Abstract
Colistin resistance is rare in Acinetobacter baumannii, and little is known about its mechanism. We investigated the role of PmrCAB in this trait, using (i) resistant and susceptible clinical strains, (ii) laboratory-selected mutants of the type strain ATCC 19606 and of the clinical isolate ABRIM, and (iii) a susceptible/resistant pair of isogenic clinical isolates, Ab15/133 and Ab15/132, isolated from the same patient. pmrAB sequences in all the colistin-susceptible isolates were identical to reference sequences, whereas resistant clinical isolates harbored one or two amino acid replacements variously located in PmrB. Single substitutions in PmrB were also found in resistant mutants of strains ATCC 19606 and ABRIM and in the resistant clinical isolate Ab15/132. No mutations in PmrA or PmrC were found. Reverse transcriptase (RT)-PCR identified increased expression of pmrA (4- to 13-fold), pmrB (2- to 7-fold), and pmrC (1- to 3-fold) in resistant versus susceptible organisms. Matrix-assisted laser desorption ionization-time of flight (MALDI-TOF) mass spectrometry showed the addition of phosphoethanolamine to the hepta-acylated form of lipid A in the resistant variants and in strain ATCC 19606 grown under low-Mg2+ induction conditions. pmrB gene knockout mutants of the colistin-resistant ATCC 19606 derivative showed >100-fold increased susceptibility to colistin and 5-fold decreased expression of pmrC; they also lacked the addition of phosphoethanolamine to lipid A. We conclude that the development of a moderate level of colistin resistance in A. baumannii requires distinct genetic events, including (i) at least one point mutation in pmrB, (ii) upregulation of pmrAB, and (iii) expression of pmrC, which lead to addition of phosphoethanolamine to lipid A.
INTRODUCTION
Acinetobacter baumannii is a Gram-negative pathogen often associated with nosocomial infections and outbreaks (8). It has a great capacity to acquire antibiotic resistance, and this has necessitated numerous successive changes to therapeutic strategies. The increasingly frequent isolation of strains resistant to antimicrobials, such as imipenem, sulbactam, rifampin, or tigecycline (18, 48), has recently driven the use of polymyxins as treatment. These antibiotics, colistin and polymyxin B, were used in the 1960s and 1970s but then largely abandoned owing to reports of toxicity, though recent studies have demonstrated that revised dosing regimens can minimize this problem (25).
Colistin and polymyxin B are rapidly bactericidal for Gram-negative bacteria, interacting with the lipid A moiety of lipopolysaccharide (LPS) to cause disorganization of the outer membrane (16). Although they remain active against most otherwise extremely resistant A. baumannii isolates, this activity is not universal, and colistin heteroresistance has been described in A. baumannii (24), as has the development of full resistance both in vitro and in vivo (17, 41). Outbreaks of polymyxin-resistant A. baumannii isolates have been reported (12, 35, 47). Acquired polymyxin resistance in Gram-negative bacteria is most often mediated by replacement of lipid A with aminoarabinose, which requires the products of the ugd and pbg loci and ethanolamine, mediated by pmrC. These modifications remove negative charges, lowering the affinity of LPS for polymyxins (21, 34, 44). In this context, it is known that the two-component response regulator and sensor kinase PmrA/B, which allows bacteria to sense and respond to various environmental conditions, including pH or Fe3+ and Mg2+ levels, also affects expression of genes implicated in lipid A modification and thereby influences susceptibility to colistin (13).
Point mutations in the PmrA/B two-component system in colistin-resistant A. baumannii, Pseudomonas aeruginosa, and Salmonella have been reported (2, 31, 32, 40), and also, increased expression of pmrA in this event in A. baumannii seems to be implicated (2). Modifications in the PhoP/Q two-component regulatory system in the resistance to polymyxins due to modifications in LPS in P. aeruginosa (7) and Salmonella (14, 15) are implicated, but this system appears to be absent from A. baumannii genomes (3). An A. baumannii ATCC 19606 colistin-resistant laboratory derivate with mutations in PmrB showed remodeling of the bacterial outer membrane and a decreased membrane permeability compared with those of its susceptible counterpart, highlighting the importance of this gene in colistin resistance.
Recently, Moffatt et al. have reported resistance to colistin in this species mediated by loss of LPS production (30). Laboratory derivates carrying mutations in the genes implicated in lipid A biosynthesis, lpxA, lpxC, and lpxD, showed increased MICs of colistin (30). An LPS-deficient colistin-resistant LpxA mutant showed changes in the membrane potential probably due to outer membrane modification and loss of affinity by colistin (38). Indeed, colistin exposure causes differences in the cell morphology of A. baumannii, particularly a more spherical appearance and increased cell surface roughness, leading to outer membrane damage (39). Although the precise molecular basis of polymyxin resistance remains unknown, it seems clear that LPS modifications and loss of affinity for anionic polymyxins are necessary to the development of resistance to these antibiotics in A. baumannii (26).
In the present study, we investigated the role of PmrA/B in colistin resistance in clinical isolates and laboratory mutants of A. baumannii and analyzed the lipid A moiety among these organisms.
MATERIALS AND METHODS
Clinical isolates and mutants.
Isolates of A. baumannii were identified by phenotype (API 20NE; bioMérieux, La Balme-les-Grottes, France) and PCR for blaOXA-51 (46). They included five colistin-resistant and five colistin-susceptible clinical isolates (Table 1). The five susceptible isolates and three of the resistant isolates were collected in United Kingdom between 2006 and 2009, one resistant isolate was collected during an outbreak in Spain in 2000 (47), and the remaining resistant isolate was collected in Saudi Arabia in 2009. One resistant/susceptible pair of clinical isolates (Ab15/132 and Ab15/133, respectively) was from the same patient. Colistin-resistant mutants were selected from susceptible type strain ATCC 19606 (12) and from susceptible clinical isolate ABRIM (9) by serial passage in LB broth with increasing concentrations of colistin (1, 2, 4, and 8 μg/ml).
Table 1.
A. baumannii strains used
| Strain | Colistin MIC (μg/ml)a | Colistin susceptibilityb | Pulsed-field profile | Source (country, year, type of infection)c | Parent straind | pmrB genotype |
|---|---|---|---|---|---|---|
| Ab14/144 | 4 | R | OXA-24 European clone II | UK, 2008, wound infection | N/A | Met145Lys |
| Ab804 | 16 | R | Unique | Saudi Arabia, 2009, blood infection | N/A | Pro233Ser |
| Ab15/132 | 32 | R | Strain ac-1 | UK, 2009, respiratory infection | Same clone as Ab15/133 | Leu87Phe |
| Ab10/30 | 128 | R | NW strain | UK, 2006, blood infection | N/A | Ser14Leu |
| Ab208628 | 32 | R | Unique | Spain, 2009 (47) | N/A | Phe387Tyr Ser403Phe |
| Ab15/133 | 1 | S | Strain ac-1 | UK, 2009, respiratory infection | Same clone as Ab15/132 | Wild type |
| Ab1/137 | 1 | S | OXA 23 clone 1 | UK, 2009, colonization | N/A | Wild type |
| Ab15/88 | 0.5 | S | Unique | UK, 2009, wound infection | N/A | Wild type |
| Ab15/24 | 0.5 | S | Strain 00AC-2 | UK, 2008, colonization | N/A | Wild type |
| Ab15/120 | 0.5 | S | Unique | UK, 2009, urinary infection | N/A | Wild type |
| ATCC 19606 ColS | 1 | S | Unique | 2009 (12) | N/A | Wild type |
| ATCC 19606 ColR | 32 | R | 2009 (12) | ATCC 19606 | Ala227Val | |
| ATCC 19606 ColS ΔpmrB | 0.125 | S | This study | ATCC 19606 ColS | ΔpmrB | |
| ATCC 19606 ColR ΔpmrB | 0.125 | S | This study | ATCC 19606 ColR | ΔpmrB | |
| ABRIM ColS | 0.5 | S | Unique | Spain, 2000 (9) | N/A | Wild type |
| ABRIM ColR | 16 | R | This study | ABRIM | Asn353Tyr |
MICs were obtained by the agar dilution method and by Etest.
R, resistant; S, susceptible.
UK, United Kingdom.
N/A, not applicable.
Characterization of isolates.
Pulsed-field gel electrophoresis (PFGE) was performed on ApaI-digested genomic DNA as described previously (42). MICs of colistin were determined by agar dilution (27) and Etest (AB Biodisk, Solna, Sweden) and interpreted using British Society for Antimicrobial Chemotherapy (BSAC) criteria (4).
Sequencing of pmrCAB.
Based on the reference sequence of A. baumannii ATCC 17978 (GenBank accession number CP000521.1), primers Full pmrCAB-F and Full pmrCAB-R (Sigma-Genosys Ltd., Cambridge, United Kingdom) were designed to amplify a 3,699-bp product containing the pmrCAB coding region (Table 2). Primers pmrC-F, pmrC-R, pmrC2-F, pmrC2-R, pmrA-F, pmrA-R, pmrB-F, pmrB-R, pmrB2-F, and pmrB2-R were used for sequencing (Table 2). The Expand high-fidelity PCR system (Roche Diagnostics, Lewes, United Kingdom) was used for amplification, and the resulting amplicons were purified with the GeneClean Turbo kit (MD Biomedicals, Strasbourg, France). Sequences were determined using an ABI genetic analyzer capillary platform 3130XL automatic DNA sequencer (ABI, Warrington, United Kingdom).
Table 2.
DNA sequences of oligonucleotides used in real-time RT-PCR in this study
| Oligonucleotidea | Sequence | Reference |
|---|---|---|
| Full pmrCAB-F | 5′-GCATCATAAAAAGATTGTAGTCAC-3′ | This study |
| Full pmrCAB-R | 5′-GCGATTTGTATTCATCGTTTTGAG-3′ | This study |
| pmrA-F | 5′-ATGACAAAAATCTTGATGATTGAAGAT-3′ | This study |
| pmrA-R | 5′-TTATGATTGCCCCAAACGGTAG-3′ | This study |
| pmrB-F | 5′-GTGCATTATTCATTAAAAAAAC-3′ | This study |
| pmrB-R | 5′-TCACGCTCTTGTTTCATGTA-3′ | This study |
| pmrB2-F | 5′-GGTTCGTGAAGCTTTCG-3′ | This study |
| pmrB2-R | 5′-CCTAAATCGATTTCTTTTTG-3′ | This study |
| pmrC-F | 5′-ATGTTTAATCTCATTATAGCCA-3′ | This study |
| pmrC-R | 5′-TTAGTTTACATGGGCACAA-3′ | This study |
| pmrC2-F | 5′-GGTTGTTATTGAAGAAAGTAT-3′ | This study |
| pmrC2-R | 5′-TCAATCCAAGTCACTTGGTAAC-3′ | This study |
| PMRBINTFW | 5′-CGCTCAAGGTATAGTCAG-3′ | This study |
| PMRBINTRV | 5′-CAGTAGGCTCGACCATAC-3′ | This study |
| PMRBEXTFW | 5′-CAGTGTCATCTTAGGTTG-3′ | This study |
| PMRBEXTRV | 5′-CGTAGTGATCTGGATCGG-3′ | This study |
| RT-PCR pmrA-F | Same as that of pmrA-F | This study |
| RT-PCR pmrA-R | 5′-CCATCATAGGCAATCCTAAATCCA-3′ | This study |
| RT-PCR pmrB-F | 5′-GAACAGCTGAGCACCCTTTAA-3′ | This study |
| RT-PCR pmrB-R | 5′-ACAGGTGGAACCAGCAAATG-3′ | This study |
| RT-PCR pmrC-F | 5′-CTCTTTACGCTTTGTTTTATGGAC-3′ | This study |
| RT-PCR pmrC-R | 5′-GTAAAAAGTAAAACACCGACCA-3′ | This study |
| RT-PCR 16S rRNA-F | 5′-TCAGCTCGTGTCGTGAGATG-3′ | 19 |
| RT-PCR 16S rRNA-R | 5′-CGTAAGGGCCATGATG-3′ | 19 |
| blaOXA-51like-F | 5′-TAATGCTTTGATCGGCCTTG-3′ | 46 |
| blaOXA-51like-R | 5′-TGGATTGCACTTCATCTTGG-3′ | 46 |
F, forward oligonucleotide; R, reverse oligonucleotide.
Insertional inactivation of pmrB in A. baumannii ATCC 19606 ColR and ColS.
Gene inactivation was carried out as previously described by Aranda et al. (5). Briefly, the target gene was amplified by PCR with the oligonucleotide pair pmrBintFW-pmrBintRV, and genomic DNA from A. baumannii ATCC 19606 was used as the template. The 539-bp PCR product obtained was cloned into the kanamycin and zeocin resistance plasmid pCR-BluntII-TOPO (Invitrogen Ltd., Paisley, United Kingdom), which is unable to replicate in A. baumannii, and propagated in Escherichia coli strain TG1. The recombinant plasmid (0.1 μg) was introduced into A. baumannii ATCC 19606 ColR and ColS, which are susceptible to kanamycin, by electroporation. Candidate transformant clones with ΔpmrB were then selected on kanamycin-containing plates (50 μg/ml) and confirmed by sequencing of the PCR products obtained using primers pmrBextFW-T7 and pmrBextRV-SP6 and genomic DNA from A. baumannii mutants.
Real-time RT-PCR of pmrCAB expression.
Real-time reverse transcriptase (RT)-PCR was used to monitor the expression of pmrCAB. Bacteria were grown on LB broth to an optical density at 600 nm (OD600) of 0.4 at 37°C, and total RNA was extracted with an RNeasy minikit (Qiagen, West Sussex, United Kingdom). This RNA was treated with RNase-free DNase (Invitrogen) and quantified with a NanoDrop 1000 spectrophotometer (Thermo Scientific, Winsford, United Kingdom). The QuantiFast SYBR green RT-PCR kit (Qiagen) was used for analysis, with 30 ng of total RNA and the primers shown in Table 2. The 16S rRNA gene was used as an internal control for quantification of relative gene expression (19), which was described as the mean of three independent experiments. Negative controls without reverse transcriptase were included to detect DNA contamination.
Real-time RT-PCR was used to monitor the expression of pmrCAB under induction conditions in the presence of Fe3+ and low Mg2+ (20). Bacterial RNA was extracted from cells grown in LB with 2 mM ferric chloride and without ferric chloride (2), and also, RNA was extracted from cells grown in Acinetobacter minimal medium (AMM) supplemented with 2 μM and 2 mM magnesium sulfate (36). RNA extraction and RT-PCR were performed as described above.
Analysis of lipid A structure.
Lipid A was extracted by an ammonium hydroxide-isobutyric acid method and subjected to negative-ion matrix-assisted laser desorption ionization-time of flight (MALDI-TOF) mass spectrometry analysis (10, 28). Briefly, lyophilized crude cells (10 mg) were resuspended in 400 μl isobutyric acid-1 M ammonium hydroxide (5:3, vol/vol) and incubated in a screw-cap test tube at 100°C for 2 h, with occasional vortexing. Samples were then cooled in ice water and centrifuged (2,000 × g for 15 min), with supernatants transferred to new tubes, diluted with equal volumes of water, and lyophilized. These samples were then washed twice with 400 μl methanol and centrifuged (2,000 × g for 15 min) to pellet the insoluble lipid A, which subsequently was solubilized in 100 to 200 μl chloroform-methanol-water (3:1.5:0.25, vol/vol/vol). Analyses were performed on a Bruker Autoflex II MALDI-TOF mass spectrometer (Bruker Daltonics, Coventry, United Kingdom) in negative reflective mode with delayed extraction. Each spectrum was an average of 300 shots. The ion-accelerating voltage was set at 20 kV. Dihydroxybenzoic acid (Sigma Chemical Co., St. Louis, MO) was used as a matrix. A few microliters of lipid A suspension (1 mg/ml) was desalted with a few grains of ion-exchange resin (H+; Dowex 50WX8) in an Eppendorf tube, and 1 μl was deposited on the target and covered with the same amount of the matrix suspended at 10 mg/ml in 0.1 M citric acid. Different ratios between the samples and dihydroxybenzoic acid were used when necessary. A peptide calibration standard (Bruker Daltonics) was used to calibrate the MALDI-TOF mass spectrometry. Further calibration was performed using lipid A extracted from Escherichia coli strain MG1655 grown in LB at 37°C. Detailed mass spectrometry analysis of E. coli lipid A has been described previously (50).
Nucleotide sequence accession numbers.
The nucleotide sequences presented in this study have been submitted to the GenBank database, as follows: pmrCAB genes, GenBank accession numbers HM134921 (ABRIM ColR), HM134922 (ABRIM ColS), HM149342 (Ab15/132 ColR), HM149343 (Ab15/133 ColS), HM149344 (ATCC 19606 ColS), and HM149345 (ATCC 19606 ColR); pmrAB genes, GenBank accession numbers HM138195 (Ab208628 ColR), HM138196 (Ab804 ColR), HM138197 (Ab14/144 ColR), and HM138198 (Ab10/30 ColR).
RESULTS
Isolate characterization.
All clinical isolates had distinct PFGE banding profiles, except isolates Ab15/133 and Ab15/132, which were both from the same patient and were confirmed to be identical.
The MICs for the colistin-resistant isolates ranged from 4 to 128 μg/ml, while those for the susceptible isolates were ≤1 μg/ml; those for the paired isolates Ab15/133 and Ab15/132 were 1 and 32 μg/ml, respectively. The polymyxin-resistant mutants of A. baumannii strains ATCC 19606 and ABRIM showed 32-fold increases in colistin MICs, which rose from 1 to 32 and 0.5 to 16 μg/ml, respectively. The pandrug-resistant A. baumannii Ab208628 clinical strain showed a resistant phenotype to colistin (64 μg/ml) (47) (Table 1).
Analysis of pmrCAB nucleotide sequences.
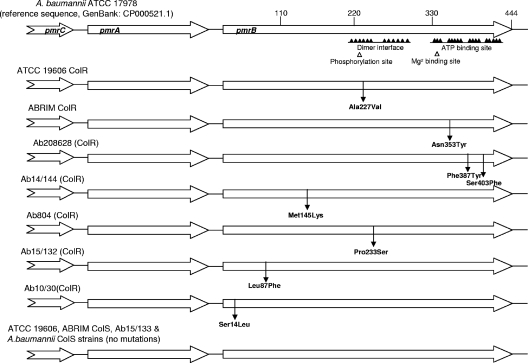
The PmrAB system is implicated in the resistance to colistin in A. baumannii (2), and the pmrCAB operon is implicated in Salmonella (21); so, the pmrCAB genes of the colistin-resistant and -susceptible isolates and mutant pairs, and the pmrAB genes of the other isolates, were amplified and sequenced on both strands. The pmrB sequences of all the colistin-susceptible isolates were identical to that of a reference sequence (GenBank accession number CP000521.1), whereas all the colistin-resistant isolates harbored mutations that resulted in at least one substitution in PmrB (Table 1). Three isolates had single amino acid changes (Ser14Leu, Met145Lys, or Pro233Ser), and one had changes at two amino acids (Phe387Tyr and Ser403Phe). Isolate Ab15/132 harbored a Leu87Phe change as opposed to its colistin-susceptible counterpart, Ab15/133. Single replacements in PmrB were likewise found in the two laboratory-derived colistin-resistant mutants, Ala227Val in ATCC 19606 ColR and Asn353Tyr in ABRIM ColR, compared with their susceptible parent strains. No differences were found in the pmrA or pmrC gene of any of the isolates (Table 1; Fig. 1).
Fig. 1.
Schematic representation of the pmrCAB genes. No amino acid changes were found in PmrA or PmrC. Amino acid replacements predicted in PmrB are identified with the arrows. No substitutions were found in these proteins of ATCC 19606 ColS, ABRIM ColS, and Ab15/133 or in the colistin-susceptible A. baumannii strains. The main domains in PmrB are identified, specifically, (i) the dimerization/phosphoacceptor domain, with the conserved histidine 228, and (ii) the ATP binding site.
Expression of pmrCAB.
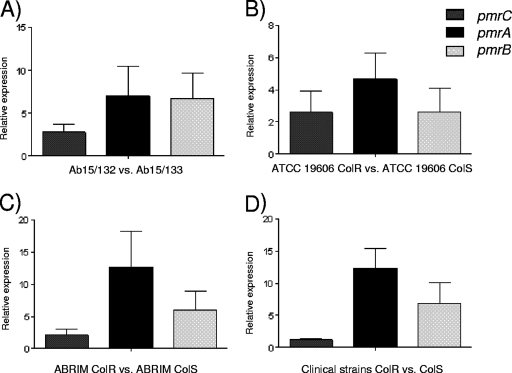
Expression of the pmr genes was investigated to determine whether differential expression was associated with colistin resistance. RT-PCR identified upregulation of pmrA and pmrB in all the resistant clinical isolates, compared with that in the susceptible ones: mean increases were 12.4-fold for pmrA expression and 6.8-fold for pmrB expression in colistin-resistant versus -susceptible isolates, whereas expression of pmrC was not significantly changed (mean increase of 1.1-fold) (Fig. 2). The polymyxin-resistant mutants of ATCC 19606 and ABRIM and clinical isolate Ab15/132 had increased expression of pmrA (4.7-, 12.7-, and 7.0-fold, respectively) and pmrB (2.6-, 6.0-, and 6.8-fold, respectively) compared with that of their colistin-susceptible counterparts, whereas increases in pmrC expression were present but smaller (2.6-, 2.1-, and 2.8-fold, respectively). Such differences were seen regardless of whether RNA was prepared from cells grown in the presence or absence of 2 μg/ml colistin. No significant differences of expression in the pmrA, pmrB, or pmrC gene were identified by RT-PCR in the ATCC 19606 ColS and ATCC 19606 ColS ΔpmrB strains in the presence of high Fe3+. However, low upregulation of pmrC and pmrB (1.77- and 1.46-fold, respectively) was observed when the strain was grown in AMM supplemented with 2 μM magnesium sulfate compared with that when the strain grown in the presence of 2 mM magnesium sulfate. No significant differences in pmrA expression under these conditions were found; likewise, neither pmrA nor pmrC of the ATCC 19606 ColS ΔpmrB strain showed expression differences (0.72- and 1.02-fold, respectively).
Fig. 2.
Relative expression of the pmrCAB genes in the colistin-resistant and -susceptible strains (expression level of pmrCAB in the ColS strains equal to 1). (A) Ab15/132 ColR compared with Ab15/133 ColS; (B) ATCC 19606 ColR compared with ATCC 19606 ColS; (C) ABRIM ColR compared with ABRIM ColS; (D) means for five clinical ColR strains compared with those for five clinical ColS strains. Values are the means and the standard deviations from three independent experiments.
Analysis of lipid A composition.
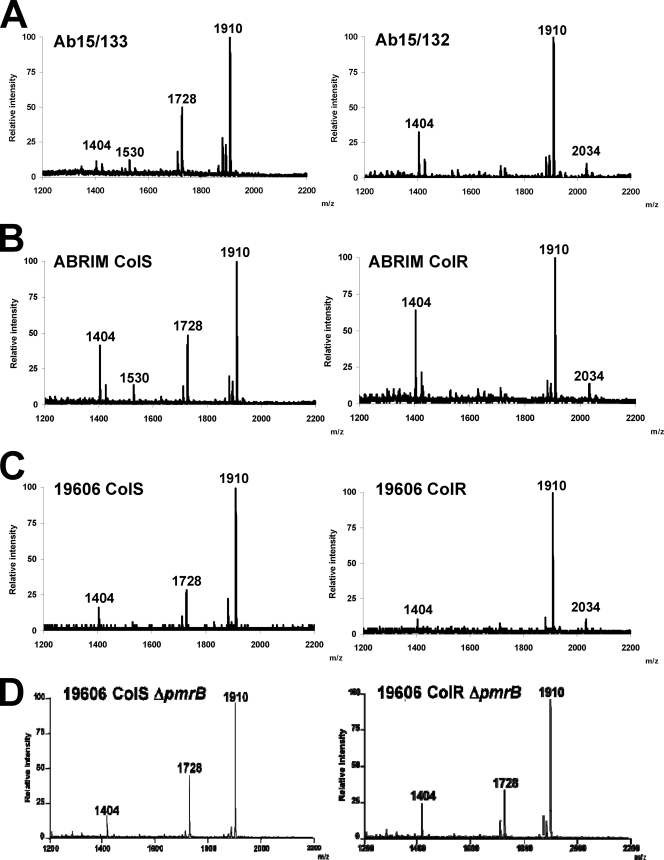
Modification of lipid A has been associated with polymyxin resistance in other genera (21, 44). We therefore characterized and compared by MALDI-TOF mass spectrometry the lipid A moieties of three colistin-susceptible and -resistant pairs of isolates, as follows: Ab15/133 and Ab15/132, ATCC19606 ColS and ColR, and ABRIM ColS and ColR (Fig. 3). Previous studies have unambiguously shown that the lipid A disaccharide backbone of Acinetobacter spp. is composed of two β-(1-6)-linked 2-amino-2-deoxyglucose residues (GlcNI and GlcNII) phosphorylated at positions 1 and 4′ (22, 23, 49). The lipid A fraction of A. baumannii contains two major species which may correspond to a hepta-acylated lipid A with two 2-amino-2-deoxyglucose residues, two phosphates, three 12:0(3-OH) fatty acids, two 14:0(3-OH) fatty acids, and two 12:0 (m/z 1,910) fatty acids attached or to a hexa-acylated lipid A with two 2-amino-2-deoxyglucose residues, two phosphates, and three 12:0(3-OH) fatty acids, two 14:0(3-OH) fatty acids, and one 12:0 (m/z 1,728) fatty acid attached (22) (Fig. 4). The colistin-susceptible Ab15/133, ABRIM, and ATCC 19606 ColS isolates all contained both of these (Fig. 3A to C; Table 3). Other ion peaks detected were m/z 1,530, also previously found (28), which may represent a penta-acylated lipid A lacking one 12:0 fatty acid moiety, and m/z 1,404, which may correspond to a tetra-acyl lipid A with three 12:0(3-OH) fatty acids and one 14:0(3-OH) fatty acid attached to the phosphorylated disaccharide (Fig. 4). In the colistin-resistant isolates, only three ion peaks were detected, corresponding to m/z 1404, 1,910, and 2,034. Critically, the latter species is consistent with the addition of phosphoethanolamine (theoretical m/z 124) to the hepta-acylated form of lipid A (m/z 1,910).
Fig. 3.
Negative-ion MALDI-TOF mass spectra of lipid A isolated from colistin-resistant and -susceptible A. baumannii isolates. (A) Ab15/133 and Ab15/132; (B) ABRIM ColS and ABRIM ColR; (C) ATCC 19606 ColS and ATCC 19606 ColR; (D) ATCC 19606 ColS ΔpmrB and ATCC 19606 ColR ΔpmrB.
Fig. 4.
Proposed structures of the main molecular lipid A species.
Table 3.
Composition of lipid A in colistin-resistant and -susceptible A. baumannii clinical pair Ab15/133 and Ab15/32 and in ATCC 19606 and ABRIM and their derivatives
| Peak (m/z) | Chemical inference | Chemical compositiona | Presence/absence of chemical interference in: |
|||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Ab15/133 (colistin susceptible) | Ab15/132 (colistin resistant) | ABRIM ColS (colistin susceptible) | ABRIM ColR (colistin resistant) | ATCC 19606 ColS (colistin susceptible) | ATCC 19606 ColR (colistin resistant) | ATCC 19606 ColS ΔpmrB (colistin susceptible) | ATCC 19606 ColR ΔpmrB (colistin susceptible) | |||
| 1,404 | Tetra-acylated lipid A | 2 glucose residues | + | + | + | + | + | + | + | + |
| 2 phosphates | ||||||||||
| 3 acyl groups, 12:0(3-OH) | ||||||||||
| 1 acyl group, 14:0(3-OH) | ||||||||||
| 1,530 | Penta-acylated lipid A | 2 glucose residues | + | − | + | − | − | − | − | − |
| 2 phosphates | ||||||||||
| 2 acyl groups, 12:0(3-OH) | ||||||||||
| 2 acyl groups, 14:0(3-OH) | ||||||||||
| 1 acyl group, 12:0 | ||||||||||
| 1,728 | Hexa-acylated lipid A | 2 glucose residues | + | − | + | − | + | − | + | + |
| 2 phosphates | ||||||||||
| 3 acyl groups, 12:0(3-OH) | ||||||||||
| 2 acyl groups, 14:0(3-OH) | ||||||||||
| 1 acyl group, 12:0 | ||||||||||
| 1,910 | Hepta-acylated lipid A | 2 glucose residues | + | + | + | + | + | + | + | + |
| 2 phosphates | ||||||||||
| 3 acyl groups, 12:0(3-OH) | ||||||||||
| 2 acyl groups, 14:0(3-OH) | ||||||||||
| 2 acyl groups, 12:0 | ||||||||||
| 2,034 | Phosphoethanolamine-modified | 2 glucose residues | − | + | − | + | − | + | − | − |
| hepta-acylated lipid A | 2 phosphates | |||||||||
| 3 acyl groups, 12:0(3-OH) | ||||||||||
| 2 acyl groups, 14:0(3-OH) | ||||||||||
| 2 acyl group, 12:0 | ||||||||||
| 1 phosphoethanolamine | ||||||||||
Role of pmrB in colistin resistance.
To examine whether the PmrA/B two-component system was implicated in the development of colistin resistance, we generated ΔpmrB mutants of the ATCC 19606 ColR and ColS strains. These proved to be more susceptible than their corresponding parents (MICs of 0.19 μg/ml versus 32 μg/ml in ATCC 19606 ColR derivatives and MICs of 0.125 μg/ml versus 1 μg/ml in ATCC 19606 ColS derivatives), showing the importance of this gene. In addition, 5.3-fold downregulation of pmrC was observed by RT-PCR in the ATCC 19606 ColR ΔpmrB mutant compared with that in its parent. These results accord with the similar pmrAB-regulated PmrC model proposed for Salmonella by Lee et al., in which a ΔpmrC mutant and a null pmrA mutant resulted in a lipid A species without the addition of phosphoethanolamine (21).
Lipid A from the ΔpmrB mutant of ATCC 19606 ColR contained the major molecular species with m/z 1,910, 1,728, and 1,404 but lacked the ion peak (m/z 2,034), seen in its parent and consistent with the addition of phosphoethanolamine (theoretical m/z 124) to hepta-acylated lipid A (m/z 1,910) (Fig. 3D). Lipid A from the ATCC 19606 ColS ΔpmrB strain was identical to those of the ATCC 19606 wild type and ATCC 19606 ColR ΔpmrB (Fig. 3C and D).
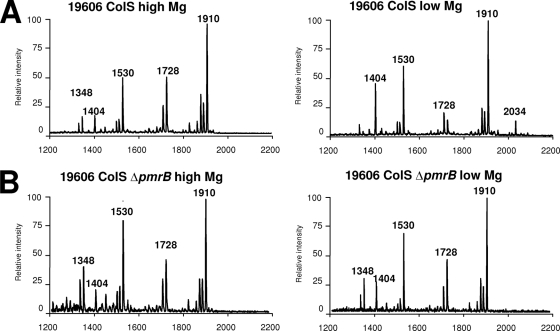
Lipid A analysis of ATCC 19606 grown in low Mg2+ (Fig. 5A) revealed the presence of the ion peak (m/z 2,034), which was absent in lipid A from the ATCC 19606 ColS ΔpmrB strain grown under the same conditions (Fig. 5B). Ion peaks (m/z 1,404, 1,728, and 1,910) were found in strain ATCC 19606 grown in LB (Fig. 3), whereas another ion peak (m/z 1,530) was identified in the Ab15/133 and ABRIM ColS strains. Molecular species with m/z 1,348 may correspond to m/z 1,530 lacking one 12:0 fatty acid moiety. The absence of molecular species with m/z 1,530 and 1,348 in bacteria grown in LB (Fig. 3C) but presence in lipid A from bacteria grown in minimal medium with either low or high Mg2+ (Fig. 5) suggests that they are dependent on the growth medium and not on the cation content.
Fig. 5.
Negative-ion MALDI-TOF mass spectra of lipid A isolated from strains grown under high-Mg2+ conditions (2 mM magnesium sulfate) and low-Mg2+ conditions (2 μM magnesium sulfate). (A) ATCC 19606 ColS wild type; (B) ATCC 19606 ColS ΔpmrB.
DISCUSSION
Colistin and tigecycline are often the only treatment options for multiresistant A. baumannii infections, but resistance to both agents has recently been described, with colistin resistance scattered worldwide. A. baumannii Ab208628, used here (Table 1), was resistant to all antibiotics tested and was associated with a nosocomial outbreak (47). We sought to define the mechanism(s) of colistin resistance in this species. Our data show that at least one mutation in PmrB and pmrAB upregulation seems necessary to confer this resistance.
Several mutations were found in PmrB, located at widely scattered sites. Ala227Val lies adjacent to the conserved histidine at the site of phosphorylation (His228), and Pro233Ser lies at an essential position for dimer formation (43). Both of these were found previously in colistin-resistant clinical strains or laboratory mutants (2), and the positions seem critical for the phosphatase activity. Mutations Asn353Tyr, Phe387Tyr, and Ser403Phe lie inside the ATP binding site and may have an effect on the phosphorylation of His228 of PmrB and thereby the phosphorylation levels of Asp52 of PmrA, the response regulator, which is the final acceptor of the phosphate group, affecting the expression levels of target genes, such as pmrC (45). The other mutations found in the nonconserved N-terminal domains of PmrB (Table 1) may be responsible for regulating the activity of the C-terminal kinase-phosphatase conserved domain, typical of the two-component regulatory systems (6, 33). Single substitutions in pmrB of P. aeruginosa at positions 243, 248, and 292, affect the function of PmrB, possibly by increasing phosphatase activity, thereby activating the PmrA regulon and increasing the resistance to colistin by LPS modifications (1, 11). Similar effects in A. baumannii may apply.
We observed increased expression of pmrA and pmrB in all the colistin-resistant clinical isolates and laboratory mutants. Adams et al. also reported increased pmrA expression in laboratory-selected colistin-resistant mutants of A. baumannii (2). The pmrA and pmrB genes seem to interact as an operon, as they showed similar expression levels in the colistin-susceptible strains, whereas inactivation of pmrB led to clear downregulation of pmrC, also highlighting the interaction between these components and the possible implication of pmrB in colistin resistance.
Negative-ion MALDI-TOF mass spectra revealed the presence of phosphoethanolamine (m/z 2,034) in lipid A isolated from the three colistin-resistant strains examined, a change previously associated with resistance to polymyxins in Salmonella (13). This phosphoethanolamine was lost when pmrB was inactivated in a colistin-resistant strain, confirming its role. Analysis also revealed the absence of hexa- and penta-acylated lipid A species in colistin-resistant strains. This is in good agreement with the idea that increased lipid A acylation (in the dominant hepta-acylated material) is associated with antimicrobial peptide resistance (11). An interesting finding is the relative increase in the levels of the tetra-acylated lipid A species (m/z 1,404) in colistin-resistant clinical isolates (Fig. 3A and B). This type of lipid A is associated with the lowest level of activation of the human immune system (37), leading to the tantalizing hypothesis that colistin-resistant Acinetobacter strains may lead to less of an inflammatory response than a colistin-susceptible one. Studies are ongoing to test this speculation.
Low-Mg2+ conditions in Pseudomonas aeruginosa can regulate the resistance to colistin in response to environmental conditions (29). In this study, reverse transcriptase (RT)-PCR showed a slight increase of expression in pmrC, and MALDI-TOF mass spectra revealed lipid A modification; thus, in a similar way, the development of a moderate level of colistin resistance in A. baumannii seems to be induced by the environmental conditions.
Our results indicate a different mechanism of colistin resistance in A. baumannii from that described by Moffatt et al. (30), who noted the total loss of LPS production via inactivation of the biosynthesis pathway genes lpxA, lpxC, and lpxD. Differences in selection methods may explain the development of different mechanisms: our mutants were selected in LB broth supplemented with stepwise increased colistin concentrations from 1 to 8 mg/liter, whereas Moffat et al. selected with colistin at a fixed concentration of 10 μg/ml in agar. These LPS-negative mutants became susceptible to cefepime, teicoplanin, and azithromycin, apparently reflecting defects to membrane integrity. No such traits in the present isolates and mutants were seen, which retained LPS.
In summary, we report the analysis of pmrCAB in a diverse collection of clinical isolates and laboratory mutants of A. baumannii and its relationship with colistin resistance. We also analyzed the composition of lipid A from resistant and susceptible isolates. Our data suggest that resistance to colistin in A. baumannii requires at least two distinct genetic events, as follows: (i) at least one amino acid change in PmrB, although these changes were diverse and not localized to a specific domain, and (ii) upregulated expression of pmrA and pmrB. The precise genetic events that cause pmrAB upregulation remain to be defined. These genetic changes lead, in turn, to the addition of phosphoethanolamine to hepta-acylated lipid A, leading to the LPS modifications that directly confer the colistin resistance.
ACKNOWLEDGMENTS
We thank J. Pachón for the gift of strain Ab208628 and L. Rivas for the gift of the ATCC 19606 ColS and ColR strains. We also thank J. Turton for her help with the typing experiments and M. de Mar Tomás for her help with the RT-PCR experiments.
This work was supported by travel grants from the Ministerio de Sanidad y Consumo, Instituto de Salud Carlos III, through the Spanish Network for Research in Infectious Diseases (grant REIPI RD 06/0008); the Fondo de Investigaciones Sanitarias (grant PS09/00687), the Ministerio de Educación y Ciencia (program “Jose Castillejo”); the Spanish Society of Infectious Diseases and Clinical Microbiology (SEIMC); and the Xunta de Galicia through the Dirección Xeral de I+D+i and Fondo Social Europeo (FSE). CIBERES is an initiative from Instituto de Salud Carlos III. A.B. is supported by the Consellería de Economia e Industria, Xunta de Galicia (program “Ángeles Alvariño”).
D.M.L. and N.W. have each received grants and conference support from most major pharmaceutical companies. D.M.L. holds shares in AstraZeneca, Pfizer, Schering-Plough, Dechra, and GlaxoSmithKline, amounting to under 10% of a diversified portfolio. The other authors have no conflicts of interest to declare.
Footnotes
Published ahead of print on 16 May 2011.
REFERENCES
- 1. Abraham N., Kwon D. H. 2009. A single amino acid substitution in PmrB is associated with polymyxin B resistance in clinical isolate of Pseudomonas aeruginosa. FEMS Microbiol. Lett. 298:249–254 [DOI] [PubMed] [Google Scholar]
- 2. Adams M. D., et al. 2009. Resistance to colistin in Acinetobacter baumannii associated with mutations in the PmrAB two-component system. Antimicrob. Agents Chemother. 53:3628–3634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Adams M. D., et al. 2008. Comparative genome sequence analysis of multidrug-resistant Acinetobacter baumannii. J. Bacteriol. 190:8053–8064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Andrews J. M. 2004. BSAC standardized disc susceptibility testing method (version 3). J. Antimicrob. Chemother. 53:713–728 [DOI] [PubMed] [Google Scholar]
- 5. Aranda J., et al. 2010. A rapid and simple method for constructing stable mutants of Acinetobacter baumannii. BMC Microbiol. 10:279–291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Atkinson M. R., Ninfa A. J. 1992. Characterization of Escherichia coli glnL mutations affecting nitrogen regulation. J. Bacteriol. 174:4538–4548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Barrow K., Kwon D. H. 2009. Alterations in two-component regulatory systems of phoPQ and pmrAB are associated with polymyxin B resistance in clinical isolates of Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 53:5150–5154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Bergogne-Berezin E., Towner K. J. 1996. Acinetobacter spp. as nosocomial pathogens: microbiological, clinical, and epidemiological features. Clin. Microbiol. Rev. 9:148–165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Bou G., Oliver A., Martínez-Beltrán J. 2000. OXA-24, a novel class D beta-lactamase with carbapenemase activity in an Acinetobacter baumannii clinical strain. Antimicrob. Agents Chemother. 44:1556–1561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. El Hamidi A., Tirsoaga A., Novikov A., Hussein A., Caroff M. 2005. Microextraction of bacterial lipid A: easy and rapid method for mass spectrometric characterization. J. Lipid Res. 46:1773–1778 [DOI] [PubMed] [Google Scholar]
- 11. Ernst R. K., et al. 1999. Specific lipopolysaccharide found in cystic fibrosis airway Pseudomonas aeruginosa. Science 286:1561–1565 [DOI] [PubMed] [Google Scholar]
- 12. Fernández-Reyes M., et al. 2009. The cost of resistance to colistin in Acinetobacter baumannii: a proteomic perspective. Proteomics 9:1632–1645 [DOI] [PubMed] [Google Scholar]
- 13. Gunn J. S. 2008. The Salmonella PmrAB regulon: lipopolysaccharide modifications, antimicrobial peptide resistance and more. Trends Microbiol. 16:284–290 [DOI] [PubMed] [Google Scholar]
- 14. Gunn J. S., et al. 1998. PmrA-PmrB-regulated genes necessary for 4-aminoarabinose lipid A modification and polymyxin resistance. Mol. Microbiol. 27:1171–1182 [DOI] [PubMed] [Google Scholar]
- 15. Guo L., et al. 1997. Regulation of lipid A modifications by Salmonella typhimurium virulence genes phoP-phoQ. Science 276:250–253 [DOI] [PubMed] [Google Scholar]
- 16. Hancock R. E. 1997. Peptide antibiotics. Lancet 349:418–422 [DOI] [PubMed] [Google Scholar]
- 17. Hawley J. S., Murray C. K., Jorgensen J. H. 2008. Colistin heteroresistance in Acinetobacter and its association with previous colistin therapy. Antimicrob. Agents Chemother. 52:351–352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Heritier C., Poirel L., Lambert T., Nordmann P. 2005. Contribution of acquired carbapenem-hydrolyzing oxacillinases to carbapenem resistance in Acinetobacter baumannii. Antimicrob. Agents Chemother. 49:3198–3202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Higgins P. G., Wisplinghoff H., Stefanik D., Seifert H. 2004. Selection of topoisomerase mutations and overexpression of adeB mRNA transcripts during an outbreak of Acinetobacter baumannii. J. Antimicrob. Chemother. 54:821–823 [DOI] [PubMed] [Google Scholar]
- 20. Kato A., Groisman E. A. 2004. Connecting two-component regulatory systems by a protein that protects a response regulator from dephosphorylation by its cognate sensor. Genes Dev. 18:2302–2313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Lee H., Hsu F. F., Turk J., Groisman E. A. 2004. The PmrA-regulated pmrC gene mediates phosphoethanolamine modification of lipid A and polymyxin resistance in Salmonella enterica. J. Bacteriol. 186:4124–4133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Leone S., et al. 2006. Structural elucidation of the core-lipid A backbone from the lipopolysaccharide of Acinetobacter radioresistens S13, an organic solvent tolerant Gram-negative bacterium. Carbohydr. Res. 341:582–590 [DOI] [PubMed] [Google Scholar]
- 23. Leone S., et al. 2007. Detailed characterization of the lipid A fraction from the nonpathogen Acinetobacter radioresistens strain S13. J. Lipid Res. 48:1045–1051 [DOI] [PubMed] [Google Scholar]
- 24. Li J., et al. 2006. Heteroresistance to colistin in multidrug-resistant Acinetobacter baumannii. Antimicrob. Agents Chemother. 50:2946–2950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Li J., Nation R. L., Milne R. W., Turnidge J. D., Coulthard K. 2005. Evaluation of colistin as an agent against multi-resistant Gram-negative bacteria. Int. J. Antimicrob. Agents 25:11–25 [DOI] [PubMed] [Google Scholar]
- 26. López-Rojas R., et al. 2011. Impaired virulence and in vivo fitness of colistin-resistant Acinetobacter baumannii. J. Infect. Dis. 203:545–548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Lo-Ten-Foe J. R., de Smet A. M., Diederen B. M., Kluytmans J. A., van Keulen P. H. 2007. Comparative evaluation of the VITEK 2, disk diffusion, Etest, broth microdilution, and agar dilution susceptibility testing methods for colistin in clinical isolates, including heteroresistant Enterobacter cloacae and Acinetobacter baumannii strains. Antimicrob. Agents Chemother. 51:3726–3730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. March C., et al. 2010. Dissection of host cell signal transduction during Acinetobacter baumannii-triggered inflammatory response. PLoS One 5:e10033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. McPhee J. B., Lewenza S., Hancock R. E. 2003. Cationic antimicrobial peptides activate a two-component regulatory system, PmrA-PmrB, that regulates resistance to polymyxin B and cationic antimicrobial peptides in Pseudomonas aeruginosa. Mol. Microbiol. 50:205–217 [DOI] [PubMed] [Google Scholar]
- 30. Moffatt J. H., et al. 2010. Colistin resistance in Acinetobacter baumannii is mediated by complete loss of lipopolysaccharide. Antimicrob. Agents Chemother. 54:4971–4977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Moskowitz S. M., Ernst R. K., Miller S. I. 2004. PmrAB, a two-component regulatory system of Pseudomonas aeruginosa that modulates resistance to cationic antimicrobial peptides and addition of aminoarabinose to lipid A. J. Bacteriol. 186:575–579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Murray S. R., Ernst R. K., Bermudes D., Miller S. I., Low K. B. 2007. pmrA confers pmrHFIJKL-dependent EGTA and polymyxin resistance on msbB Salmonella by decorating lipid A with phosphoethanolamine. J. Bacteriol. 189:5161–5169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Ninfa A. J. 1991. Protein phosphorylation and the regulation of cellular processes by the homologous two-component regulatory systems of bacteria. Genet. Eng. 13:39–72 [DOI] [PubMed] [Google Scholar]
- 34. Nummila K., Kilpeläinen I., Zähringer U., Vaara M., Helander I. M. 1995. Lipopolysaccharides of polymyxin B-resistant mutants of Escherichia coli are extensively substituted by 2-aminoethyl pyrophosphate and contain aminoarabinose in lipid A. Mol. Microbiol. 16:271–278 [DOI] [PubMed] [Google Scholar]
- 35. Quinteira S., Grosso F., Ramos H., Peixe L. 2007. Molecular epidemiology of imipenem-resistant Acinetobacter haemolyticus and Acinetobacter baumannii isolates carrying plasmid-mediated OXA-40 from a Portuguese hospital. Antimicrob. Agents Chemother. 51:3465–3466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Rauch P. J., et al. 1996. The expression of the Acinetobacter calcoaceticus recA gene increases in response to DNA damage independently of RecA and of development of competence for natural transformation. Microbiology 142:1025–1032 [DOI] [PubMed] [Google Scholar]
- 37. Seydel U., Oikawa M., Fukase K., Kusumoto S., Brandenburg K. 2000. Intrinsic conformation of lipid A is responsible for agonistic and antagonistic activity. Eur. J. Biochem. 267:3032–3039 [DOI] [PubMed] [Google Scholar]
- 38. Soon R. L., et al. 2011. Different surface charge of colistin-susceptible and -resistant Acinetobacter baumannii cells measured with zeta potential as a function of growth phase and colistin treatment. J. Antimicrob. Chemother. 66:126–133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Soon R. L., Nation R. L., Hartley P. G., Larson I., Li J. 2009. Atomic force microscopy investigation of the morphology and topography of colistin-heteroresistant Acinetobacter baumannii strains as a function of growth phase and in response to colistin treatment. Antimicrob. Agents Chemother. 53:4979–4986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Sun S., Negrea A., Rhen M., Andersson D. I. 2009. Genetic analysis of colistin resistance in Salmonella enterica serovar Typhimurium. Antimicrob. Agents Chemother. 53:2298–2305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Tan C. H., Li J., Nation R. L. 2007. Activity of colistin against heteroresistant Acinetobacter baumannii and emergence of resistance in an in vitro pharmacokinetic/pharmacodynamic model. Antimicrob. Agents Chemother. 51:3413–3415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Tenover F. C., et al. 1995. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J. Clin. Microbiol. 33:2233–2239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Tomomori C., et al. 1999. Solution structure of the homodimeric core domain of Escherichia coli histidine kinase EnvZ. Nat. Struct. Biol. 6:729–734 [DOI] [PubMed] [Google Scholar]
- 44. Trent M. S., Ribeiro A. A., Lin S., Cotter R. J., Raetz C. R. 2001. An inner membrane enzyme in Salmonella and Escherichia coli that transfers 4-amino-4-deoxy-l-arabinose to lipid A: induction on polymyxin-resistant mutants and role of a novel lipid-linked donor. J. Biol. Chem. 276:43122–43131 [DOI] [PubMed] [Google Scholar]
- 45. Tuckerman J. R., Gonzalez G., Gilles-Gonzalez M. A. 2001. Complexation precedes phosphorylation for two-component regulatory system FixL/FixJ of Sinorhizobium meliloti. J. Mol. Biol. 308:449–455 [DOI] [PubMed] [Google Scholar]
- 46. Turton J. F., Gabriel S. N., Valderrey C., Kaufmann M. E., Pitt T. L. 2007. Use of sequence-based typing and multiplex PCR to identify clonal lineages of outbreak strains of Acinetobacter baumannii. Clin. Microbiol. Infect. 13:807–815 [DOI] [PubMed] [Google Scholar]
- 47. Valencia R., et al. 2009. Nosocomial outbreak of infection with pan-drug-resistant Acinetobacter baumannii in a tertiary care university hospital. Infect. Control Hosp. Epidemiol. 30:257–263 [DOI] [PubMed] [Google Scholar]
- 48. Vila J., Pachon J. 2008. Therapeutic options for Acinetobacter baumannii infections. Expert Opin. Pharmacother. 9:587–599 [DOI] [PubMed] [Google Scholar]
- 49. Vinogradov E. V., Bock K., Petersen B. O., Holst O., Brade H. 1997. The structure of the carbohydrate backbone of the lipopolysaccharide from Acinetobacter strain ATCC 17905. Eur. J. Biochem. 243:122–127 [DOI] [PubMed] [Google Scholar]
- 50. Vorachek-Warren M. K., Ramirez S., Cotter R. J., Raetz C. R. 2002. A triple mutant of Escherichia coli lacking secondary acyl chains on lipid A. J. Biol. Chem. 277:14194–14205 [DOI] [PubMed] [Google Scholar]