Kingdom). The internal standard was anidulafungin (0.1 mg/liter). A gradient method was used with initial concentrations of 70% 0.02 M potassium dihydrogen phosphate and 30% acetonitrile (Fisher Scientific, Loughborough, United Kingdom), changing to 30% and 70%, respectively, over 12 min, with an overall run time of 16 min. A flow rate of 1 ml/min was used. Micafungin and the internal standard were detected using fluorescence with excitation at 273 nm and emission at 464 nm and eluted after 10 and 13.4 min, respectively. For plasma and kidney, the coefficient of variation was <10% over the concentration range 0.05 to 25 mg/liter. The limit of detection was 0.05 mg/liter. The intra- and interday variations were <10%.
Mathematical modeling.
The pharmacokinetic and pharmacodynamic data were comodeled using a population methodology with the Big version of the program nonparametric adaptive grid (BIG NPAG) (13). The same structural model was fitted to the data from the wild-type and data from the heterozygous mutant (A15). Because the exposure-response relationships for the three mutants (36S, 20S, and 22S) were comparable, a single mathematical model was fitted to the pooled data from these strains. The structural mathematical model used for these analyses consisted of the following five ordinary differential equations:
| (1) |
| (2) |
| (3) |
| (4) |
| (5a) |
| (5b) |
| (5c) |
where X1, X2, X3, and X4 are the amounts of micafungin (in milligrams) in the peritoneum, serum, kidney, and peripheral compartment, respectively. R(1) is the amount of drug administered to the peritoneum; SCL is the clearance; Vc and Vkid are the volumes of the central compartment and kidney, respectively; Ka is the first-order rate constant connecting the peritoneum with the central compartment; Kcp and Kpc are the first-order rate constants connecting the central and peripheral compartments; and Kck and Kkc are the first-order rate constants connecting the central compartment and kidney. N is the density (number of organisms/gram kidney) of Candida albicans; Kgmax is the growth constant describing maximal growth; POPMAX is the theoretical maximum C. albicans burden in the kidney; Hg is the sigmoidicity constant for the drug effect on C. albicans growth; C50g is the concentration of micafungin in the kidney required to produce a 50% effect on the maximal rate of growth; Kkmax is the maximal rate of micafungin-induced kill; Hk is the sigmoidicity constant for the drug effect on C. albicans kill; and C50k is the concentration of micafungin in the kidney that produces half-maximal killing.
Equation 1 describes the flux of micafungin out of the peritoneum (where it has been injected), equation 2 describes the rate of change of micafungin in the central compartment (plasma), equation 3 describes the rate of change of micafungin in the kidney, equation 4 describes the rate of change of micafungin in the peripheral compartment (i.e., everything other than the blood and the kidney), and equation 5 describes the rate of change of fungal burden in the kidney that contains terms describing the capacity-limited growth of Candida (equation 5a), the drug-associated suppression of growth (equation 5b), and the drug-associated fungal kill (equation 5c).
The area under the concentration-time curve (AUC) in plasma and kidney was calculated using integration after implementation of the mathematical model in the program ADAPT (version 5) (6). The extent of penetration of micafungin into the kidney relative to plasma was expressed as a ratio.
Humanization of murine pharmacokinetics.
To further place the experimental results in a clinical context, the murine pharmacokinetics were humanized. The simulation module of the pharmacokinetic program ADAPT (version 5) (6) was used to construct a dosing regimen in mice (in silico) that resulted in a concentration-time profile that approximated the mean concentration-time profile of a human receiving 100 mg/day (the currently used clinical regimen). The population pharmacokinetic model for micafungin in adult patients of Gumbo et al. (11) was used to define the mean concentration-time profile for micafungin in the population. The mean parameter vector and their associated variances were inserted into subroutine PRIOR of ADAPT (version 5) (6). A 5,000-patient simulation was then performed to identify the 2.5th and 97.5th centiles of the population.
RESULTS
Isolates: MICs and genotype.
The MICs for the wild type and the Fks1 mutants determined using CLSI and EUCAST methodologies are summarized in Table 1. The heterozygous isolate had an MIC (0.5 mg/liter) that was intermediate between the MICs of the wild type (0.06 mg/liter) and each of the homozygous mutants (2 mg/liter). The MICs for isolates obtained from the kidneys of treated mice after treatment were not different from those prior to inoculation. Sequencing of both hot-spot regions of the FKS1 gene confirmed that the recovered strain had the same genotype as that used for inoculation, thereby excluding the possibility of inadvertent mixing of strains, superinfection with an unrelated Candida strain, or the occurrence of progressive mutational events.
Table 1.
Strains used in this study
| Isolate | Mode (range) MIC (mg/liter) by method |
Genotype | |
|---|---|---|---|
| CLSI | EUCAST | ||
| SC5314 | 0.06 (0.016–0.06) | 0.03 (<0.015–0.03) | Wild type |
| A15 | 0.5 (0.25–2) | 1 (0.125–1) | T1933C heterozygote |
| 36S | 1 (1–2) | 2 (1–2) | T1933C homozygote |
| 22S | 2 (1–2) | 2 (1–2) | C1934A homozygote |
| 20S | 1 (1–2) | 2 (1–2) | C1934T homozygote |
Pharmacokinetics and pharmacodynamics.
The estimates for the mean parameter values and the standard deviation from the mathematical model are detailed in Table 2. The fit of the mathematical model to each of the data sets was highly acceptable, with observed-predicted values that were all >99% after the Bayesian step. Each model was associated with satisfactory measures of precision and bias.
Table 2.
Means and standard deviations for parameter values from mathematical model for wild-type and isogenic mutants
| Parameter | Wild type |
Heterozygous mutantb |
Homozygous mutantsc |
|||
|---|---|---|---|---|---|---|
| Mean | SDd | Mean | SD | Mean | SD | |
| Ka (h−1) | 13.23 | 5.21 | 14.11 | 8.35 | 23.22 | 4.28 |
| SCL (liter/h) | 0.0005 | 0.0002 | 0.0004 | 0.0003 | 0.0005 | 0.0003 |
| Vc (liter) | 0.0029 | 0.0002 | 0.006 | 0.006 | 0.0079 | 0.004 |
| Kcp (h−1) | 5.20 | 5.36 | 12.22 | 10.98 | 5.00 | 11.01 |
| Kpc (h−1) | 33.15 | 6.66 | 25.78 | 9.83 | 27.98 | 10.80 |
| Kck (h−1) | 31.82 | 5.14 | 24.41 | 8.36 | 17.71 | 5.34 |
| Kkc (h−1) | 19.46 | 7.71 | 14.71 | 7.96 | 25.7 | 5.41 |
| Vkid (liter) | 0.007 | 0.0053 | 0.01 | 0.008 | 0.006 | 0.002 |
| Kgmax (log10 CFU/g/h) | 0.11 | 0.038 | 0.14 | 0.043 | 0.133 | 0.028 |
| Hg | 3.70 | 2.00 | 4.23 | 1.90 | 4.38 | 2.50 |
| C50g (mg/liter) | 1.54 | 0.26 | 21.74 | 12.20 | 52.66 | 33.36 |
| POPMAX (CFU/g) | 1,230,960 | 2,003,520 | 231,528 | 225,722 | 179,371 | 108,849 |
| Kkmax (log10 CFU/g/h) | 0.118 | 0.028 | 0.128 | 0.05 | 0.15 | 0.044 |
| Hk | 3.08 | 1.27 | 3.65 | 2.34 | 4.66 | 2.66 |
| C50k (mg/liter) | 2.01 | 1.14 | 28.81 | 14.39 | 42.06 | 26.57 |
| Initial conditiona (CFU/g) | 1,262 | 821.6 | 74.01 | 21.75 | 134.08 | 166.36 |
Initial condition, fungal burden at the time of systemic drug administration.
The heterozygous mutant is A15.
The homozygous mutants include 36S, 22S, and 22S.
SD, standard deviation.
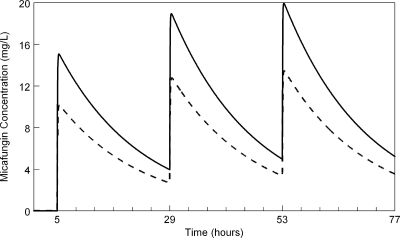
The concentrations of micafungin in the kidney were slightly lower than those in serum (Fig. 1). The AUCkidney/AUCplasma ratio determined in the third dosing interval from the mathematical model fitted to the wild-type strain was 67.8%. Furthermore, the mathematical model showed that micafungin tracked rapidly into and out of the kidney with no evidence of hysteresis, as has previously been described with caspofungin (14) (Fig. 1).
Fig. 1.
Simulated concentration-time profiles of micafungin in the kidney (broken line) and serum (solid line) for mice receiving 5 mg/kg every 24 h.
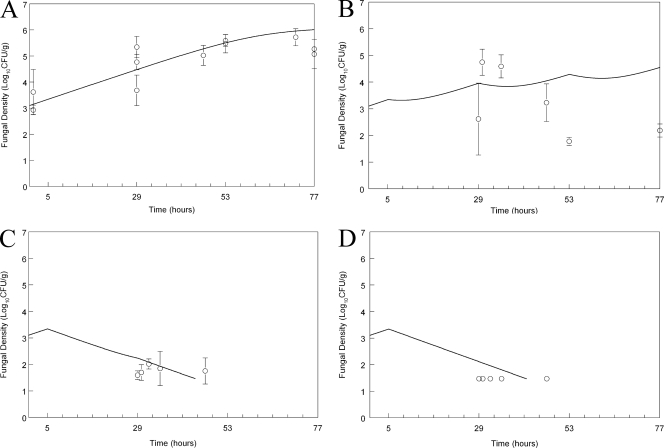
Inoculation of the wild-type strain resulted in an estimated initial density of 1,262 (log10 CFU/g 3.10) organisms/g kidney. There was logarithmic growth throughout the experimental period, with an estimated maximum rate of growth (i.e., Kgmax; see mathematical model and Table 2) of 0.11 log units per hour and a final density of log10 CFU/g 6.01 (Fig. 2 A). The administration of micafungin at 0.1 mg/kg had no appreciable antifungal effect (data not shown), whereas the administration of 1 mg/kg had a moderate antifungal effect (Fig. 2B). In contrast, the administration of 5 and 20 mg/kg resulted in rapid fungicidal activity, with the attainment of a near maximal effect at the end of the first dosing interval.
Fig. 2.
Effect of micafungin on the density of Candida albicans SC5314 wild type in the kidneys of mice as a function of time. (A) Controls; (B) micafungin at 1 mg/kg administered at 5, 29, and 53 h; (C) micafungin at 5 mg/kg administered at 5, 29, and 53 h; (D) micafungin at 20 mg/kg administered at 5, 29, and 53 h. Data are means ± SDs. The solid line is the fit of the population mathematical model.
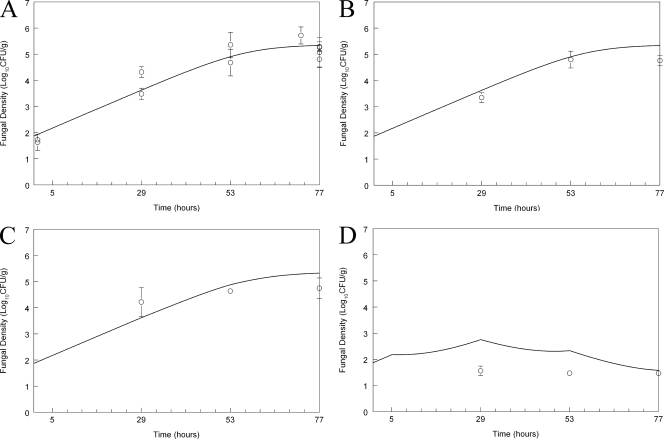
Inoculation of the heterozygous mutant resulted in an estimated initial density of 74.01 (log10 CFU/g 1.87) CFU/g kidney (Fig. 3 A), which was slightly lower than that of the wild type (see above). The estimated maximum rate of growth was 0.14 log unit per hour, which was comparable to that of the wild type. The fungal density at the end of the experiment was log10 CFU/g 5.34, which was less than that of the wild type. The antifungal effect of micafungin against the heterozygote was different from that against the wild type. While there was no discernible antifungal effect following administration of 0.1 (data not shown), 1, or 5 mg/kg compared with that against untreated controls (Fig. 3B and C), the administration of 20 mg/kg resulted in rapid fungicidal activity (Fig. 3D).
Fig. 3.
Effect of micafungin on the density of Candida albicans SC5313 A15 heterozygous mutant in the kidneys of mice as a function of time. (A) Controls; (B) micafungin at 1 mg/kg administered at 5, 29, and 53 h; (C) micafungin at 5 mg/kg administered at 5, 29, and 53 h; (D) micafungin at 20 mg/kg administered at 5, 29, and 53 h. Data are means ± SDs. The solid line is the fit of the population mathematical model.
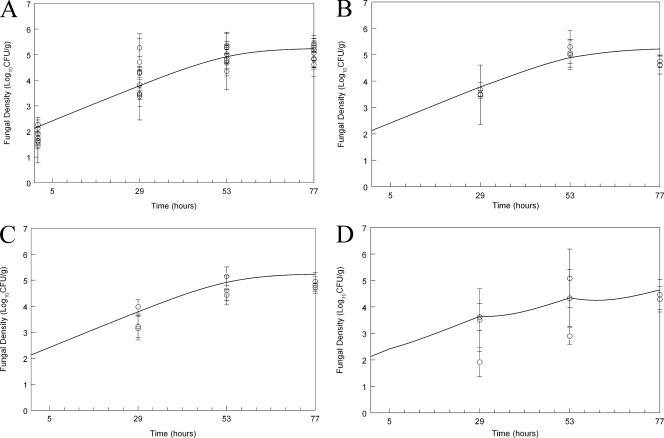
Inoculation of the homozygous mutants resulted in an initial estimated fungal density in the kidney of log10 CFU/g 2.13 (Fig. 4 A). All 3 mutants exhibited logarithmic growth, with some tailing of growth toward the end of the experimental period and an overall estimated maximal rate of growth of 0.13 log unit per hour. The estimated fungal burden after 77 h was log10 CFU/g 5.23 (Fig. 4A), which was comparable to that of the heterozygote but lower than that of the wild type. There was no demonstrable antifungal effect for micafungin following administration of 0.1 mg/kg (data not shown) or 1 or 5 mg/kg (Fig. 4B and C) against any of the three homozygous mutants, but there was marginal activity following the administration of 20 mg/kg/day (Fig. 4D).
Fig. 4.
Effect of micafungin on the density of Candida albicans SC5314 36S, 20S, and 22S homozygous mutants in the kidneys of mice as a function of time. (A) Controls; (B) micafungin at 1 mg/kg administered at 5, 29, and 53 h; (C) micafungin at 5 mg/kg administered at 5, 29, and 53 h; (D) micafungin at 20 mg/kg administered at 5, 29, and 53 h. Data are means ± SDs. The solid line is the fit of the population mathematical model.
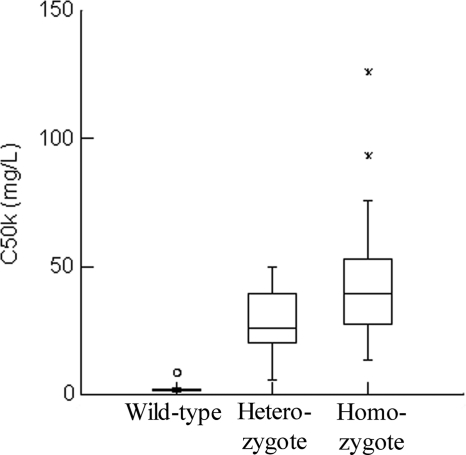
The differential activity of micafungin against the wild type and the heterozygous and homozygous mutants can be further appreciated from the mean parameter estimates for C50k, which is the concentration of micafungin in the kidney resulting in a half-maximal rate of cell kill (Table 2). The individual Bayesian estimates from the various strains are displayed using box plots (Fig. 5).
Fig. 5.
Box-and-whisker plots for the Bayesian estimates of C50k for the wild type, heterozygote, and homozygous mutants. The median (range) values for the wild type, heterozygous mutant, and homozygous mutants are 2.14 mg/liter (0.93 to 8.63 mg/liter), 26.24 mg/liter (5.86 to 49.99 mg/liter), and 39.46 mg/liter (13.57 to 125.87 mg/liter), respectively.
Humanization of murine pharmacokinetics.
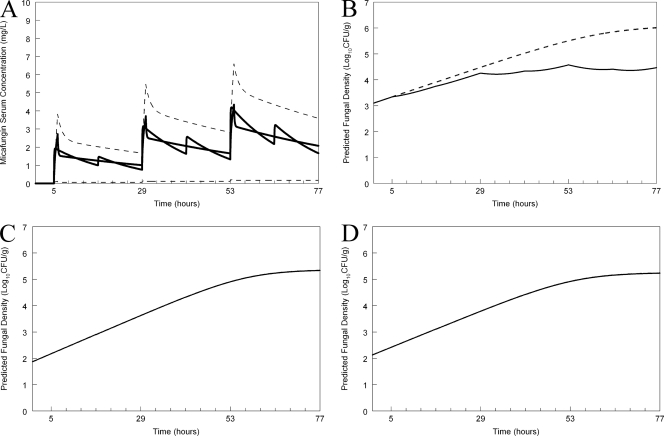
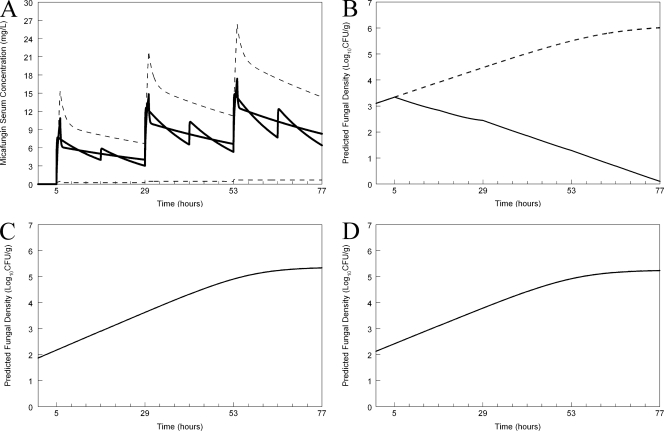
A regimen administered to mice that simulated the mean concentration-time profile for humans receiving 100 mg (Fig. 6 A) who were also infected with the wild type resulted in suppression of fungal growth (Fig. 6B). In contrast, however, the same humanized regimen, when administered to the heterozygote and the homozygous mutants, did not result in any predicted clinical antifungal activity (Fig. 6C and D, respectively). A regimen administered to mice that simulated the mean concentration-time profile for humans receiving 400 mg (Fig. 7 A) and infected with the wild type resulted in rapid fungicidal activity. The same humanized regimen had no effect on humans infected with the heterozygote or homozygous mutants (Fig. 7C and D, respectively).
Fig. 6.
Predicted outcomes for humans receiving micafungin at 100 mg/day who are infected with the various strains used in the murine experiments. (A) Humanization of micafungin in the mouse. Micafungin has been administered in silico to a mouse every 12 h to simulate the mean concentration-time profile of micafungin in the serum for humans receiving 100 mg/day. The dashed line represents the 2.5th and 97.5th centiles for the population. (B) Predicted antifungal effect of the humanized regimen for the wild type. The broken line represents the growth in untreated hosts. (C) Predicted antifungal effect of the humanized regimen for the heterozygous mutant. (D) Predicted antifungal effect of the humanized regimen for the mutant strains.
Fig. 7.
Predicted outcomes for humans receiving micafungin at 400 mg/day who are infected with the various strains. (A) Humanization of micafungin in the mouse. Micafungin has been administered in silico to a mouse every 12 h to simulate the mean concentration-time profile of micafungin in the serum for humans receiving 400 mg/day. The dashed line represents the 2.5th and 97.5th centiles for the population. (B) Predicted antifungal effect of the humanized regimen for the wild type. The broken line represents the growth in untreated hosts. (C) Predicted antifungal effect of the humanized regimen for the heterozygous mutant. (D) Predicted antifungal effect of the humanized regimen for the mutant strains.
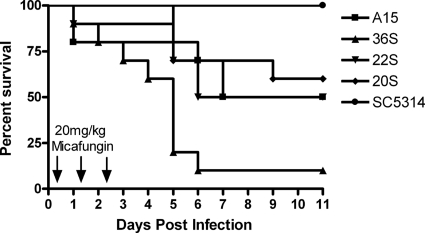
Survival.
The survival of mice infected with the various C. albicans strains and treated for 3 dosages of micafungin at 20 mg/kg is shown in Fig. 8. There was 100% survival of mice infected with the wild type for at least 14 days postinoculation. In contrast, there was a statistically significant decline in the fraction of surviving mice infected with the mutant strains. The rates of survival of mice infected with isolates 36S, 22S, and 20S (homozygous mutants) and treated with three dosages of 20 mg/kg i.p. were 10, 50, and 60%, respectively. The survival of mice infected with isolate A15 (heterozygous mutant) treated with three dosages of 20 mg/kg i.p. was 50%.
Fig. 8.
Survival of mice infected with the wild type (SC5314) and the Candida albicans mutant strains. All mice received three dosages of micafungin at 20 mg/kg administered at 5, 29, and 53 h postinoculation. The survival in mutants was statistically different from that in the wild type (P < 0.05; Kruskall-Wallis test).
DISCUSSION
The echinocandins are first-line agents for the treatment of disseminated candidiasis (15). Their fungicidal activity, paucity of drug interactions, and wide therapeutic index facilitate their use in critically ill patients with this disease. Increasingly, however, there are reports of isolates with reduced echinocandin susceptibility, which usually occurs in the context of prolonged therapy and at sites where drug penetration may be compromised (e.g., esophagus, respiratory tract, and abdomen) (20). Under these circumstances, cross-resistance between other echinocandin agents is frequently observed (2).
The molecular mechanism of reduced susceptibility for the majority of isolates has been defined and is related to nonsynonymous nucleotide substitutions in the gene FKS1, which encodes the putative echinocandin target, Fks1 (17). Amino acid substitutions occur in two well-defined areas within the target protein: (i) hot spot 1, which is an 8-amino-acid region between Phe641 and Asp648, and (ii) hot spot 2, at amino acid positions 1345 to 1365 (17). The in vitro echinocandin exposure-response relationships can be defined using partially purified glucan synthase with tritiated substrate (17). The concentration of drug that produces half-maximal inhibition of enzyme activity (50% effective concentration [EC50]) in glucan synthase assays from Fks1 mutants is several orders of magnitude higher than that for the wild type (17). Importantly, however, the enzyme complex can still be ultimately inhibited by high drug concentrations (17). Because the echinocandins have such wide therapeutic ratios, a clinically relevant question is whether any Fks1 mutant can be treated with either a conventional or an elevated dosage of drug.
The pharmacokinetic-pharmacodynamic analyses in this study suggest that infections with the wild type can be treated with micafungin at 100 mg/day, which is consistent with clinical experience. In contrast, a variety of amino acid substitutions (i.e., proline [P], phenylalanine [F], and tyrosine [Y]) for the serine residue at position 645 within Fks1 render these strains untreatable in the kidneys of mice even with the use of dosages of micafungin as high as 20 mg/kg. This murine dosage corresponds to a human dosage of ∼1,165 mg/day, which is significantly more than the currently employed 100 to 150 mg/day and the highest reported human dosage of 900 mg/day (21). Our bridging study suggests that the concentrations of micafungin that are required to have any antifungal effect are significantly in excess of those used in currently approved regimens. Our results suggest that Candida albicans isolates with amino acid substitutions in Fks1 at Ser645 should be deemed resistant and that this applies to both homozygous and heterozygous strains.
Our results confirm a view that echinocandin breakpoints should be set to exclude the vast majority of Fks1 mutants. The CLSI originally defined isolates with MICs of ≤2 mg/liter to be susceptible at a time when experience with resistant isolates was relatively limited and there was a desire not to categorize C. parapsilosis as nonsusceptible (19). More recently, concern has been expressed that these breakpoints may be too high because of reports that some Fks1 mutants have MICs that are currently classified as susceptible. For example, a recent study of both wild-type and Fks1 mutants revealed that some of these Fks1 mutants have MICs that overlap the tail of the wild-type distribution (i.e., have MICs that are within a mode and 2 doubling dilutions) (2). These findings have prompted CLSI to propose a downward revision of breakpoints for all three echinocandin agents, and this process is under way (see www.clsi.org). The EUCAST committee is currently formulating echinocandin breakpoints, and a final decision will be available in the near future.
Our study also has implications for critically ill patients receiving echinocandins who have breakthrough Candida infection. An increasing clinical conundrum is the interpretation of a Candida isolate that has grown in the context of prolonged echinocandin therapy. While there may be a number of explanations for this phenomenon (e.g., a retained infected catheter or an unresolved deep focus of infection), the emergence of drug resistance is an increasing concern. Our results indicate that echinocandin therapy is contraindicated for Ser645 Fks1 mutants. Because the prior probability of an Fks1 mutation is much higher in an echinocandin-exposed patient, clinical microbiology laboratories require a rapid and accurate way of determining the genotype to inform the decision on further echinocandin therapy. This is probably best achieved by sequencing both hot-spot regions in Fks1. While this is technically straightforward, it may not be always possible or practical, especially for smaller regional laboratories. In these cases, alternative phenotypic approaches may suffice, and potential approaches might include (i) measuring an MIC with 50% serum (9) and (ii) testing isolates against anidulafungin using epidemiological cutoff values until breakpoints that exclude resistant mutants have been established (2, 18). Regardless, there are increasing concerns that the MIC may not be a universally reliable tool to screen for echinocandin resistance and may not always provide correct information to guide appropriate antifungal therapy for critically ill patients.
A number of additional factors deserve further discussion and future study: (i) we studied relatively few strains, and further preclinical and clinical studies are warranted; (ii) the exposure-response relationships of Candida albicans isolates with amino acid substitutions at positions other than Ser645 are not known but could be readily determined using the in vivo-to-clinical bridging methods used in this study; in the absence of specific experimental pharmacokinetic-pharmacodynamic and/or clinical data, the most conservative approach is to classify these isolates as resistant, but this may ultimately prove to be too conservative; (iii) the PK-PD relationships of the other echinocandins against Candida albicans with Fks1 substitutions need to be defined; (iv) the consequences of analogous substitutions in Fks1 in other species such as Candida glabrata need to be determined; (v) the response of isolates with elevated echinocandin MICs without substitutions in Fks1 is of interest and could also be determined using the same methods used in this study; and (vi) we used a well-validated model of disseminated candidiasis using the fungal burden in the kidney as the pharmacodynamic readout. We acknowledge that the pharmacodynamic relationships may be different for Candida infections at other sites (e.g., bloodstream and brain) and that this could be addressed in future studies.
Finally, this study demonstrates the advantages of using modern pharmacokinetic-pharmacodynamic modeling techniques in combination with molecular microbiology to address clinically relevant questions. The bridging component of our study enables the experimental data to be firmly placed in a clinical context. The question of echinocandin activity against Fks1 mutants is extremely difficult to answer from clinical trial data because of the paucity of resistant isolates and confounding by other factors that may also have an impact upon clinical outcome (e.g., age, APACHE II score, management of deep infectious foci, and timing of initiation of therapy). Well-designed clinically relevant laboratory animal models and mathematical modeling techniques can and should be used to investigate many remaining questions related to antifungal drug resistance and derivation of in vitro susceptibility breakpoints.
ACKNOWLEDGMENTS
This work was funded, in part, by Astellas Pharma and The Fungal Research Trust. D.S.P. is supported by National Institutes of Health (NIH) grant AI069397. W.W.H. is supported by a National Institute of Health Research (NIHR) Clinician Scientist Fellowship.
Footnotes
Published ahead of print on 18 April 2011.
REFERENCES
- 1. Arendrup M. C., et al. 2009. Breakthrough Aspergillus fumigatus and Candida albicans double infection during caspofungin treatment: laboratory characteristics and implication for susceptibility testing. Antimicrob. Agents Chemother. 53:1185–1193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Arendrup M. C., et al. 2010. Echinocandin susceptibility testing of Candida species: comparison of EUCAST EDef 7.1, CLSI M27-A3, Etest, disk diffusion, and agar dilution methods with RPMI and Isosensitest media. Antimicrob. Agents Chemother. 54:426–439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Baixench M. T., et al. 2007. Acquired resistance to echinocandins in Candida albicans: case report and review. J. Antimicrob. Chemother. 59:1076–1083 [DOI] [PubMed] [Google Scholar]
- 4. Balashov S. V., Park S., Perlin D. S. 2006. Assessing resistance to the echinocandin antifungal drug caspofungin in Candida albicans by profiling mutations in FKS1. Antimicrob. Agents Chemother. 50:2058–2063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Clinical and Laboratory Standards Institute 2008. Reference method for broth dilution antifungal susceptibility testing of yeasts, 3rd ed. Approved standard M27-A3(28). Clinical and Laboratory Standards Institute, Wayne, PA [Google Scholar]
- 6. D'Argenio D. Z., Schumitzky A., Wang X. 2009. ADAPT 5 user's guide: pharmacokinetic/pharmacodynamic systems analysis software. Biomedical Simulations Resource, Los Angeles, CA [Google Scholar]
- 7. Denning D. W. 2003. Echinocandin antifungal drugs. Lancet 362:1142–1151 [DOI] [PubMed] [Google Scholar]
- 8. European Committee for Antimicrobial Susceptibility Testing 2008. EUCAST definitive document EDef 7.1: method for the determination of broth dilution MICs of antifungal agents for fermentative yeasts. Clin. Microbiol. Infect. 14:398–405 [DOI] [PubMed] [Google Scholar]
- 9. Garcia-Effron G., Park S., Perlin D. S. 2009. Correlating echinocandin MIC and kinetic inhibition of fks1 mutant glucan synthases for Candida albicans: implications for interpretive breakpoints. Antimicrob. Agents Chemother. 53:112–122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Gudlaugsson O., et al. 2003. Attributable mortality of nosocomial candidemia, revisited. Clin. Infect. Dis. 37:1172–1177 [DOI] [PubMed] [Google Scholar]
- 11. Gumbo T., et al. 2008. Population pharmacokinetics of micafungin in adult patients. Diagn. Microbiol. Infect. Dis. 60:329–331 [DOI] [PubMed] [Google Scholar]
- 12. Kuse E. R., et al. 2007. Micafungin versus liposomal amphotericin B for candidaemia and invasive candidosis: a phase III randomised double-blind trial. Lancet 369:1519–1527 [DOI] [PubMed] [Google Scholar]
- 13. Leary R., Jelliffe R., Schumitzky A., van Guilder M. 2001. An adaptive grid, non-parametric approach to pharmacokinetic and dynamic (PK/PD) models, p. 389–394 Proc. 14th IEEE Computer Soc. [Google Scholar]
- 14. Louie A., et al. 2005. Pharmacodynamics of caspofungin in a murine model of systemic candidiasis: importance of persistence of caspofungin in tissues to understanding drug activity. Antimicrob. Agents Chemother. 49:5058–5068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Pappas P. G., et al. 2009. Clinical practice guidelines for the management of candidiasis: 2009 update by the Infectious Diseases Society of America. Clin. Infect. Dis. 48:503–535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Pappas P. G., et al. 2007. Micafungin versus caspofungin for treatment of candidemia and other forms of invasive candidiasis. Clin. Infect. Dis. 45:883–893 [DOI] [PubMed] [Google Scholar]
- 17. Park S., et al. 2005. Specific substitutions in the echinocandin target Fks1p account for reduced susceptibility of rare laboratory and clinical Candida sp. isolates. Antimicrob. Agents Chemother. 49:3264–3273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Pfaller M. A., et al. 2010. Wild-type MIC distributions and epidemiological cutoff values for the echinocandins and Candida spp. J. Clin. Microbiol. 48:52–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Pfaller M. A., et al. 2008. Correlation of MIC with outcome for Candida species tested against caspofungin, anidulafungin, and micafungin: analysis and proposal for interpretive MIC breakpoints. J. Clin. Microbiol. 46:2620–2629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Pfeiffer C. D., et al. 2010. Breakthrough invasive candidiasis in patients on micafungin. J. Clin. Microbiol. 48:2373–2380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Sirohi B., et al. 2006. A study to determine the safety profile and maximum tolerated dose of micafungin (FK463) in patients undergoing haematopoietic stem cell transplantation. Bone Marrow Transplant. 38:47–51 [DOI] [PubMed] [Google Scholar]
- 22. Smith P. B., et al. 2009. Pharmacokinetics of an elevated dosage of micafungin in premature neonates. Pediatr. Infect. Dis. J. 28:412–415 [DOI] [PMC free article] [PubMed] [Google Scholar]