Abstract
Aminoglycoside-modifying enzymes (AMEs) constitute the most prevalent mechanism of resistance to aminoglycosides by bacteria. We show that aminoglycosides can be doubly modified by the sequential actions of AMEs, with the activity of the second AME in most cases unaffected, decreased, or completely abolished. We demonstrate that the bifunctional enzyme AAC(3)-Ib/AAC(6′)-Ib′ can diacetylate gentamicin. Since single acetylation does not always inactivate the parent drugs completely, two modifications likely provide more-robust inactivation in vivo.
INTRODUCTION
The extensive use of aminoglycosides (AGs) in the treatment of serious bacterial infections has resulted in the emergence of bacterial strains resistant to these broad-spectrum antibiotics (13). The most prevalent mechanism of resistance to AGs is the enzymatic modification of the AGs by three types of AG-modifying enzymes (AMEs): AG acetyltransferases (AACs), AG phosphotransferases (APHs), and AG nucleotidyltransferases (ANTs) (see Fig. 1A). The inactive AG is unable to bind to its target, the bacterial ribosome. Due to their constant quest for survival, bacteria have further evolved bifunctional AMEs, including AAC(3)-Ib/AAC(6′)-Ib′ (9, 15), AAC(6′)/APH(2″)-Ia (2), ANT(3″)-Ii/AAC(6′)-IId (7, 14), and AAC(6′)-30/AAC(6′)-Ib (33). One biological reason for these bifunctional AMEs could be the broadening of the resistance profile through the ability to inactivate a greater variety of AGs, with each enzymatic module targeting its own set of AG substrates [as observed with ANT(3″)-Ii/AAC(6′)-IId (14)]. Another reason could be the inability of a single modification by either subunit to fully inactivate the parent AG, in which case the second modification could be required to abolish the residual activity of the modified AG. We previously reported a methodology for the chemoenzymatic synthesis of 3- and 6′-N-acylated AGs and showed that 6′-N-acetyl-neomycin B (NEO) and 6′-N-n-propionyl-NEO retain antibacterial activity against Bacillus subtilis (12). Here, by using synthetic 6′-N-acetylated AGs, we first confirm that acetylation does not always result in complete inactivation of AGs. It is well known that acylation of AGs can decrease the binding of the AG to the enzyme, as exemplified by the 20-fold and 5-fold increases in the Km for kanamycin A (KAN) compared to amikacin (AMK) with AAC(6′)/APH(2″)-Ia (8) and AAC(3)-IV (16), respectively. We have recently demonstrated that 6′-N-glycinyl-tobramycin and 6‴-N-glycinyl-paromomycin show reduced reaction rates with AAC(3)-IV (22). The combination of these results warrants the investigation of the role that AGs modified at a variety of positions play in AME reactivity.
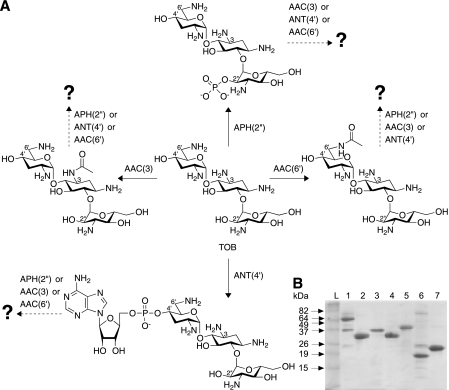
Fig. 1.
(A) Schematic representation of the sequential modifications performed by the AMEs APH(2″), AAC(3), ANT(4′), and AAC(6′), used in this study, with TOB as a representative AG. (B) Coomassie blue-stained 15% Tris-HCl SDS-PAGE gel showing the purified AMEs AAC(6′)/APH(2″)-Ia (56,992 Da) (lane 1), AAC(3)-IV (31,882 Da) (lane 2), APH(2″) (34,791 Da) (lane 3), ANT(4′) (33,299 Da) (lane 4), AAC(3)-Ib/AAC(6′)-Ib′ (37,582 Da) (lane 5), AAC(3)-Ib (19,707 Da) (lane 6), and AAC(6′)-Ib′ (21,344 Da) (lane 7). Lane L, BenchMark prestained protein ladder (Invitrogen). Six micrograms of each protein was loaded onto the gel.
To gain insight into the potential of N-acylated AGs to overcome resistance by AMEs and the potential of bifunctional AMEs to doubly modify AGs, one needs to understand if and how one chemical modification on an AG scaffold affects another, at a different position. Here we report the investigation of such effects by subjecting a variety of AGs (AMK, KAN, NEO, paromomycin [PAR], ribostamycin [RIB], sisomicin [SIS], and tobramycin [TOB]) to sequential alterations by the AMEs AAC(6′)/APH(2″) [used only for its AAC(6′) activity], AAC(3)-IV, ANT(4′), and APH(2″) from the bifunctional enzyme AAC(6′)/APH(2″) (Fig. 1A). We also discuss the study of AAC(3)-Ib/AAC(6′)-Ib′ and its individually expressed components, and we demonstrate, for the first time, the ability of a bifunctional AME to diacetylate an AG, gentamicin (GEN).
MATERIALS AND METHODS
Bacterial strains, plasmids, materials, and instrumentation.
B. subtilis 168 was obtained from the Bacillus Genetic Stock Center (Columbus, OH). The C-terminally His tagged enzyme AAC(6′)/APH(2″)(CHis) (12), the N-terminally His tagged enzyme AAC(3)-IV(NHis) (from the Int-pET19b-pps vector) (12), ANT(4′)(NHis) (21), and APH(2″)(CHis) [amino acids 175 to 479 of the bifunctional AME AAC(6′)/APH(2″)] (22) were purified as described previously. The chemically competent bacteria Escherichia coli TOP10 and E. coli BL21(DE3) were obtained from Invitrogen (Carlsbad, CA). The DNA primers for PCR amplification were obtained from Integrated DNA Technologies (Coralville, IA). The pET28a and pET22b vectors were purchased from Novagen (Gibbstown, NJ). DNA sequencing was done at the University of Michigan DNA Sequencing Core. Reagents for cloning (restriction endonucleases, T4 DNA ligase, and Phusion DNA polymerase) were purchased from New England Biolabs (Ipswich, MA). DTDP (4,4′-dithiodipyridine), acetyl coenzyme A (acetyl-CoA), ATP, GTP, a pyruvate kinase-lactic dehydrogenase mixture (catalog no. P0294), NADH, phosphoenolpyruvate (catalog no. P0564), and AGs (AMK, GEN, KAN, NEO, PAR, RIB, SIS, and TOB) (see Fig. S1 in the supplemental material) were obtained from Sigma-Aldrich and were used without further purification. The UV-visible (UV-Vis) spectrophotometric assays were monitored on a multimode SpectraMax M5 plate reader using flat-bottom 96-well plates (Fisher Scientific). Mass spectrometry (MS) analyses were performed on the Shimadzu LCMS-2019EV mass spectrometer equipped with an LC-20AD liquid chromatograph and an SPD-20AV UV-Vis detector.
Preparation of the pAAC(3)-Ib/AAC(6′)-Ib′-pET28a, pAAC(3)-Ib-pET28a, and pAAC(6′)-Ib′-pET28a overexpression constructs.
The genes encoding AAC(3)-Ib/AAC(6′)-Ib′, AAC(3)-Ib, and AAC(6′)-Ib′ were PCR amplified from the pA3A6 plasmid (9) (kindly provided by Véronique Dubois, Université de Bordeaux) containing the fused AAC(3)-Ib/AAC(6′)-Ib′ gene in the pGEM-T vector. PCRs were carried out using Phusion high-fidelity DNA polymerase with primers identical to those reported previously (15). The amplified genes were inserted into linearized pET28a via the corresponding NdeI- and EcoRI-cut sites. All proteins were expressed following transformation into competent E. coli TOP10 cells. The plasmids were sequenced (University of Michigan DNA Sequencing Core) and showed perfect alignment with the reported sequence (GenBank accession number AF355189.1).
Overproduction and purification of AAC(3)-Ib/AAC(6′)-Ib′(NHis), AAC(3)-Ib(NHis), and AAC(6′)-Ib′(NHis).
The purified plasmids pAAC(3)-Ib/AAC(6′)-Ib′-pET28a(NHis), pAAC(3)-Ib-pET28a(NHis), and pAAC(6′)-Ib′-pET28a(NHis) were transformed into chemically competent E. coli BL21(DE3) cells for protein expression and purification. Luria-Bertani (LB) medium (1 liter) supplemented with kanamycin (50 μg/ml) was inoculated with an overnight culture (10 ml) of the transformants harboring the pAAC(3)-Ib/AAC(6′)-Ib′-pET28a, pAAC(3)-Ib-pET28a, and pAAC(6′)-Ib′-pET28a overexpression constructs. The cultures were grown (at 37°C and 200 rpm) to an absorbance of ∼0.6 at 600 nm, induced with isopropyl-β-thiogalactopyranoside (IPTG) (final concentration, 1.0 mM; 1 ml of a 1 M stock), and then grown (200 rpm) for an additional 16 h at 20°C. Cells were then harvested by centrifugation (at 6,000 rpm for 5 min at 4°C in a Beckman Coulter Avanti JE centrifuge with an F10 rotor) and were resuspended in buffer A (10% glycerol, 200 mM NaCl, and 25 mM Tris, adjusted to pH 8.0 at room temperature [rt]). The resuspended cells were lysed (1 pass at 10,000 to 15,000 lb/in2 in an Avestin EmulsiFlex-C3 high-pressure homogenizer), and the cell debris was removed by centrifugation (at 16,000 rpm for 45 min at 4°C in a Beckman Coulter Avanti JE centrifuge with a JA-17 rotor). Imidazole (final concentration, 2 mM) was added to the supernatant prior to incubation (at 4°C for 2 h with gentle rocking) with Ni-nitrilotriacetic acid (NTA) agarose resin (1.5 ml; Qiagen). The resin was loaded onto a column and was washed with buffer A (10 ml) containing 5 mM imidazole followed by buffer A (10 ml) containing 20 mM imidazole. The desired protein was eluted from the column in a stepwise imidazole gradient (a 10-ml fraction of 20 mM imidazole [1×] and 5-ml fractions of 20 mM [2×], 40 mM [3×], and 250 mM [3×] imidazole). Fractions containing the pure desired proteins, as determined by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), were combined and dialyzed at 4°C for 3 to 12 h against a total of 6 liters of buffer B (10% glycerol, 300 mM NaCl, and 25 mM Tris adjusted to pH 8.0 at rt). All proteins were concentrated using an Amicon Ultra PL-10 centrifugal filter device. Protein concentrations were determined using a NanoDrop spectrometer (Thermo Scientific). Protein yields were 2.4 [AAC(3)-Ib/AAC(6′)-Ib′], 0.8 [AAC(3)-Ib], and 1.8 [AAC(6′)-Ib′] mg/liter of culture. All proteins were flash-frozen using liquid nitrogen and were stored at −80°C.
Determination of MIC values.
MIC values were determined using the double-dilution method against E. coli BL21(DE3) (strain A) and B. subtilis 168 (strain B), using a starting concentration of 125 μg/ml as reported previously (22). The negative and positive controls consisted of omission of the AG or of bacteria, respectively. All experiments were performed in triplicate.
BioTLC assays.
To establish the activity, or lack thereof, of N-acylated AGs, the following bioTLC (thin-layer chromatography) protocols were followed.
(i) BioTLC assay for acylation of GEN.
Enzymatic reactions (10- to 20-μl reaction mixture) were carried out in morpholineethanesulfonic acid (MES) (50 mM; adjusted to pH 5.7 at rt) using AAC(3)-IV (0.5 μM) in the presence of GEN (60 nmol) and acyl-CoA (acetyl coenzyme A or n-propionyl coenzyme A) (80 nmol). The progress of the reactions was monitored by thin-layer chromatography (TLC), and upon completion, reactions were quenched using methanol (MeOH) (10 to 20 μl), and reaction mixtures were kept at −20°C for at least 20 min. After removal of the precipitated protein by centrifugation (at 14,000 rpm for 10 min at rt), the reaction mixture was loaded onto a TLC and was run in MeOH-NH4OH (ratio, 6:1; ∼30% in H2O). B. subtilis was grown overnight at 30°C in LB medium (with no antibiotic). The culture was diluted (100 μl in 10 ml of soft agar [0.75%]-LB medium at 37°C), and the diluted culture was poured over the TLC plate in a sterile petri dish. The plate was then incubated at 30°C (10 h to overnight) until clear zones of inhibition were observed.
(ii) BioTLC assay for confirmation of inactivation of 6′-N-acylated AGs by addition of an acetyl group at position 3.
Enzymatic sequential diacetylation reactions were carried out as described above in MES (50 mM; adjusted to pH 6.6 at rt) by first incubating AAC(6′)/APH(2″) (0.5 μM) with NEO (15 nmol) and acetyl-CoA (60 nmol) for 1 h at 37°C. TLC was used to monitor the progress of the reaction. After the completion of the first acetylation reaction, AAC(3)-IV (0.5 μM) was added, and the mixture was incubated at rt until the reaction was complete (∼1 h) as indicated by TLC. The mixture was treated as described above by using MeOH-NH4OH at a 3:1 ratio as the TLC eluent system. Bacteria were added as described above and were grown overnight prior to visualization for zones of inhibition. The position of ring II of GEN compared to that of ring I could explain the residual activity for 3-N-acetyl-GEN and 3-N-n-propionyl-GEN (see Fig. S2 in the supplemental material).
UV-Vis spectrophotometric assay for determining sequential modifications.
To establish if sequential modifications can be achieved, the assays described below were performed. In all of these sequential assays, the first reaction (100-μl reaction mixture) was followed by addition of the second enzyme in combination with the appropriate indicator and buffer (100 μl) for a total volume of 200 μl. All concentrations reported are the final concentrations in the 200-μl total reaction volume.
(i) AACs followed by AACs.
AGs (50 μM) were first acetylated at 37°C using acetyl-CoA (150 μM) and AAC(6′)/APH(2″) (0.5 μM) in MES buffer (50 mM; pH 6.6). After 45 min of incubation, a mixture of AAC(3)-IV (0.5 μM) and DTDP (2 mM) was added, and the reaction was monitored (at 324 nm and 25°C) using a plate reader by taking measurements every 30 s for 30 min. This procedure was also used to monitor the sequential AAC(3)-IV (25°C) and AAC(6′)/APH(2″) (37°C) reactions.
(ii) APH(2″) followed by AACs.
AGs (50 μM) were first phosphorylated at 37°C using GTP (0.75 mM) (GTP was previously reported to be a cosubstrate of APH, and we found that it yielded better experimental data than ATP [8, 25]) and APH(2″) (1 μM) in HEPES buffer (50 mM; pH 8.0), MgCl2 (5 mM), and KCl (20 mM). After 1 h of incubation, the pH of the solution was lowered to 6.8 by addition of HCl (1.12 μl of a 1 M stock) to accommodate the reactivity of the AAC enzymes. Acetyl-CoA (100 μM) was then added, and the acetylation reactions were initiated by addition of a solution containing AAC(3)-IV (0.25 μM) or AAC(6′)/APH(2″) (0.5 μM) and DTDP (2 mM). The reaction mixtures were incubated at 25°C [AAC(3)-IV] or 37°C [AAC(6′)/APH(2″)], and measurements were taken at 324 nm every 30 s for 30 min.
(iii) ANT(4′) followed by AACs.
AGs (50 μM) were first nucleotidylated at 25°C using ATP (150 μM) and ANT(4′) (1 μM) in HEPES buffer (50 mM; pH 7.5). After 16 h, the nucleotidylation reactions were stopped by acidification to pH 6.8 by the addition of HCl (1.12 μl of a 1 M stock). Acetyl-CoA (150 μM) was then added, and the acetylation reactions were initiated by the addition of a solution containing AAC(3)-IV (0. 5 μM) or AAC(6′)/APH(2″) (0.5 μM) and DTDP (2 mM). The reactions were monitored at 25°C [AAC(3)-IV] or 37°C [AAC(6′)/APH(2″)], and measurements were taken at 324 nm every 30 s for 30 min.
(iv) ANT(4′) followed by APH(2″).
AGs (50 μM) were first nucleotidylated at 25°C using ATP (150 μM) and ANT(4′) (1 μM) in HEPES buffer (50 mM; pH 7.5). After overnight incubation, a mixture of APH(2″) (2.5 μM), NADH (0.5 mg/ml), phosphoenolpyruvate (2.5 mM), GTP (1.25 mM), and the pyruvate kinase-lactic dehydrogenase mixture (5 μl) was added to initiate the reaction. The phosphorylation reaction was monitored at 37°C, and measurements were taken at 340 nm every 30 s for 30 min.
(v) AACs followed by APH(2″).
AGs (100 μM) were first acetylated using acetyl-CoA (150 μM) and AAC(6′)/APH(2″) (0.25 μM) at 37°C or AAC(3)-IV (0.125 μM) at 25°C in HEPES buffer (50 mM; pH 6.8). After 45 min of incubation, the acetylation reactions were stopped by basification to pH 8.0 by addition of KOH (4.2 μl of a 4 M stock). A mixture of APH(2″) (2.5 μM), MgCl2 (10 mM), KCl (40 mM), NADH (0.5 mg/ml), phosphoenolpyruvate (2.5 mM), GTP (1.25 mM), and the pyruvate kinase-lactic dehydrogenase mixture (5 μl) was added to initiate the reaction. The phosphorylation reaction was monitored at 37°C, and measurements were taken at 340 nm every 30 s for 30 min.
Mass spectrometric analyses. (i) AACs followed by AACs.
For the first reaction, a 10-μl solution containing AGs (2 mM), acetyl-CoA (3 mM), and AAC(6′)/APH(2″) (9 μM) [or AAC(3)-IV (22 μM)] in HEPES buffer (50 mM; pH 6.8) was incubated at rt overnight. Then a solution (10 μl) containing acetyl-CoA (3 mM) and AAC(3)-IV (22 μM) [or AAC(6′)/APH(2″) (9 μM)] in HEPES buffer (50 mM; pH 6.8) was added, and the mixture was incubated at rt overnight. Proteins were precipitated by addition of ice-cold methanol (20 μl) and were kept at −20°C for at least 10 min prior to centrifugation (at 13,000 rpm for 10 min). Seventy microliters of H2O was added to 10 μl of the supernatant, and at least 20 μl of the diluted sample was loaded onto a Shimadzu LCMS-2010EV mass spectrometer system for mass analysis using H2O (with 0.1% formic acid) under positive-ionization conditions. The results were in perfect agreement with the spectrophotometric data (see Table S1 in the supplemental material).
(ii) AACs followed by APH(2″).
For the first reaction, a 10-μl solution containing AGs (2 mM), acetyl-CoA (3 mM), and AAC(6′)/APH(2″) (9 μM) [or AAC(3)-IV (22 μM)] in HEPES buffer (50 mM; pH 6.8) was incubated at rt overnight. The pH of the reaction mixture was adjusted to 8.0 by the addition of KOH (1 μl of a 4 M stock) to accommodate the reactivity of APH(2″). After the first incubation, a solution (10 μl) containing GTP (3 mM) and APH(2″) (20 μM) in HEPES buffer (50 mM; pH 8.0) was added, and the mixture was incubated at 37°C overnight. Proteins were precipitated, and samples were analyzed, as described above.
(iii) APH(2″) followed by AACs.
For the first reaction, a 10-μl solution containing AGs (2 mM), GTP (3 mM), and APH(2″) (20 μM) in HEPES buffer (50 mM; pH 8.0) was incubated at rt overnight. The pH of the reaction mixture was adjusted to 6.8 by the addition of HCl (0.1 μl of a 1 M stock) to accommodate the reactivity of the AAC enzymes. After the first incubation, a solution (10 μl) containing acetyl-CoA (3 mM) and AAC(3)-IV (22 μM) [or AAC(6′)/APH(2″) (9 μM)] in HEPES buffer (50 mM; pH 6.8) was added, and the mixture was incubated at rt overnight. Proteins were precipitated, and samples were analyzed, as described above.
(iv) ANT(4′) followed by AACs.
For the first reaction, a 10-μl solution containing AGs (2 mM), ATP (3 mM), and ANT(4′) (4.5 μM) in HEPES buffer (50 mM; pH 7.5) was incubated at rt overnight. Then a solution (10 μl) containing acetyl-CoA (3 mM) and AAC(3)-IV (22 μM) [or AAC(6′)/APH(2″) (9 μM)] in HEPES buffer (50 mM; pH 6.8) was added, and the mixture was incubated at rt overnight. Proteins were precipitated, and samples were analyzed, as described above.
(v) ANT(4′) followed by APH(2″).
For the first reaction, a 10-μl solution containing AGs (2 mM), ATP (3 mM), and ANT(4′) (4.5 μM) in HEPES buffer (50 mM; pH 7.5) was incubated at rt overnight. Then a solution (10 μl) containing GTP (3 mM) and APH(2″) (20 μM) in HEPES buffer (50 mM; pH 8.0) was added, and the mixture was incubated at 37°C overnight. Proteins were precipitated, and samples were analyzed, as described above.
In vivo analyses of AG modifications.
The procedure used was analogous to a method reported previously (1). Bacteria containing AAC(3)-Ib/AAC(6′)-Ib′ (pET82a) were grown (at 37°C overnight) in the presence of GEN (10 μM, which equates to 4.7 μg/ml). The cultures were centrifuged (at 3,500 rpm for 20 min at 4°C), and the spent medium was set aside for analysis. The cell pellets were resuspended in buffer C (150 mM NaCl and 25 mM Tris, adjusted to pH 8.0 at rt) and were boiled for 15 min. The suspension was centrifuged (at 3,500 rpm for 20 min at 4°C) and the lysate collected. The lysate and spent medium were lyophilized separately. The residues were resuspended in MeOH (1 ml) and were centrifuged (at 3,500 rpm for 20 min at 4°C). The solvent was removed by decantation, and the resulting residue was resuspended in an H2O-ethyl acetate (EtOAc)-MeOH mixture (1:1:1) (1.5 ml). After centrifugation (at 3,500 rpm for 20 min at 4°C) and removal of the supernatant, the residue was dissolved in H2O (250 μl) and was analyzed by mass spectrometry using H2O (with 0.1% formic acid) under positive-ionization conditions.
RESULTS
Antimicrobial studies.
To evaluate the effect of acetylation on the antibacterial activities of AGs, the parent AGs KAN and TOB, as well as their synthetic 6′-N-acetylated counterparts, were tested using the double-dilution method against E. coli BL21(DE3) (strain A) and B. subtilis 168 (strain B). MIC values of 1.0 μg/ml (strain A) and 2.0 μg/ml (strain B) were determined for KAN, while MICs of 64 μg/ml (strain A) and 125 μg/ml (strain B) were established for 6′-N-acetyl-KAN. Even though an approximately 60-fold MIC increase was observed in both cases, 6′-N-acetyl-KAN still displayed antibacterial activity. Similar observations were made with TOB and 6′-N-acetyl-TOB, where MICs of 0.5 μg/ml (strain A) and 3.9 μg/ml (strain B) were determined for the parent AG, and MICs of 16 μg/ml (strain A) and 62.5 μg/ml (strain B) were observed for the 6′-N-acetylated drug, representing 32-fold and 16-fold decreases in antibacterial activity, respectively. The 6′-N-acetylated AGs may retain activity due to many factors, including alteration of cellular uptake or cellular metabolism. Modified AGs are compared to their parent compounds for reference without considering such possible alterations. These results strongly suggest that N acetylation alone may not be sufficient to inactivate AGs completely. Using bioTLC, we demonstrated that these 6′-N-acetylated compounds can be completely inactivated by addition of another acetyl moiety by AAC(3)-IV (data not shown).
Alteration of enzymatic activities.
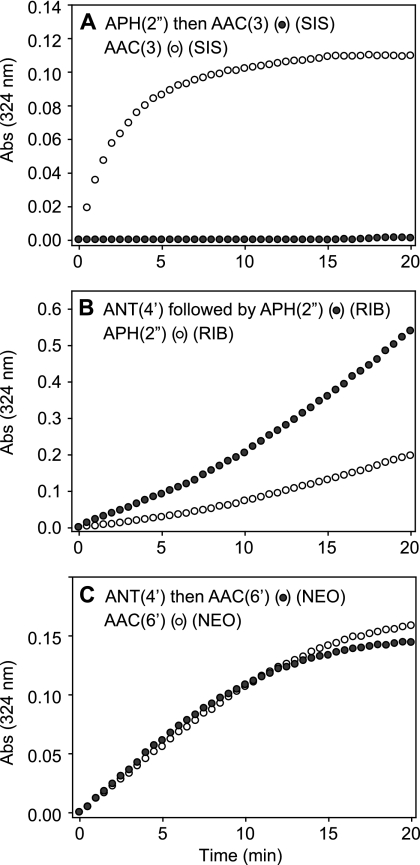
In order to establish the effects of one modification on the second modification, all enzymes were cloned and purified as reported previously (2, 12, 15, 21, 22), except for AAC(3)-Ib/AAC(6′)-Ib′, for which cloning into pET28a instead of pET22b resulted in the production of N-terminally His6 tagged proteins instead of the previously reported untagged proteins (Fig. 1B). The AGs were tested at concentrations of 100 and 50 μM for the first and second AME reactions, respectively. The initial rates of the reactions of the unmodified compounds were then compared to the initial rates of the enzymatic modifications of the singly modified AGs (Fig. 2).
Fig. 2.
Representative graphs comparing the activities of AMEs on an unmodified parent AG (given in parentheses above each graph) (open circles) with the activities of the same AMEs following AG modification (filled circles). The graphs show examples of complete abolition (A), an increase (B), or no change (C) in the activity of the second AME following the action of the first AME. Abs, absorbance.
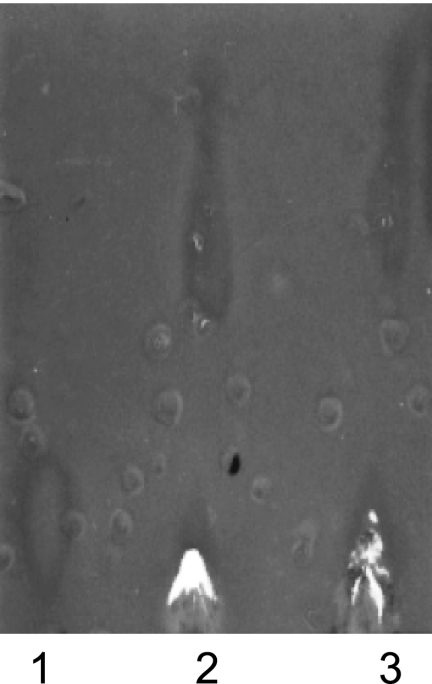
We found that the rates of 3-acetylation of 6′-N-acetylated, 4,6-disubstituted deoxystreptamine (DOS) AGs were significantly lower than those observed for the unmodified drugs (Table 1). No difference in the rates of 3-acetylation was observed between 6′-N-acetylated 4,5-DOS AGs and their unmodified counterparts (Table 2). It should be noted that acetylation at the 6′ position does not occur after acetylation at position 3, consistent with our previous report (12). This suggests that biologically active 3-N-acylated AGs could overcome the resistance caused by AAC(6′) enzymes. When tested using bioTLC, 3-N-acetyl-GEN and 3-N-n-propionyl-GEN both displayed antibacterial activity against B. subtilis (Fig. 3). Based on the available structural information for GEN bound to the bacterial ribosome small-subunit target (3, 6), ring I of GEN inserts into the RNA helix and causes the universally conserved 16S rRNA residues A1492 and A1493 to flip out, resulting in an increase in tRNA binding affinity, which ultimately leads to high error rates in translation (see Fig. S2 in the supplemental material). Compared to the 6-amino moiety, the 3-amino group of GEN is positioned further away from A1492 and A1493, and the addition of the small acetyl and n-propionyl groups at this position is less likely to interfere with the binding of the AG to the target. Therefore, N-3-modified GEN may still be an active antibacterial compound, as indicated by our bioTLC, and the need for a second modification to abolish its activity is a highly plausible scenario. With regard to the sequential activities of AACs and APH(2″), we did not observe 2″-phosphorylation of 6′-N-acetylated or 3-N-acetylated 4,5-DOS AGs. It has been reported previously that the 4,5-DOS AGs are phosphorylated at the 3′ and 3‴ positions in the presence of cofactor and APH(2″) enzymes (8). This suggests that phosphorylation at the 3′ and 3‴ positions of 4,5-DOS AGs is not possible when the AGs are first acetylated at position 6′ or 3. We found that the rates of 3-acetylation were significantly reduced by prior 2″-phosphorylation for all AGs tested. Subsequent to 2″-phosphorylation, the rates of 6′-acetylation were unchanged for KAN and NEO but were reduced for the remaining AGs. Adenylation lowered the rates of 6′- and 3-acetylation, as well as the rates of 2″-phosphorylation, of 4,5-DOS AGs. Interestingly, in the case of 3-acetylation followed by 2″-phosphorylation of 4,6-DOS AGs, a unique increase in the initial rate was observed. Finally, we observed that ANT(4′) was, in almost all cases, inactive on AGs first modified by AACs and APH(2″), with only 6′-N-acetyl-AMK, 6′-N-acetyl-SIS, and 6′-N-acetyl-TOB acting as substrates of ANT(4′) by mass analysis. To confirm the production of the doubly modified AGs by sequential action of AMEs, mass spectrometric analyses were performed (see Table S1 in the supplemental material). The results of mass analyses were in perfect agreement with the spectrophotometric data.
Table 1.
Analysis of sequential modifications of the 4,6-disubstituted DOS AGsa
| First AME | Activity of the following second AMEb: |
||
|---|---|---|---|
| APH(2″) | AAC(3)-IV | AAC(6′) | |
| APH(2″) | − | ↓ | ↓ |
| AAC(3)-IV | ↑ | − | x |
| ANT(4′) | Mix | ↓ | ↓, = |
| AAC(6′) | Mix | ↓ | − |
AMK, KAN, SIS, and TOB.
x, no second modification observed; ↓, decreased activity of the second enzyme; ↑, increased activity of the second enzyme; =, same activity for the second enzyme as for the first; −, reaction not tested; Mix, mixed results (in some cases, the activity of the second enzyme increases, whereas in others cases, it decreases).
Table 2.
Analysis of sequential modifications of the 4,5-disubstituted DOS AGsa
| First AME | Activity of the following second AMEb: |
||
|---|---|---|---|
| APH(2″) | AAC(3)-IV | AAC(6′) | |
| APH(2″) | − | ↓ | ↓, = |
| AAC(3)-IV | x | − | x |
| ANT(4′) | ↓ | ↓ | ↓ |
| AAC(6′) | x | = | − |
NEO, PAR, and RIB.
x, no second modification observed; ↓, decreased activity of the second enzyme; ↑, increased activity of the second enzyme; =, same activity for the second enzyme as for the first; −, reaction not tested.
Fig. 3.
BioTLC of GEN (lane 1), 3-N-acetyl-GEN (lane 2), and 3-N-propionyl-GEN (lane 3), showing antibacterial activity against B. subtilis.
DISCUSSION
In order to rationalize the observed relative effects of modifications at various locations of AG scaffolds on the binding of these singly modified AGs to subsequent resistance enzymes, we closely examined the structures of a variety of AMEs. Nearly a dozen crystal structures of AMEs, including five AACs, six APHs, and one ANT, have been reported (4, 5, 10, 11, 17–20, 23, 26–32). Unfortunately, the structures of the exact AMEs tested on singly modified AGs in this work have not been determined, limiting our rationale to analogous AMEs performing acetylation and phosphorylation of AGs at the 2″, 3, and 6′ positions studied. For 4,5- and 4,6-DOS AGs, we observed that 3-acetylation prevented 6′-acetylation by AAC(6′)/APH(2″). In the structures of a similar enzyme, AAC(6′)-Ib (Protein Data Bank [PDB] structures 2QIR [18] and 2VQY [29]), the 3-amino group of KAN (or PAR) is in close proximity to the catalytic residues in the enzyme active site. Therefore, the addition of an acetyl group at position 3 could disrupt the proper orientation of the AGs at the enzyme active site, resulting in the enzyme's inability to acetylate the 6′ position of the singly modified AGs. For all AGs tested, we observed that the addition of a 4′-AMP group or a 2″-phospho group resulted in either a decrease or no change in the 6′-acetylating activity of AAC(6′)/APH(2″). The AAC(6′)-Ib structures show that the AG 4′-hydroxyl moiety is positioned away from the active site and that the 2″-hydroxyl moiety is environmentally exposed. It is therefore expected that modifications on either the 2" or the 4′ position would not have detrimental effects on the acetylation activity of AAC(6′)/APH(2″), which is in good agreement with our data.
For 4,6-DOS AGs, we observed that the rate of phosphorylation at the 2" position increased as a result of 3- or 6′-acetylation. In the structure of APH(2″)-IIa (PDB structure 3HAM [32]), the 6′-amine of GEN is positioned away from the active site. However, position 3 is in close proximity to acidic residues (Asp or Glu) that are proposed to play a role in the binding of the AG substrates. The addition of an acetyl group at position 3 could bring GEN closer to the catalytic Asp200 [corresponding to Asp192 in APH(2″)-IIa based on amino acid sequence alignment] and closer to the proposed location of the γ-phosphate at ATP, leading to an increase in the observed 2″-phosphorylation rate. The 6′-N-acetylation of GEN has the potential to disrupt the nonpolar stacking interaction with the aromatic ring of Tyr274 [Trp265 in APH(2″)-IIa is found in place of Tyr274 in APH(2″), used in this study], potentially bringing ring III of GEN, bearing the 2″-hydroxyl group to be phosphorylated, closer to the catalytic residues, thereby facilitating the observed increase in the phosphorylation rate. As for AAC(3) activity on singly modified AGs, the lack of availability of a structure of AAC(3) in complex with AG substrates prevented a direct rationale of the results obtained with this enzyme.
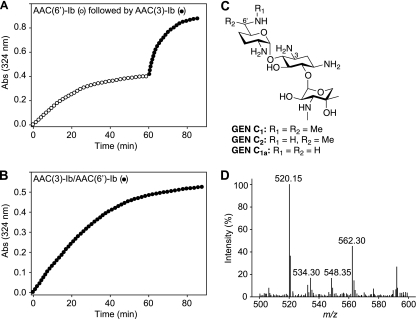
Having established the ability of AMEs to doubly modify AGs by the sequential action of isolated enzymes, we set out to determine if the bifunctional AAC(3)-Ib/AAC(6′)-Ib′ enzyme could diacetylate AGs. By comparing the activities of the individual components of AAC(3)-Ib/AAC(6′)-Ib′ to that of the full-length bifunctional enzyme, we showed by MS that AAC(3)-Ib/AAC(6′)-Ib′ can indeed yield 3,6′-N,N′-diacetyl-GEN (Fig. 4D). Double modification was previously reported for the bifunctional AAC(6′)/APH(2″) enzyme with KAN (1), as well as for the APH(3′)-IIIa enzyme with butirosin, NEO, and RIB (24). The GEN mixture that we used is composed of GEN C1, GEN C2, and GEN C1a (Fig. 4C). Diacetylation of all GEN compounds was observed by MS either by use of the bifunctional enzyme (Fig. 4B) or through the sequential action of the individual enzymes AAC(3)-Ib and AAC(6′)-Ib′ (Fig. 4A). Interestingly, diacetylation of GEN by AAC(3)-Ib/AAC(6′)-Ib′ was pH dependent: diacetylation was observed at pH 7.5 or 8.0, whereas monoacetylation was observed at pHs of <7.0.
Fig. 4.
(A) Spectrophotometric assay demonstrating the diacetylation of GEN via sequential reactions using the individual components AAC(6′)-Ib′ (open circles) and AAC(3)-Ib (filled circles) from the bifunctional enzyme AAC(3)-Ib/AAC(6′)-Ib′. (B) Spectrophotometric assay demonstrating the diacetylation of GEN using the bifunctional enzyme AAC(3)-Ib/AAC(6′)-Ib′ (filled circles). (C) Structures of GEN C1, GEN C2, and GEN C1a. (D) Mass spectra for the reaction of GEN with acetyl-CoA using the bifunctional enzyme AAC(3)-Ib/AAC(6′)-Ib′, showing the production of N-acetyl-GEN C1 (m/z 520.15), N,N′-diacetyl-GEN C1a (m/z 534.30), N,N′-diacetyl-GEN C2 (m/z 548.35), and N,N′-diacetyl-GEN C1 (m/z 562.30). The same masses were observed when AAC(6′)-Ib′ and AAC(3)-Ib were used sequentially.
To corroborate the data from the in vitro diacetylation experiments, we analyzed the spent medium and cell lysate of E. coli BL21(DE3) harboring the AAC(3)-Ib/AAC(6′)-Ib′ gene grown in the presence of GEN (10 μM). The spent medium from these bacteria was analyzed by mass spectrometry. Masses corresponding to diacetylated GEN C2 (m/z 548.40) and diacetylated GEN C1 (m/z 562.25) were found along with those corresponding to GEN C1 (m/z 478.35), GEN C2 (m/z 464.40), GEN C1a (m/z 450.10), and a small peak of phosphorylated GEN C1 (m/z 558.50) from the APH(3″) resistance enzyme contained in the pET28a vector utilized. These data confirm the ability of bifunctional AMEs to doubly modify AGs in vivo so as to confer resistance.
In summary, we have demonstrated that by their sequential actions, AMEs can doubly modify AGs. In many cases, we have observed that after the modification of one position, the ability of AMEs to perform a second modification is unchanged, decreased, or completely abolished. Not only are these observations useful, in that they will help anticipate the effect of a modification on the subsequent activity of AMEs and will guide the design of novel AG antibiotics; they are also promising, in that they indicate the potential of biologically active N-acylated AGs to combat important factors responsible for the ever-growing resistance problem. We have shown that a single acetylation is not always powerful enough to inactivate AGs completely. Even though the reasons for the selection of fused genes rather than the retention of the individual counterparts remain to be defined, we have demonstrated in vitro and in vivo, for the first time, the ability of a bifunctional AME to diacetylate the same AG scaffold at two positions, complementing the previously reported dual modification (acetylation and phosphorylation) of KAN in vitro (1). Guided by these results, efforts toward the development of novel AG antibiotics are under way in our group.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by start-up funds from the Life Sciences Institute and the College of Pharmacy at the University of Michigan, as well as by a grant from the United States-Israel Binational Science Foundation (BSF), Jerusalem, Israel (grant 2008017, to S.G.-T.).
We thank Véronique Dubois (Université de Bordeaux, France), Timor Baasov (Technion, Israel Institute of Technology, Israel), John S. Blanchard (Albert Einstein College of Medicine), and Juan L. Asensio (Instituto de Química Bio-orgánica General [CSIC], Spain) for the generous gifts of the plasmids harboring the genes encoding AAC(3)-Ib/AAC(6′)-Ib′, AAC(6′)/APH(2″)-Ia, AAC(3)-IV, and ANT(4′), respectively, which we used for the cloning of the proteins used in this work. We thank Micha Fridman (Tel Aviv University, Israel) for the generous gift of the synthetic drugs 6′-N-acetyl-KAN and 6′-N-acetyl-TOB. Vanessa R. Porter is acknowledged for providing purified ANT(4′).
Footnotes
Supplemental material for this article may be found at http://aac.asm.org/.
Published ahead of print on 2 May 2011.
REFERENCES
- 1. Azucena E., Grapsas I., Mobashery S. 1997. Properties of a bifunctional bacterial antibiotic resistance enzyme that catalyzes ATP-dependent 2″-phosphorylation and acetyl-CoA-dependent 6′-acetylation of aminoglycosides. J. Am. Chem. Soc. 119:2317–2318 [Google Scholar]
- 2. Boehr D. D., Daigle D. M., Wright G. D. 2004. Domain-domain interactions in the aminoglycoside antibiotic resistance enzyme AAC(6′)-APH(2″). Biochemistry 43:9846–9855 [DOI] [PubMed] [Google Scholar]
- 3. Borovinskaya M. A., et al. 2007. Structural basis for aminoglycoside inhibition of bacterial ribosome recycling. Nat. Struct. Mol. Biol. 14:727–732 [DOI] [PubMed] [Google Scholar]
- 4. Burk D. L., Ghuman N., Wybenga-Groot L. E., Berghuis A. M. 2003. X-ray structure of the AAC(6′)-Ii antibiotic resistance enzyme at 1.8 Å resolution; examination of oligomeric arrangements in GNAT superfamily members. Protein Sci. 12:426–437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Burk D. L., Hon W. C., Leung A. K., Berghuis A. M. 2001. Structural analyses of nucleotide binding to an aminoglycoside phosphotransferase. Biochemistry 40:8756–8764 [DOI] [PubMed] [Google Scholar]
- 6. Carter A. P., et al. 2000. Functional insights from the structure of the 30S ribosomal subunit and its interactions with antibiotics. Nature 407:340–348 [DOI] [PubMed] [Google Scholar]
- 7. Centrón D., Roy P. H. 2002. Presence of a group II intron in a multiresistant Serratia marcescens strain that harbors three integrons and a novel gene fusion. Antimicrob. Agents Chemother. 46:1402–1409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Daigle D. M., Hughes D. W., Wright G. D. 1999. Prodigious substrate specificity of AAC(6′)-APH(2″), an aminoglycoside antibiotic resistance determinant in enterococci and staphylococci. Chem. Biol. 6:99–110 [DOI] [PubMed] [Google Scholar]
- 9. Dubois V., et al. 2002. Molecular characterization of a novel class 1 integron containing blaGES-1 and a fused product of aac(3)-Ib/aac(6[dprime])-Ib″ gene cassettes in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 46:638–645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Fong D. H., Berghuis A. M. 2009. Structural basis of APH(3′)-IIIa-mediated resistance to N1-substituted aminoglycoside antibiotics. Antimicrob. Agents Chemother. 53:3049–3055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Fong D. H., Berghuis A. M. 2002. Substrate promiscuity of an aminoglycoside antibiotic resistance enzyme via target mimicry. EMBO J. 21:2323–2331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Green K. D., Chen W., Houghton J. L., Fridman M., Garneau-Tsodikova S. 2010. Exploring the substrate promiscuity of drug-modifying enzymes for the chemoenzymatic generation of N-acylated aminoglycosides. Chembiochem 11:119–126 [DOI] [PubMed] [Google Scholar]
- 13. Houghton J. L., Green K. D., Chen W., Garneau-Tsodikova S. 2010. The future of aminoglycosides: the end or renaissance? Chembiochem 11:880–902 [DOI] [PubMed] [Google Scholar]
- 14. Kim C., Hesek D., Zajicek J., Vakulenko S. B., Mobashery S. 2006. Characterization of the bifunctional aminoglycoside-modifying enzyme ANT(3″)-Ii/AAC(6′)-IId from Serratia marcescens. Biochemistry 45:8368–8377 [DOI] [PubMed] [Google Scholar]
- 15. Kim C., Villegas-Estrada A., Hesek D., Mobashery S. 2007. Mechanistic characterization of the bifunctional aminoglycoside-modifying enzyme AAC(3)-Ib/AAC(6′)-Ib′ from Pseudomonas aeruginosa. Biochemistry 46:5270–5282 [DOI] [PubMed] [Google Scholar]
- 16. Magalhães M. L., Blanchard J. S. 2005. The kinetic mechanism of AAC3-IV aminoglycoside acetyltransferase from Escherichia coli. Biochemistry 44:16275–16283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Magalhães M. L., et al. 2008. Kinetic and structural analysis of bisubstrate inhibition of the Salmonella enterica aminoglycoside 6′-N-acetyltransferase. Biochemistry 47:579–584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Maurice F., et al. 2008. Enzyme structural plasticity and the emergence of broad-spectrum antibiotic resistance. EMBO Rep. 9:344–349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Nurizzo D., et al. 2003. The crystal structure of aminoglycoside-3′-phosphotransferase-IIa, an enzyme responsible for antibiotic resistance. J. Mol. Biol. 327:491–506 [DOI] [PubMed] [Google Scholar]
- 20. Pedersen L. C., Benning M. M., Holden H. M. 1995. Structural investigation of the antibiotic and ATP-binding sites in kanamycin nucleotidyltransferase. Biochemistry 34:13305–13311 [DOI] [PubMed] [Google Scholar]
- 21. Porter V. R., Green K. D., Zolova O. E., Houghton J. L., Garneau-Tsodikova S. 2010. Dissecting the cosubstrate structure requirements of the Staphylococcus aureus aminoglycoside resistance enzyme ANT(4′). Biochem. Biophys. Res. Commun. 403:85–90 [DOI] [PubMed] [Google Scholar]
- 22. Shaul P., et al. 2011. Assessment of 6′- and 6‴-N-acylation of aminoglycosides as a strategy to overcome bacterial resistance. Org. Biomol. Chem. 9:4057–4063 [DOI] [PubMed] [Google Scholar]
- 23. Stogios P. J., Shakya T., Evdokimova E., Savchenko A., Wright G. D. 2011. Structure and function of APH(4)-Ia, a hygromycin B resistance enzyme. J. Biol. Chem. 286:1966–1975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Thompson P. R., Hughes D. W., Wright G. D. 1996. Regiospecificity of aminoglycoside phosphotransferase from enterococci and staphylococci (APH(3′)-IIIa). Biochemistry 35:8686–8695 [DOI] [PubMed] [Google Scholar]
- 25. Toth M., Chow J. W., Mobashery S., Vakulenko S. B. 2009. Source of phosphate in the enzymic reaction as a point of distinction among aminoglycoside 2″-phosphotransferases. J. Biol. Chem. 284:6690–6696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Toth M., Frase H., Antunes N. T., Smith C. A., Vakulenko S. B. 2010. Crystal structure and kinetic mechanism of aminoglycoside phosphotransferase-2″-IVa. Protein Sci. 19:1565–1576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Vetting M. W., Hegde S. S., Javid-Majd F., Blanchard J. S., Roderick S. L. 2002. Aminoglycoside 2′-N-acetyltransferase from Mycobacterium tuberculosis in complex with coenzyme A and aminoglycoside substrates. Nat. Struct. Biol. 9:653–658 [DOI] [PubMed] [Google Scholar]
- 28. Vetting M. W., Magnet S., Nieves E., Roderick S. L., Blanchard J. S. 2004. A bacterial acetyltransferase capable of regioselective N-acetylation of antibiotics and histones. Chem. Biol. 11:565–573 [DOI] [PubMed] [Google Scholar]
- 29. Vetting M. W., et al. 2008. Mechanistic and structural analysis of aminoglycoside N-acetyltransferase AAC(6′)-Ib and its bifunctional, fluoroquinolone-active AAC(6′)-Ib-cr variant. Biochemistry 47:9825–9835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Wolf E., et al. 1998. Crystal structure of a GCN5-related N-acetyltransferase: Serratia marcescens aminoglycoside 3-N-acetyltransferase. Cell 94:439–449 [DOI] [PubMed] [Google Scholar]
- 31. Wybenga-Groot L. E., Draker K., Wright G. D., Berghuis A. M. 1999. Crystal structure of an aminoglycoside 6′-N-acetyltransferase: defining the GCN5-related N-acetyltransferase superfamily fold. Structure 7:497–507 [DOI] [PubMed] [Google Scholar]
- 32. Young P. G., et al. 2009. The crystal structures of substrate and nucleotide complexes of Enterococcus faecium aminoglycoside-2″-phosphotransferase-IIa [APH(2″)-IIa] provide insights into substrate selectivity in the APH(2″) subfamily. J. Bacteriol. 191:4133–4143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Zhang W., Fisher J. F., Mobashery S. 2009. The bifunctional enzymes of antibiotic resistance. Curr. Opin. Microbiol. 12:505–511 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.