Abstract
The resistance of Acinetobacter baumannii strains to carbapenems is a worrying problem in hospital settings. The main mechanism of carbapenem resistance is the expression of β-lactamases (metalloenzymes or class D enzymes). The mechanisms of the dissemination of these genes among A. baumannii strains are not fully understood. In this study we used two carbapenem-resistant clinical strains of A. baumannii (AbH12O-A2 and AbH12O-CU3) expressing the plasmid-borne blaOXA-24 gene (plasmids pMMA2 and pMMCU3, respectively) to demonstrate that A. baumannii releases outer membrane vesicles (OMVs) during in vitro growth. The use of hybridization studies enabled us to show that these OMVs harbored the blaOXA-24 gene. The incubation of these OMVs with the carbapenem-susceptible A. baumannii ATCC 17978 host strain yielded full resistance to carbapenems. The presence of the original plasmids harboring the blaOXA-24 gene was detected in strain ATCC 17978 after the transformation of OMVs. New OMVs harboring blaOXA-24 were released by A. baumannii ATCC 17978 after it was transformed with the original OMV-mediated plasmids, indicating the universality of the process. We present the first experimental evidence that clinical isolates of A. baumannii may release OMVs as a mechanism of horizontal gene transfer whereby carbapenem resistance genes are delivered to surrounding A. baumannii bacterial isolates.
INTRODUCTION
Acinetobacter baumannii exhibits an impressive panoply of mechanisms of antibiotic resistance (20, 21). The upregulation of innate resistance mechanisms and the acquisition of foreign determinants are observed for these bacteria. The acquisition of antibiotic resistance mediated by gene acquisition is frequently derived from the horizontal transfer of genes between different species in the environment or in clinical settings (19).
Outer membrane vesicles (OMVs) are used by bacteria in a secretion mechanism that leads to the delivery of various bacterial proteins and lipids into host cells, thus eliminating the need for bacterial contact with the host cell (2, 16, 18).
As A. baumannii is broadly distributed in the hospital environment and is a reservoir of antibiotic resistance genes, we wanted to know whether the OMVs released by A. baumannii could be vectors for the spread of antibiotic resistance genes, specifically for carbapenems. As is the case for many other Gram-negative bacteria, A. baumannii produces OMVs. The OMVs are typically spherical, are 50 to 200 nm in diameter, and are composed of outer membrane proteins, lipopolysaccharides (LPSs), periplasmic proteins, phospholipids, DNA, and RNA (1, 16, 27). From an energetic point of view, the production of these structures is so costly that it is difficult to consider the release of OMVs as a purposeless process. These vesicles support a common function, as they are a means by which bacteria interact with prokaryotic and eukaryotic cells in their environment (16). OMVs have an important role in several bacterial activities as carriers for quorum-sensing molecules (18), toxin delivery (8, 26), the inhibition of the maturation of phagosomes in macrophages (11), and the formation of biofilms (28). The results of previous studies suggested that vesicles may be involved in the transfer of genetic material among similar bacterial species (6, 9, 14, 15). Yaron et al. previously demonstrated the transfer of virulence genes between Escherichia coli and other enteric bacteria and also demonstrated that intravesicle DNA was protected from DNase digestion, suggesting that DNA is packaged within vesicles (27).
On the other hand, carbapenem resistance in A. baumannii is due mainly to the presence of β-lactamases (class B metallo-β-lactamases or class D OXA-type β-lactamases) as well as altered permeability and penicillin binding protein (PBP) modifications (21). The process by which β-lactamase genes are transferred cell to cell has not been studied in detail.
In the present study, we demonstrated for the first time that OMVs are vectors of plasmids carrying carbapenem resistance genes and that they may act by transferring a functional blaOXA-24 carbapenem resistance gene between different strains of A. baumannii.
MATERIALS AND METHODS
Bacterial strains and antibiotic susceptibility testing.
A. baumannii clinical strains AbH12O-A2 and AbH12O-CU3 harboring plasmids pMMA2 and pMMCU3, respectively, and both carrying the blaOXA-24 gene (4, 19) were used. These clinical strains were isolated during a large nosocomial outbreak in Madrid, Spain (19), and showed a broad antibiotic resistance profile, including resistance to carbapenems. The strains were identified to the species level by nucleotide sequencing of the 16S rRNA genes. Antibiotic susceptibility testing was performed by an Etest (bioMérieux) according to the manufacturer's instructions.
A. baumannii ATCC 17978 (24, 25) was used as a host for transformation experiments. A. baumannii strain ATCC 17978 is fully susceptible to carbapenems and produces biofilms moderately.
Purification of OMVs.
Outer membrane vesicles (OMVs) were isolated from exponential-growth-phase cultures of A. baumannii clinical strains AbH12O-A2 and AbH12O-CU3 and from A. baumannii strain ATCC 17978. In brief, 500 ml of Müeller-Hinton (MH) broth was inoculated with 5 ml of a culture grown overnight and was incubated at 37°C overnight at 150 rpm. The cells were pelleted by centrifugation (14,000 × g for 10 min), and the supernatant was filtered through a 0.22-μm membrane (Millipore Corporation, Bedford, MA) and subjected to ultracentrifugation (200,000 × g for 90 min at 4°C with a 70 Ti rotor [Beckman]). The vesicle pellet was resuspended in phosphate-buffered saline (PBS) (pH 7.4). The suspension was filtered again through a 0.22-μm membrane (Millexgp) and spread onto agar plates to test for any bacterial growth.
Electron microscopy.
The vesicle suspension was fixed with 2.5% cold glutaraldehyde in 0.2 M sodium cacodylate buffer (pH 7.4) for 2 h at 4°C and postfixed with 1% osmium tetroxide in 0.1 M sodium cacodylate buffer (pH 7.4) for 1 h at 4°C. The sample was observed and photographed with a Jeol JEM 1010 transmission electron microscope (80 kV).
Determination of DNA, proteins, and lipopolysaccharides in OMVs.
OMV proteins were quantified by a Bradford assay (5). For the quantification of the intravesicle DNA, OMV samples (88 μl) were treated with DNase (Invitrogen) to hydrolyze surface-associated DNA and free DNA in suspension. Reactions were stopped by the heat treatment of the mixtures for 10 min at 65°C. DNase-treated vesicles were then lysed with 0.125% Triton X-100 solution for 30 min at 37°C, and DNA was purified by the use of a High Pure PCR product purification kit (Roche). The DNA was quantified by using a Nanodrop ND-1000 instrument. The ratio of DNA to μg of protein vesicle was determined (Table 1). The presence of lipopolysaccharides (LPSs) in the outer membrane vesicle samples was tested by using the ToxinSensor Chromogenic LAL endotoxin assay kit (GenScript) according to the manufacturer's instructions.
Table 1.
DNA contents associated with outer membrane vesicles from different isolates of Acinetobacter baumannii
| Source | Species | Plasmid | μg of vesicle protein/μl | ng of DNA/μg of vesicle protein |
|---|---|---|---|---|
| AbH12O-A2 | A. baumannii | pMMA2 | 0.35 | 24.2 |
| AbH12O-CU3 | A. baumannii | pMMCU3 | 0.16 | 146.5 |
OMV-mediated transformation.
Transformation experiments were performed according to a method described previously by Yaron et al. (27). In brief, A. baumannii strain ATCC 17978 was cultured in MH broth at 37°C at 150 rpm for 3 to 4 h to an optical density at 600 nm (OD600) of 0.4. Cells were pelleted and resuspended in cold SOC medium (2% tryptone, 0.5% yeast extract, 0.4% glucose, 10 mM NaCl, 2.5 mM KCl, 5 mM MgCl2+, 5 mM MgSO4) at 107 CFU/ml. This cell suspension (60 μl) was mixed with different amounts of purified vesicles (containing 1, 5, 10, and 20 μg of protein) obtained from A. baumannii clinical strains H12O-A2 and H12O-CU3 (Table 1). The suspensions were incubated statically for 4 h at 37°C and then for another 4 h with shaking (150 rpm). Ten milliliters of MH broth was then added to the suspension, and the incubation continued at 37°C overnight with shaking (150 rpm). The broth culture grown overnight was pelleted at 10,000 × g and resuspended in 100 μl of MH broth. A. baumannii ATCC 17978 transformants were selected on LB agar plates with 500 μg/ml of ampicillin. Negative controls included a transformation experiment performed under identical conditions but with approximately the same amount of free plasmid DNA instead of DNA packed into 20 μg of OMV protein (1,000 ng and 2,300 ng of free plasmid of pMMA2 and pMMCU3, respectively). All experiments were carried out in duplicate and repeated twice. To further demonstrate that DNA transfer was vesicle mediated, separate experiments were conducted with 20 μg of OMVs previously lysed with Triton X-100 for 30 min at 37°C, as previously described (27), and with OMVs previously treated with 50 μg of proteinase K (Sigma)/ml for 30 min at 37°C and with 2 U of DNase I (Invitrogen Life Technologies) according to the manufacturer's instructions. Proteinase K was used to hydrolyze phage coats and release phage contents. It was then removed from treated samples by using microconcentrators that exclude molecules with an Mr of <30,000 (Amicon Ultra-0.5; Millipore). DNase was added to treated vesicle preparations to degrade released free DNA.
To demonstrate that OMV concentrations normally found in culture supernatants are able to mediate plasmid transfer, additional experiments were performed by using the supernatant obtained from a fresh bacterial culture of clinical strain AbH12O-A2 at the stationary phase of growth in the presence of 100 μg/ml of ampicillin. The cells were pelleted by centrifugation (4,000 rpm for 15 min), and the supernatant was filtered through a 0.22-μm membrane (Millipore Corporation, Bedford, MA). Twenty milliliters of supernatant was inoculated with 2 ml of a culture of A. baumannii ATCC 17978 grown overnight. Next, after incubation at 3, 6, 24, and 36 h, 200 μl was plated onto LB agar plates supplemented with 500 μg/ml of ampicillin. Controls included the supernatant treated with Triton X-100, DNase, and free plasmid DNA (in a concentration similar to that for OMVs).
Dot blotting.
The blaOXA-24 gene was used as a probe for hybridization procedures. The DNA probe was made by amplifying the blaOXA-24 gene from clinical strain AbH12O-A2 with the primers described in Table 2. The amplified fragment was labeled with digoxigenin (DIG)-11-dUTP (Roche Applied Science) at dilutions of 1/3 for DIG-UTP and 2/3 for TTP. Excess DIG-dUTP was removed with a Sephadex G-50 column (Roche).
Table 2.
Oligonucleotides used in this work
| Primer | Sequence (5′-3′) | Use(s) |
|---|---|---|
| REP Forw | TTTGCGCCGTCATCAGGC | REP-PCR |
| REP Rev | ACGTCTTATCAGGCCTAC | REP-PCR |
| OXA24 Forw | AAAACTAAATCTGAAGATAATTTT | Dot blot and plasmid PCR fingerprinting |
| OXA24 Rev | GAGCGAAAAGGGGATTTTT | Plasmid PCR fingerprinting |
| IS4 Forw | GCATTAGTTTCATGAATTGC | Plasmid PCR fingerprinting |
| IS4 Rev | GATCAGAAAGTCTATGCCC | Plasmid PCR fingerprinting |
| IS4 Forw bis | CACTCGCTCTATGCATCTA | Plasmid PCR fingerprinting |
The DNA from the OMVs was denatured for 10 min at 96°C and cooled on ice before dot blotting onto a Hybond-N+ membrane (Amersham Bioscience). The membrane was cross-linked by UV irradiation at 254 nm and 120 J/cm2 in a Biolink BLX 254 UV cross-linker (Vilbert Lourmat). The membrane was prehybridized in a solution containing 7% SDS, 1 mM EDTA, and 0.25 M disodium phosphate for 30 min at 60°C. The oligonucleotide digoxigenin probe was then added to the solution and incubated overnight at 60°C. The membrane was then washed twice with 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) plus 1% (wt/vol) SDS for 15 min at room temperature and twice with 0.1× SSC plus 0.1% (wt/vol) SDS for 30 min at 60°C and was then blocked for 30 min with 1% (wt/vol) blocking reagent and incubated for 30 min at 37°C with an anti-DIG-alkaline phosphatase conjugate (Roche diagnostic) diluted 1:10,000 in blocking solution. The unbound antibody conjugate was removed by washing three times with 100 ml of washing buffer containing 1× SSC and 1% (wt/vol) SDS. The membrane was placed between two cellulose acetate sheets, and 0.5 ml/100 cm2 of CSPD [disodium 3-(4-methoxyspiro{1,2-dioxetane-3,2′-(5′-chloro)tricyclo[3,3.1.13.7]decane}-4-yl)phenyl phosphate] was added to the membrane, between the acetate sheet sandwiches. The air bubbles were removed carefully, the acetate sheets were sealed, and the membrane was incubated for 5 min at room temperature. Finally, the membrane was exposed to Kodak XAR-5 X-ray film for 60 min.
REP-PCR.
The primers used to perform repetitive extragenic palindromic PCR (REP-PCR) are described in Table 2. The procedure was performed as previously reported (3). Two strains were considered to belong the same genotype when they shared the same pattern of bands, and a maximum difference of two bands was considered acceptable.
Plasmid analysis.
PCR assays were performed to confirm the presence of plasmids pMMA2 and pMMCU3 in the A. baumannii ATCC 17978 transformants. The oligonucleotides were designed from the sequences of pMMA2 and pMMCU3 deposited in the GenBank database (accession numbers GQ377752 and GQ904227, respectively). The sequences of the oligonucleotides are described in Table 2. The PCR was performed under the following conditions: a denaturation step for 10 min at 94°C; an amplification step consisting of 30 cycles of 1 min at 94°C, 1 min at 52°C, and 4 min at 72°C; and an elongation step for 16 min at 72°C. The samples were visualized on a 0.5% agarose gel.
RESULTS
OMVs harboring the blaOXA-24 gene are released from Acinetobacter baumannii cells.
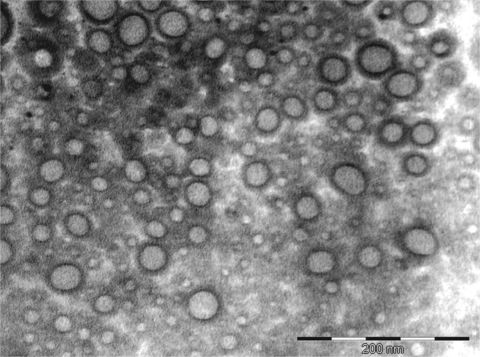
Transmission electron microscopy analysis revealed outer membrane vesicles (OMVs) from A. baumannii cells collected at the exponential phase of growth, thus suggesting that this bacterium actively releases OMVs during growth in broth culture (Fig. 1). The vesicles had an intact bilayer membrane, contained electron-dense material, and ranged in diameter from 20 to 100 nm (with vesicles with a diameter of 40 nm being the most abundant). The vesicles were uniform and spherical and did not have flagella. Moreover, the presence of LPS was detected in the OMVs (data not shown), confirming that they were derived from outer membrane. In all assays, the LPS concentration obtained from OMV samples was out of the measurement range (>2 units of endotoxin/ml). The OMVs were well purified, since no bacteria were visualized by electron microscopy, and contamination controls on culture plates did not show any growth.
Fig. 1.
Electron microscopy micrograph of OMVs released by A. baumannii clinical strain AbH12O-A2. Vesicles were purified from broth cultures by ultracentrifugation and filtered through a 0.22-μm filter. The average diameter of the vesicles was 40 nm. The OMVs were free of contaminating bacteria.
Transformation experiments.
Colonies were obtained on LB agar plates at each dose of purified OMVs when A. baumannii ATCC 17978 was transformed after incubation with OMVs purified from both A. baumannii clinical strains (Table 3 ). The amplification of blaOXA-24 from all transformants was achieved by PCR (data not shown).
Table 3.
OMV-mediated transformants obtained after 24 h of incubation with 10 μg of OMVs in plates of LB agar plates supplemented with 500 μg/ml of ampicillin
| Treatment | No. of transformants | % of transformantsd |
|---|---|---|
| ATCC 17978 + OMVs | 440 | 100 |
| ATCC 17978 + OMVs treated with DNase and proteinase Ka | 316 | 72 |
| ATCC 17978 + OMVs treated with Triton X-100a | 0 | |
| ATCC 17978 + pMMA2b | 0 | |
| ATCC 17978c | 0 |
See Materials and Methods.
Free plasmid at concentrations similar to those found in OMVs.
Negative control, A. baumannii ATCC 17978 incubated alone.
Data are the mean values from triplicate experiments.
A. baumannii clinical isolates AbH12O-A2 and AbH12O-CU3 showed a broad profile of β-lactam resistance, including resistance to carbapenems (Table 4). A. baumannii ATCC 17978 transformed with OMVs from clinical strains AbH12O-A2 and AbH12O-CU3 showed a profile of MIC values compatible with the expression of blaOXA-24 (4), thus indicating resistance to penicillin, a slight increase in the cefepime MIC, and a high degree of resistance to carbapenems. In all cases, imipenem, meropenem, and doripenem MICs for A. baumannii isolate ATCC 17978 increased from 0.38, 0.75, and 0.19 μg/ml, respectively, to >32 μg/ml after transformation with OMVs (Table 4).
Table 4.
Antibiotic susceptibility profiles of A. baumannii clinical strains AbH12O-A2 and AbH12O-CU3, the host strain A. baumannii ATCC 17978, and A. baumannii ATCC 17978 transformed with OMVs of the indicated clinical strains
| Antibiotic | MIC (μg/ml) |
||||
|---|---|---|---|---|---|
| AbH12O-A2 (pMMA2) | AbH12O-CU3 (pMMCU3) | ATCC 17978 | ATCC 17978a (OMV-pMMA2) | ATCC 17978a (OMV-pMMCU3) | |
| Amoxicillin | >256 | >256 | 96 | >256 | >256 |
| Amoxicillin-clavulanic acid | >256 | >256 | 48 | >256 | >256 |
| Piperacillin-tazobactam | >256 | >256 | 2 | >256 | >256 |
| Cefotaxime | >256 | >256 | 32 | 32 | 32 |
| Ceftazidime | >256 | >256 | 12 | 12 | 12 |
| Cefepime | >256 | >256 | 6 | 16 | 16 |
| Amikacin | >256 | >256 | 3 | 4 | 3 |
| Tobramycin | 12 | 4 | 0.5 | 0.5 | 0.5 |
| Gentamicin | >256 | >256 | 0.5 | 0.5 | 0.5 |
| Imipenem | >32 | >32 | 0.38 | >32 | >32 |
| Meropenem | >32 | >32 | 0.75 | >32 | >32 |
| Doripenem | >32 | >32 | 0.19 | >32 | >32 |
ATCC 17978 transformed with the OMVs from A. baumannii AbH12O-A2 and AbH12O-CU3 and harboring plasmids pMMA2 and pMMCU3, respectively.
In parallel, no transformants were obtained when A. baumannii ATCC 17978 was incubated with 1,000 ng and 2,300 ng of free naked plasmid obtained from clinical isolates AbH12O-A2 and AbH12O-CU3, respectively (pMMA2 and pMMCU3, respectively), or with the same amount of OMVs previously lysed with Triton X-100 for 30 min at 37°C (negative controls). Almost the same numbers of transformants were obtained when the OMVs were previously treated with 50 μg of proteinase K/ml and with 2 U of DNase I (Table 3). It is important that in the process of the inactivation of DNase at 65°C, damage to the integrity of OMVs occurs, thus reducing the transformation efficiency (data not shown). Overall, OMV-mediated A. baumannii transformation is strongly dependent on the integrity of the OMVs protecting the plasmid DNA inside OMVs.
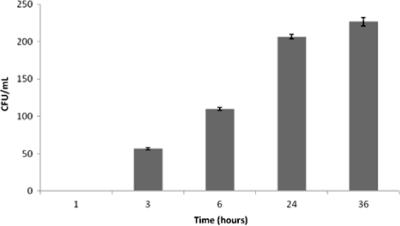
Moreover, and in order to demonstrate whether supernatants of bacterial cultures are responsible for OMV-mediated transformation, a time-response experiment was performed by using culture supernatants of AbH12O-A2 cells as a source of OMVs. Interestingly, in these cases, several transformants were obtained (Fig. 2). This revealed that OMVs under more physiological conditions are released from cells and are mobilized by transformation into the surrounding bacterial cells. The process of transformation through OMVs is detected within 3 h of incubation and increases to 24 h, then reaching a plateau. Longer incubation times did not significantly increase the number of transformants (Fig. 2).
Fig. 2.
Time-response experiment with supernatants from AbH12OA2 as a source of OMVs. Shown are numbers of colonies resistant to ampicillin (transformants) obtained at different times of incubation of A. baumannii ATCC 17978 with supernatant from AbH12OA2. At least three replicate experiments were performed, and each experiment had results similar to those shown. Error bars indicate the standard deviations for replicate samples.
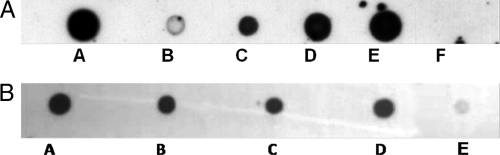
To demonstrate that the OMVs carried the pMMA2 and pMMCU3 clinical plasmids harboring blaOXA-24, a dot blot experiment was performed with the blaOXA-24 gene as a probe. The OMVs collected from A. baumannii clinical strains AbH12O-A2 and AbH12O-CU3 hybridized with the blaOXA-24 probe are shown in Fig. 3 A. These results strongly suggest that OMVs released from the A. baumannii clinical strains harbored plasmids carrying the blaOXA-24 carbapenemase gene.
Fig. 3.
(A) Dose-response experiment. Shown is a dot blot analysis for detecting the presence of the blaOXA-24 gene in OMVs released from A. baumannii clinical isolate AbH12O-A2. Blot A, 10 μl of purified plasmid pMMA2 as a positive control; blot B, 10 μl of the OMV suspension (see concentration in Table 1); blot C, 50 μl of the OMV suspension; blot D, 100 μl of the OMV suspension; blot E, 200 μl of the OMV suspension; blot F, 100 μl of the OMV suspension from A. baumannii ATCC 17978 mock transformed as a negative control (equal amount of OMVs with respect to that in blot E). All samples were concentrated to a final volume of 10 μl before the experiment, and 5 μl of each sample was analyzed by dot blotting. (B) Dot blot analysis to detect the presence of the blaOXA-24 gene in OMVs released from A. baumannii clinical isolates AbH12O-A2 and AbH12O-CU3 (blots A and B, respectively) and from A. baumannii ATCC 17978 transformed with OMVs from clinic isolates AbH12O-A2 and AbH12O-CU3 (blots C and D, respectively). The mock-transformed A. baumannii strain ATCC 17978 (blot E) was used as a negative control. Equal amounts of OMVs of each sample were analyzed in the dot blots.
To demonstrate the universality of the process, A. baumannii strain ATCC 17978 cells transformed with OMVs from clinic isolates AbH12O-A2 and AbH12O-CU3 were also analyzed by dot blot analysis. In the case of clinical strains AbH12O-A2 and AbH12O-CU3, the ATCC isolate transformed with OMVs carrying clinical plasmids pMMA2 and pMMCU3 released OMVs harboring the blaOXA-24 gene (Fig. 3B).
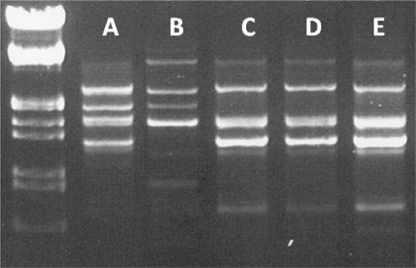
REP-PCR and plasmid analysis.
Although the purified OMVs used for the vesicle transformation experiments were free of contaminating bacterial cells (original clinical isolates AbH12O-A2 and AbH12O-CU3), A. baumannii ATCC 17978 cells transformed with the OMV-mediated clinical plasmids were genotypically characterized by REP-PCR (Fig. 4). There was no difference in the band profiles of amplified fragments between A. baumannii strain ATCC 17978 and the strains obtained after OMV transformation, thus ruling out the presence of the original clinical strains on the plates after OMV-mediated transformation experiments. To support this conclusion, OMVs purified directly from clinical strains AbH12O-A2 and AbH12O-CU3 were inoculated onto LB broth and LB agar plates. No bacterial growth was detected after 24 or 48 h of incubation, thus indicating that there were no viable microorganisms present in the purified OMVs.
Fig. 4.
REP-PCR profile of the A. baumannii strains used in this study. Blot A, AbH12O-A2; blot B, AbH12O-CU3; blot C, ATCC 17978; blot D, ATCC 17978 transformed with OMVs from AbH12O-A2; blot E, ATCC 17978 transformed with OMVs from AbH12O-CU3.
Since A. baumannii ATCC 17978 naturally carries two plasmids, pAB1 and pAB2, of 11,302 and 13,408 bp, respectively (24), the interpretation of the restriction fragment length polymorphism (RFLP) profile after OMV-mediated plasmid transformation is difficult. Therefore, OMV-mediated plasmid transfer between clinical isolates and ATCC 17978 was confirmed by PCR fingerprinting of plasmids pMMA2 and pMCU3 by using a set of oligonucleotides (Table 2).
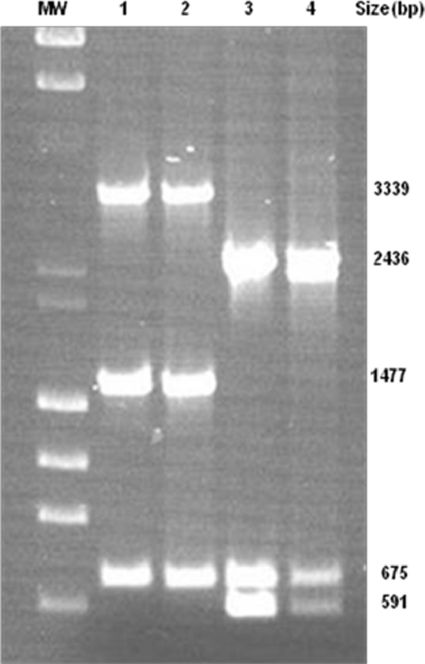
For plasmid pMMA2, oligonucleotides IS4 forw bis and IS4 rev were used to amplify the 3,339-bp band, IS4 forw and IS4 rev were used to amplify the 1,477-bp band, and OXA24 forw and OXA24 rev were used to amplify the 651-bp band. For pMMCU3, oligonucleotide pairs IS4 forw bis and IS4 rev, OXA24 forw and OXA24 rev, and IS4 forw and IS4 rev were used to amplify bands of 2,436 bp, 675 bp, and 591 bp, respectively (Fig. 5). Data confirm the horizontal transfer of plasmids between clinical isolates and the host strain ATCC 17978.
Fig. 5.
PCR fingerprinting profile of plasmids pMMA2 and pMCU3, determined by using a set of oligonucleotides. Lane 1, PCR fingerprint of plasmids isolated from strain AbH12O-A2; lane 2, PCR fingerprint of plasmids isolated from strain ATCC 17978 transformed with OMVs from AbH12O-A2; lane 3, PCR fingerprint of plasmids isolated from strain AbH12O-CU3; lane 4, PCR fingerprint of plasmids isolated from strain ATCC 17978 transformed with OMVs from AbH12O-CU3 as a source. MW, λ DNA-HindIII and φ174 DNA-HaeIII mix.
DISCUSSION
Both pathogenic and nonpathogenic species of Gram-negative bacilli secrete vesicles. Studies of these bacterial outer membrane vesicles support a common role for the vesicles as the means by which bacteria interact with prokaryotic and eukaryotic cells in their environment (16, 18). OMV-mediated toxin delivery is a potent virulence mechanism exhibited by different Gram-negative microorganisms; i.e., proteases and leukotoxin from Actinobacillus actinomycetemcomitans, Shiga toxin from Shiga toxin-producing E. coli (STEC), VacA from Helicobacter pylori, and multiple virulence factors from Pseudomonas aeruginosa, including β-lactamase, alkaline phosphatase, hemolytic phospholipase C, and Cif (a recently characterized toxin secreted by Pseudomonas aeruginosa), have been observed to be delivered over long distances via OMVs (2). The mechanism is therefore an effective means of delivering bacterial virulence factors.
However, as well as lipids and proteins, double-stranded DNA has been recovered from the membrane vesicles of the ruminal cellulolytic Ruminococcus genus (14), suggesting that this membrane-associated transformation system may play a role in lateral gene transfer in complex microbial ecosystems such as the rumen. In addition, the vesicle-mediated transfer of virulence genes from E. coli O157:H7 to other enteric bacteria has been reported, suggesting that OMVs may act as a transfer system for the delivery of genetic material to other microorganisms (27). Moreover, there are no reports of the importance of this DNA transfer system with regard to having any clinical impact on the transfer of carbapenem resistance genes or affecting human microbiota.
DNA can be exchanged between bacteria by conjugation, transduction, or transformation (10). During transformation, DNA is taken up directly by cells. The bacteria from which the DNA was taken are called donors, and the bacteria to which the DNA has been added are called recipients. Bacteria that have taken up DNA are called transformants. In the current concept of the transformation process, bacterial donors may need to be lysed in advance to release the free DNA in order for it to be taken up by the recipient. Here we report data showing that A. baumannii strains release OMVs able to carry the blaOXA-24 carbapenem resistance gene without a loss of viability, thus constituting a possible way of disseminating plasmids with antibiotic resistance genes of clinical impact.
It is important that (i) OMVs are produced by growing cells and are not a product of cell lysis or cell death (10), (ii) pathogenic strains can produce between 10 and >25 times more vesicles than nonpathogenic strains (10), (iii) P. aeruginosa PAO1 cells grown under oxygen stress conditions demonstrate an increase in vesicle formation (23), and (iv) vesicles from both intracellular and extracellular bacterial pathogens have been identified in diverse infected host tissues, revealing the ability of vesicles to access a variety of environments within the host (12).
For the above-mentioned reasons, it may be hypothesized that OMVs are important not only for the virulence of a specific pathogen in a process of infection or host interaction but also because during this process, the release of OMVs may favor the spread of antibiotic resistance genes (including genes for resistance to carbapenems) to surrounding bacteria. This is supported by the fact that the DNA inside OMVs is somehow protected from the action of nucleases (as in the present study), which may be present in the environment or in the host tissues, thus favoring the exchange of genetic material or the process of the horizontal transfer of DNA, thus probably conferring an additional advantage for the process of gene dissemination to those microorganisms able to release OMVs.
The overall data presented here may add some novelty to the notion of horizontal DNA transfer, at least between strains of A. baumannii that may release “OMV-protected DNA” while remaining viable. This process appears to occur continuously, since once a bacterial strain is transformed with OMVs (i.e., A. baumannii isolate ATCC 17978), this bacterial isolate is also able to release OMVs harboring the plasmid that was previously taken up (Fig. 3).
To explain the presence of DNA inside OMVs, we propose two possible mechanisms. (i) OMVs contain proteins and lipids from the outer membrane and periplasm which are absent from the inner membrane and cytoplasm, as described previously by Kuehn and Kesty (16). This suggests that the plasmids have to migrate in some way to the periplasm, where they are trapped in OMVs. (ii) OMVs also have some proteins from the inner membrane, as described previously by Kwon et al. (17). Proteomic analysis of the OMVs from clinical strain H12O-A2, revealed a typical profile for the outer membrane (for example, porins), but we also detected inner membrane proteins (at least 13% of OMV proteins), e.g., tolerance to colicins E2, E, A, and K, required for outer membrane integrity; membrane-bound lytic murein transglycosylase B; an ABC-type transport system; a succinate dehydrogenase flavo subunit; the FoF1 ATP synthase beta-subunit; l-sorbosone dehydrogenase; and others (data not shown). These results suggest that some OMVs could contain both inner and outer membrane compounds, trapping cytoplasmic compounds and even plasmids, which was demonstrated previously for other species (13).
Moreover, the process of DNA transformation mediated by OMVs seems to be a dose-independent process, since increasing the amount of OMVs does not increase the number of transformant colonies. Otherwise, and importantly, the OMV concentration found in culture supernatants is enough to mediate DNA transformation, making this process time dependent (Fig. 2). To explain this paradoxical result, it may be suggested that only a small proportion of the population of OMVs contain plasmids and that both types of OMVs (with and without plasmids) will compete to enter the bacterial host cell through specific points, or “hot spots,” where OMVs merge, releasing the plasmid. This may explain that increasing the amount of OMV will also increase the proportion of OMVs without plasmids, and therefore, the rate of transformation efficiency will not be increased despite the increase of the number of OMVs. However, it seems clear that transformation through OMVs is a time-dependent process, as shown in Fig. 2, where the effectiveness of the process increases over time, reaching a plateau at approximately 24 h of incubation.
It is interesting that both plasmids pMMA2 and pMMCU3 lack the Mob-like plasmid mobilization protein (GenBank accession numbers GQ377752 and GQ904227, respectively), suggesting that OMVs act as a likely alternative mechanism for mobilization plasmids lacking this protein, such as those harboring the blaOXA-24 carbapenemase gene that we present here.
Carbapenems are so far the therapeutic alternatives of choice for the treatment of infections caused by A. baumannii. Considering that some nationwide multicenter studies from Spain have revealed that up to 43% of A. baumannii isolates are resistant to carbapenems (7) and that all 43% of these isolates carried blaOXA-24 and 20% carried blaOXA-58 (22), the notion that OMVs released by this microorganism may harbor plasmid-mediated carbapenemases is worrisome and emphasizes the need to adhere to strict infection control measures once such carbapenemase-producing A. baumannii isolates are detected in the hospital environment.
In summary, we present here experimental evidence showing that OMVs can spread carbapenem resistance genes in A. baumannii strains. This would be a new mechanism of dissemination of antibiotic resistance genes in addition to the previously known conjugation, transformation, and transduction processes.
ACKNOWLEDGMENTS
We thank Catalina Sueiro and Ada Castro for their kind help with microscopic techniques.
This work was supported by SERGAS grants (PS07/51 and PS07/90) and INCITE grant 08CSA064916PR from Xunta de Galicia, by Spanish Network for Research in Infectious Diseases grants RD06/0008/0011 and RD06/0008/0025, and by grants PI081613 and PS09/00687 from the Instituto de Salud Carlos III. Margarita Poza and Carlos Rumbo are recipients of an Isidro Parga Pondal research contract (Xunta de Galicia) and a scholarship from the Instituto de Salud Carlos III, respectively. Nelson C. Soares is a recipient of an Angeles Alvariño research contract (Xunta de Galicia). Jose A. Mendez is supported by a Río Hortega research contract from the Instituto de Salud Carlos III.
Footnotes
Published ahead of print on 25 April 2011.
REFERENCES
- 1. Beveridge T. J. 1999. Structures of gram-negative cell walls and their derived membrane vesicles. J. Bacteriol. 181:4725–4733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bomberger J. M., et al. 2009. Long-distance delivery of bacterial virulence factors by Pseudomonas aeruginosa outer membrane vesicles. PLoS Pathog. 5:e1000382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bou G., Cervero G., Dominguez M. A., Quereda C., Martinez-Beltran J. 2000. PCR-based DNA fingerprinting (REP-PCR, AP-PCR) and pulsed-field gel electrophoresis characterization of a nosocomial outbreak caused by imipenem- and meropenem-resistant Acinetobacter baumannii. Clin. Microbiol. Infect. 6:635–643 [DOI] [PubMed] [Google Scholar]
- 4. Bou G., Oliver A., Martinez-Beltran J. 2000. OXA-24, a novel class D beta-lactamase with carbapenemase activity in an Acinetobacter baumannii clinical strain. Antimicrob. Agents Chemother. 44:1556–1561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bradford M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248–254 [DOI] [PubMed] [Google Scholar]
- 6. Ciofu O., Beveridge T. J., Kadurugamuwa J., Walther-Rasmussen J., Hoiby N. 2000. Chromosomal beta-lactamase is packaged into membrane vesicles and secreted from Pseudomonas aeruginosa. J. Antimicrob. Chemother. 45:9–13 [DOI] [PubMed] [Google Scholar]
- 7. Cisneros J. M., et al. 2005. Risk-factors for the acquisition of imipenem-resistant Acinetobacter baumannii in Spain: a nationwide study. Clin. Microbiol. Infect. 11:874–879 [DOI] [PubMed] [Google Scholar]
- 8. Davis J. M., Carvalho H. M., Rasmussen S. B., O'Brien A. D. 2006. Cytotoxic necrotizing factor type 1 delivered by outer membrane vesicles of uropathogenic Escherichia coli attenuates polymorphonuclear leukocyte antimicrobial activity and chemotaxis. Infect. Immun. 74:4401–4408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Dorward D. W., Garon C. F., Judd R. C. 1989. Export and intercellular transfer of DNA via membrane blebs of Neisseria gonorrhoeae. J. Bacteriol. 171:2499–2505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Dubnau D. 1999. DNA uptake in bacteria. Annu. Rev. Microbiol. 53:217–244 [DOI] [PubMed] [Google Scholar]
- 11. Fernandez-Moreira E., Helbig J. H., Swanson M. S. 2006. Membrane vesicles shed by Legionella pneumophila inhibit fusion of phagosomes with lysosomes. Infect. Immun. 74:3285–3295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Fiocca R., et al. 1999. Release of Helicobacter pylori vacuolating cytotoxin by both a specific secretion pathway and budding of outer membrane vesicles. Uptake of released toxin and vesicles by gastric epithelium. J. Pathol. 188:220–226 [DOI] [PubMed] [Google Scholar]
- 13. Kadurugamuwa J. L., Beveridge T. J. 1995. Virulence factors are released from Pseudomonas aeruginosa in association with membrane vesicles during normal growth and exposure to gentamicin: a novel mechanism of enzyme secretion. J. Bacteriol. 177:3998–4008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Klieve A. V., et al. 2005. Naturally occurring DNA transfer system associated with membrane vesicles in cellulolytic Ruminococcus spp. of ruminal origin. Appl. Environ. Microbiol. 71:4248–4253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kolling G. L., Matthews K. R. 1999. Export of virulence genes and Shiga toxin by membrane vesicles of Escherichia coli O157:H7. Appl. Environ. Microbiol. 65:1843–1848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kuehn M. J., Kesty N. C. 2005. Bacterial outer membrane vesicles and the host-pathogen interaction. Genes Dev. 19:2645–2655 [DOI] [PubMed] [Google Scholar]
- 17. Kwon S. O., Gho Y. S., Lee J. C., Kim S. I. 2009. Proteome analysis of outer membrane vesicles from a clinical Acinetobacter baumannii isolate. FEMS Microbiol. Lett. 297:150–156 [DOI] [PubMed] [Google Scholar]
- 18. Mashburn L. M., Whiteley M. 2005. Membrane vesicles traffic signals and facilitate group activities in a prokaryote. Nature 437:422–425 [DOI] [PubMed] [Google Scholar]
- 19. Merino M., et al. 2010. OXA-24 carbapenemase gene flanked by XerC/XerD-like recombination sites in different plasmids from different Acinetobacter species isolated during a nosocomial outbreak. Antimicrob. Agents Chemother. 54:2724–2727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Peleg A. Y., Seifert H., Paterson D. L. 2008. Acinetobacter baumannii: emergence of a successful pathogen. Clin. Microbiol. Rev. 21:538–582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Poirel L., Nordmann P. 2006. Carbapenem resistance in Acinetobacter baumannii: mechanisms and epidemiology. Clin. Microbiol. Infect. 12:826–836 [DOI] [PubMed] [Google Scholar]
- 22. Ruiz M., Marti S., Fernandez-Cuenca F., Pascual A., Vila J. 2007. High prevalence of carbapenem-hydrolysing oxacillinases in epidemiologically related and unrelated Acinetobacter baumannii clinical isolates in Spain. Clin. Microbiol. Infect. 13:1192–1198 [DOI] [PubMed] [Google Scholar]
- 23. Sabra W., Lunsdorf H., Zeng A. P. 2003. Alterations in the formation of lipopolysaccharide and membrane vesicles on the surface of Pseudomonas aeruginosa PAO1 under oxygen stress conditions. Microbiology 149:2789–2795 [DOI] [PubMed] [Google Scholar]
- 24. Smith M. G., et al. 2007. New insights into Acinetobacter baumannii pathogenesis revealed by high-density pyrosequencing and transposon mutagenesis. Genes Dev. 21:601–614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Soares N. C., et al. 2009. 2-DE analysis indicates that Acinetobacter baumannii displays a robust and versatile metabolism. Proteome Sci. 7:37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Wai S. N., et al. 2003. Vesicle-mediated export and assembly of pore-forming oligomers of the enterobacterial ClyA cytotoxin. Cell 115:25–35 [DOI] [PubMed] [Google Scholar]
- 27. Yaron S., Kolling G. L., Simon L., Matthews K. R. 2000. Vesicle-mediated transfer of virulence genes from Escherichia coli O157:H7 to other enteric bacteria. Appl. Environ. Microbiol. 66:4414–4420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Yonezawa H., et al. 2009. Outer membrane vesicles of Helicobacter pylori TK1402 are involved in biofilm formation. BMC Microbiol. 9:197. [DOI] [PMC free article] [PubMed] [Google Scholar]