Abstract
3-O-(3′,3′-Dimethylsuccinyl) betulinic acid (DSB), also known as PA-457, bevirimat (BVM), or MPC-4326, is a novel HIV-1 maturation inhibitor. Unlike protease inhibitors, BVM blocks the cleavage of the Gag capsid precursor (CA-SP1) to mature capsid (CA) protein, resulting in the release of immature, noninfectious viral particles. Despite the novel mechanism of action and initial progress made in small-scale clinical trials, further development of bevirimat has encountered unexpected challenges, because patients whose viruses contain genetic polymorphisms in the Gag SP1 (positions 6 to 8) protein do not generally respond well to BVM treatment. To better define the role of amino acid residues in the HIV-1 Gag SP1 protein that are involved in natural polymorphisms to confer resistance to the HIV-1 maturation inhibitor BVM, a series of Gag SP1 chimeras involving BVM-sensitive (subtype B) and BVM-resistant (subtype C) viruses was generated and characterized for sensitivity to BVM. We show that SP1 residue 7 of the Gag protein is a primary determinant of SP1 polymorphism-associated drug resistance to BVM.
INTRODUCTION
3-O-(3′,3′-Dimethylsuccinyl) betulinic acid (DSB), also known as bevirimat (BVM), is a potent inhibitor of HIV-1 maturation (7, 8, 20, 21). BVM targets the HIV-1 Gag CA-SP1 boundary region by blocking viral protease cleavage of SP1 from the CA-SP1 precursor and inhibiting release of the mature CA protein, which is the final step required for virion maturation. Despite the novel mechanism of action and initial progress made in small-scale clinical trials (11–13, 17), further development of BVM has encountered unexpected challenges in the clinical setting, because patients whose viruses contain genetic polymorphisms in the Gag SP1 (positions 6 to 8) protein do not respond well to BVM treatment (3, 10, 14, 16). These three residues (glutamine-valine-threonine [QVT]) are referred to as the SP1 polymorphism motif. Interestingly, these naturally occurring mutations that confer intrinsic resistance to BVM were not identified by in vitro drug resistance selection experiments and are not located in the region immediately flanking the CA-SP1 cleavage site (2, 7, 8). Extensive in vitro drug selection experiments identified six amino acid changes (proximal to the CA-SP1 cleavage site) that independently confer BVM resistance (2). Three substitutions were located at the 1st and 3rd residues of SP1 (A1V, A3V, and A3T), and three substitutions were identified at the extreme C terminus of CA (H226Y, L231M, and L231F).
There are extensive data on HIV-1 subtype B viruses and their representative molecular clone pNL4-3 concerning BVM's mechanism of action and antiviral activity (2, 4, 7, 8, 20, 21). In contrast, HIV-1 non-B subtypes and derived molecular clones have received less attention, despite knowledge that these non-B subtypes are responsible for nearly 90% of the current worldwide HIV-1 pandemic (6, 19). Additionally, we noted that non-B subtypes exhibit more frequent changes in the identified SP1 polymorphism motif than B subtype viruses do. These non-B viruses therefore provide important systems for studying the impact of these naturally occurring polymorphisms on BVM antiviral activity and the mechanism of BVM action. The aims of the present study were to examine the influence of SP1 polymorphisms on BVM-mediated blockade of CA-SP1 processing (a hallmark of BVM activity) in HIV-1 subtype C and to determine which position(s) within the SP1 polymorphism motif is integral in conferring resistance to BVM in HIV-1.
MATERIALS AND METHODS
Construction of HIV-1 DNA constructs.
To investigate the influence of SP1 polymorphisms on BVM-mediated antiviral activity in subtype C viruses, we generated two chimeras (Fig. 1 B), designated NL/MJ4 and NL/1084i, that contained a representative CA-SP1-NC segment (containing the C-terminal 126 residues of CA, complete SP1, and the N-terminal 29 residues of NC) from corresponding BVM-resistant subtype C clone MJ4 (NIH AIDS Research and Reference Reagent Program) or 1084i (a gift of Ruth M. Ruprecht) in the context of the BVM-sensitive NL4-3 clone (NIH AIDS Research and Reference Reagent Program). Chimeras were made by cloning the SpeI/ApaI double-digested PCR-amplified CA-SP1-NC fragment of MJ4 or 1084i into NL4-3 which had been digested completely with SpeI/ApaI. These two chimeras, together with NL4-3, were then used as templates to carry out extensive mutagenesis to generate panels of single and double substitution mutations as shown in Fig. 2 and Fig. 3, respectively. All DNA mutagenesis was carried out using the PCR–overlapping-PCR strategy and other standard molecular cloning approaches. Each clone was sequenced to confirm the sequence of the inserted Gag CA-SP1-NC segment.
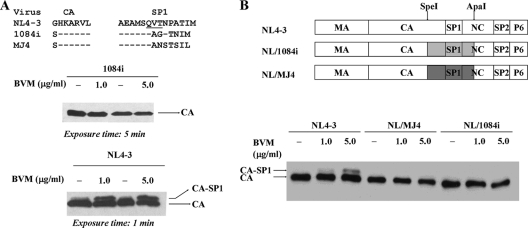
Fig. 1.
Western blot analysis of virion-associated Gag derived from COS-1 cells transfected with proviral DNA of NL4-3, 1084i, or their derivatives in the absence or presence of BVM. The top diagram in panel A shows the amino acid sequences of the CA-SP1 boundary domain for subtype B NL4-3 and the two subtype C viruses (1084i and MJ4). Gag SP1 residues 6 to 8 (glutamine-valine-threonine [QVT]), termed the SP1 polymorphism motif, are underlined. The residues previously identified as conferring BVM resistance in vitro are shown in bold (2). Dashes represent residues that are identical to the parental NL4-3 sequence. The top diagram in panel B shows the panel of chimeric Gag constructs used in this study. NL4-3 domains are in white, while 1084i and MJ4 domains are represented in gray. Two restriction enzymes (SpeI and ApaI) were used for cloning and are indicated schematically by arrows at their corresponding locations in the Gag domains. A previously published protocol was used to address the effect of BVM on Gag CA-SP1 processing (8). COS-1 cells were transfected with proviral NL4-3, 1084i, or their derivatives and cultured in the absence or presence of the indicated concentrations of BVM. BVM concentrations in μg/ml can be converted to molar concentrations by using the equation 1 μg/ml = 1.71 μM. At 2 days posttransfection, equal volumes of supernatant from each culture were collected and then concentrated through a 20% sucrose cushion in a microcentrifuge at 4°C for 120 min at 13,000 rpm. Viral pellets were resuspended in lysis buffer (150 mM Tris-HCl, 5% Triton X-100, 1% deoxycholate, 0.1% sodium dodecyl sulfate, pH 8.0). Viral proteins were separated by 12% SDS-PAGE and transferred to a nitrocellulose membrane (Pierce). Following transfer, membranes were blocked in PBS containing 0.5% Tween and 5% dry milk powder. The membrane was then incubated with HIV-Ig (obtained through the NIH AIDS Research and Reference Reagent Program, Division of AIDS, NIAID) and hybridized with goat anti-human–horseradish peroxidase (Abcam). The immune complex was visualized using an ECL system (Pierce) according to the instructions provided by the manufacturer. The positions of virally encoded proteins CA (p24) and CA-SP1 (CA-SP1) are indicated. Note the accumulation of CA-SP1 in the presence of BVM in NL4-3 but not in 1084i or strains expressing chimeric Gag between NL4-3 and MJ4 (NL/MJ4) or NL4-3 and 1084i (NL/1094i). Due to high protein expression levels by NL4-3, a gel with a 1-min exposure time is shown. The data presented in this figure are representative of at least four independent experiments.
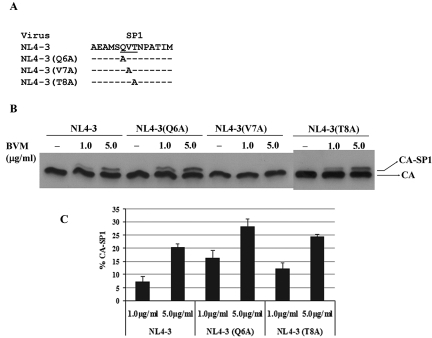
Fig. 2.
Western blot analysis of the effect of mutations within the SP1 polymorphism motif on BVM's activity in inhibiting the conversion of the capsid precursor, CA-SP1, to mature capsid protein. (A) Mutations introduced into the SP1 polymorphism motif (underlined). Dashes represent residues that are identical to the parental NL4-3 sequence. COS-1 cells were transfected with both parental HIV-1 pNL4-3 and alanine substitution mutant proviral DNAs as described in the legend to Fig. 1. BVM at the indicated concentrations or DMSO (no-drug control) was maintained throughout the period of the culture. For BVM, 1 μg/ml = 1.71 μM. (B) SDS-PAGE and Western blot analysis of viral proteins derived from these cultures were performed as described in the legend to Fig. 1. (C) Protein bands were analyzed by ImageJ software (http://rsbweb.nih.gov/ij/) to quantify the percentage of CA-SP1 relative to total CA-SP1 plus CA. All data shown are means and standard deviations for three independent experiments performed in duplicate.
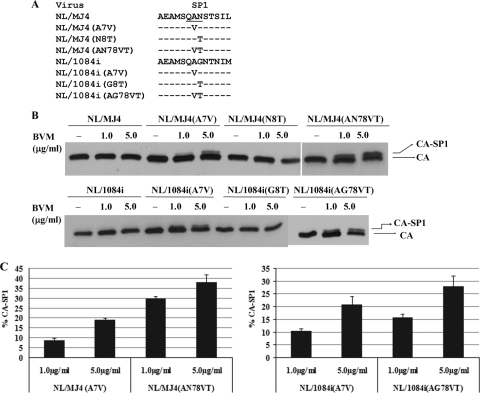
Fig. 3.
Importance of SP1 residues 7 and 8 in the modulation of BVM activity in inhibiting the conversion of the capsid precursor, CA-SP1, to mature capsid protein. (A) Panel of Gag SP1 chimeras constructed between NL4-3 and MJ4 or between NL4-3 and 1084i and containing various mutations targeting the SP1 polymorphism motif (underlined). Dashes represent residues that are identical to the parental virus (NL/MJ4 or NL/1084i) sequence. (B) COS-1 cells were transfected with the indicated mutant proviral DNAs as described in the legend to Fig. 1. BVM or DMSO (no-drug control) was maintained throughout the period of the culture, and SDS-PAGE and Western blot analysis of viral proteins obtained from the cultures were performed as described in the legend to Fig. 1. (C) Protein bands were analyzed by ImageJ software (http://rsbweb.nih.gov/ij/) to quantify the percentage of CA-SP1 relative to total CA-SP1 plus CA. All data shown are means and standard deviations for three independent experiments performed in duplicate.
SDS-PAGE and Western blotting.
To address the effects of SP1 polymorphisms on the ability of BVM to inhibit CA-SP1 processing, COS-1 cells were transfected with parental HIV-1 pNL4-3 and various mutant proviral DNAs, as indicated. Transfection was performed by using TransIT-LT1 (Mirus, Madison, WI) following the procedure outlined by the manufacturer. BVM at a concentration of 1 μg/ml or 5 μg/ml or dimethyl sulfoxide (DMSO) (no-drug control) was maintained throughout the period of the culture, and SDS-PAGE and Western blotting to analyze viral proteins derived from these cultures were performed as described previously (7, 8).
Briefly, at 48 h posttransfection, culture medium containing viral particles was collected and clarified by centrifugation at 4°C for 20 min at 2,000 rpm, using a Sorvall RT 6000B centrifuge. Virus-containing supernatants were then concentrated through a 20% sucrose cushion in a microcentrifuge at 4°C for 120 min at 13,000 rpm, and pellets were resuspended in lysis buffer (150 mM Tris-HCl, 5% Triton X-100, 1% deoxycholate, 0.1% SDS, pH 8.0). Viral proteins were then separated by 12% SDS-PAGE and transferred to a nitrocellulose membrane (Invitrogen), followed by blocking in phosphate-buffered saline (PBS) containing 0.5% Tween and 5% dry milk powder. The membrane was incubated with pooled immunoglobulin from HIV-1-infected patients (HIV-Ig) (NIH AIDS Research and Reference Reagent Program) and hybridized with goat anti-human–horseradish peroxidase (Sigma). The immune complex was visualized using an ECL system (Pierce, Rockford, IL) according to the instructions provided by the manufacturer. Protein bands were analyzed by ImageJ software (http://rsbweb.nih.gov/ij/) to quantify the percentage of CA-SP1 relative to total CA-SP1 plus CA. The same exposure time was used to develop films.
Single-cycle infectivity assay.
We used vesicular stomatitis virus glycoprotein (VSV-G)-pseudotyped HIV-1 particles coupled with the TZM-bl indicator cell line (NIH AIDS Research and Reference Reagent Program) to measure the 50% effective concentrations (EC50s) of BVM for the above chimeras. An HIV-1 Env-defective reporter clone (NIH AIDS Research and Reference Reagent Program) that is widely used for the phenotypic analysis of HIV-1 drug resistance was used in our study. This clone, designated pNL4-3-ΔENV-EGFP, is derived from the HIV-1 proviral clone pNL4-3 but encodes a form of enhanced green fluorescent protein (EGFP) in the Env open reading frame. The SpeI/ApaI cloning strategy described above was used to introduce CA-SP1-NC segments from NL4-3 and NL/MJ4 chimeras into pNL4-3-ΔENV-EGFP.
VSV-G-pseudotyped HIV-1 stocks were produced in COS-1 cells treated with or without BVM as described previously. Briefly, COS-1 cells were cotransfected with wild-type (WT) NL4-3 or derived chimeras and VSV-G by use of TransIT-LT1 (Mirus) in the presence or absence of BVM. Four 10-fold serial dilutions (0.01, 0.1, 1.0, and 10 μg/ml) were tested in triplicate. Forty-eight hours after transfection, supernatants containing VSV-G-pseudotyped HIV-1 were collected. Cell debris was removed from the supernatant by spinning at 450 × g for 5 min and filtering through filters (Millipore). Viral supernatants were then used for infection of TZM-bl indicator cells in a 96-well flat-bottom plate at 3 × 103 cells/well. Infections were carried out for 2 h in the presence of 10 μg/ml DEAE-dextran, and luciferase activity was then measured at 48 h postinfection, using a luciferase assay system (Promega). Linear regression was used to determine the EC50s of BVM.
RESULTS AND DISCUSSION
For this study, we utilized two well-characterized subtype C molecular clones (5, 15), termed 1084i and MJ4, which have SP1 polymorphisms at 7 and 8 positions, respectively, but contain no known drug resistance-conferring mutations identified previously in the subtype B NL4-3 clone. Because similar SP1 polymorphism patterns were witnessed for MJ4 and 1084i (Fig. 1A), we opted to use subtype C clone p1084i in our initial experiment. We employed a Western blot-based HIV-1 Gag processing assay previously developed by our group to assess the BVM susceptibility of these non-subtype B viruses (8). The Gag processing experiment specifically measures the effect of BVM on CA-SP1 cleavage and the ability of the compound to inhibit the release of mature CA protein. A strong correlation between the Gag processing experiment and the cell-based replication assay has been established in characterizing BVM activity against HIV-1 (8). For Gag processing experiments, COS-1 cells were transfected with the parental or SP1 variant proviral DNA in the presence of BVM or a DMSO control. At 48 h posttransfection, virus was harvested from the cell culture supernatant, and the Gag processing profile for each virus was analyzed and compared to Gag processing exhibited in the absence of compound (DMSO control).
NL4-3 CA-SP1 (p25) processing in virions was significantly blocked by BVM, similar to observations in our previous studies (Fig. 1A) (7, 8). In contrast, inhibition of CA-SP1 processing was not observed in BVM-treated 1084i virions, indicating that this subtype C virus was resistant to BVM. Because 1084i virus itself lacks any previously identified drug resistance-conferring mutations, it is likely that mutations within the SP1 polymorphism motif were responsible for the observed BVM resistance, as demonstrated previously for subtype B variants (3, 14, 18). We noticed variability in viral particle production between subtype C and subtype B viruses that may affect our assay readout. To minimize the variability and ensure that we had a comparable virus particle production system for analysis, we generated two chimeras, designated NL/MJ4 and NL/1084i, that contained a representative CA-SP1-NC segment (containing a portion of C-terminal CA, complete SP1, and a portion of N-terminal NC) from corresponding BVM-resistant viruses in the context of the NL4-3 backbone. The observation that these Gag CA-SP1 chimeras, like their parental clones, exhibited reduced BVM sensitivity supported the utilization of these chimeras for further study (Fig. 1B).
Next, two approaches, examining the loss or gain of function, were pursued in parallel to determine the effect of the mutations on BVM activity in Gag CA-SP1 chimeras. The percentage of CA-SP1 relative to total CA-SP1 plus CA observed in BVM-treated virions was calculated and used to determine differential susceptibility to BVM treatment. The first approach (loss of function) involved the creation of alanine-scanning mutations across the SP1 polymorphism motif (QVT), and the resulting mutants were characterized for sensitivity to BVM in the Gag processing experiment. In agreement with a recent study (3), only SP1 residue seven (SP1-V7), not the other two residues, was shown to be critical for determining BVM antiviral activity, as the presence of a V7A mutation sufficiently conferred BVM resistance in Gag processing experiments, while Q6A and V8A mutations had no apparent effect on BVM activity (Fig. 2). These data provide further evidence that genetic polymorphisms at the seventh position of SP1 are particularly detrimental to BVM-mediated antiviral activity.
Our second approach involved the generation of a panel of either single or multiple substitution mutants targeting SP1 residues 7 and 8 in which the SP1 residues from the BVM-sensitive NL4-3 virus were inserted into the analogous positions of the SP1 proteins of the BVM-resistant chimeras (Fig. 3). These mutants were then characterized for sensitivity to BVM in studies that measured the effect of the compound on CA-SP1 cleavage. Gain-of-function studies were used to define the specific residues responsible for drug resistance. As summarized in Fig. 3, these experiments revealed a striking similarity in compound sensitivity rendered by SP1 residues derived from BVM-sensitive NL4-3 between the two BVM-resistant NL4-3/non-subtype B chimeras. For example, the A7V substitution conferred to both Gag CA-SP1 chimeras (NL/1084i and NL/MJ4) approximately 100% of the BVM activity observed in NL4-3, as well as a similar extent of CA-SP1 precursor accumulation in the presence of 1 μg/ml BVM (∼7% CA-SP1 accumulation) (cf. Fig. 2 and 3) or 5 μg/ml BVM (∼20% CA-SP1 accumulation) (cf. Fig. 2 and 3). Although the threonine substitution at SP1 residue 8 alone was insufficient to confer drug sensitivity in our assays, double replacement of SP1 residues 7 and 8 by VT residues significantly enhanced the sensitivity of these two NL4-3/C chimeras (NL/1084i and NL/MJ4) to BVM (showing an approximate 2-fold increase) (Fig. 3C), indicating a synergetic effect of the two positions in regulating BVM activity.
The Gag processing experiments specifically measured the effect of BVM on CA-SP1 cleavage and the ability of the compound to inhibit the release of mature CA protein. To further characterize the effect of BVM on these chimeras, we employed a single-cycle infectivity assay involving VSV-G-pseudotyped HIV-1 particles and the TZM-bl indicator cell line. As summarized in Table 1, the results of this assay correlated well with those obtained in the Gag processing experiment. For example, parental EC50s of BVM were consistently observed with NL4-3(Q6A), NL4-3(T8A), and NL/MJ4(AN78VT), which exhibited WT-like BVM sensitivity in the Gag processing experiment. Additionally, three mutants, NL4-3(V7A), NL/MJ4, and NL/MJ4(N8T), identified as BVM resistant in the Gag processing experiment, lacked compound sensitivity in the assay, with EC50s of >17.1 μM. The NL/MJ4(A7V) mutant was unique in that it exhibited intermediate sensitivity to compound treatment, with an EC50 of around 3.261 ± 1.739 μM. Interestingly, NL/MJ4(AN78VT), with an additional substitution at SP1 residue 8, exhibited a WT-like EC50. This result further strengthens our previous interpretation of a synergetic effect of SP1 residues 7 and 8 in regulating BVM activity.
Table 1.
In vitro activity of BVM against different viruses in single-cycle infectivity assay
| Virus | BVM EC50 (μM)a |
|---|---|
| NL4-3 | 0.039 ± 0.005 |
| NL4-3(Q6A) | 0.031 ± 0.009 |
| NL4-3(V7A) | >17.1b |
| NL4-3(T8A) | 0.038 ± 0.009 |
| NL/MJ4 | >17.1b |
| NL/MJ4(A7V) | 3.261 ± 1.739 |
| NL/MJ4(N8T) | >17.1b |
| NL/MJ4(AN78VT) | 0.125 ± 0.053 |
Antiviral activity was determined after infection of TZM-bl indicator cells with VSV-G-pseudotyped HIV-1 as described in Materials and Methods. Values represent means ± standard deviations for two independent experiments performed in triplicate.
The highest drug concentration tested in this study.
As summarized in Table 2, we performed a detailed analysis of the prevalence of SP1 polymorphisms in both HIV-1 subtype C viruses (n = 1,174) and HIV-1 subtype B viruses (n = 2,111). The SP1 position 7-associated polymorphism was predominant in subtype C viruses and less so in subtype B viruses. For example, the V7A mutation (conferring BVM resistance) was present in 70.3% of subtype C viruses, while the same mutation was identified in only 16.8% of subtype B viruses. Thus, the naturally occurring SP1-V7A polymorphism, particularly in HIV-1 subtype C viruses, constitutes a significant obstacle to clinical testing of BVM, as viruses with this polymorphism display robust BVM resistance.
Table 2.
Prevalence of amino acid residues in Gag SP1 QVT motif in HIV-1 subtype B and C virusesa
| HIV-1 subtype (n) | Amino acid residue (prevalence [%]) in Gag SP1 QVT motifb |
||
|---|---|---|---|
| 6 (Q) | 7 (V) | 8 (T) | |
| B (2,111) | Q (96.7), H (2.3) | V (75.3), A (16.8), M (4.3), I (1.9) | T (88.0), N (8.5), S (1.4) |
| C (1,174) | Q (96.5), H (1.3), K (1.2) | A (70.3), T (17.9), V (9.4) | N (81.8), G (8.0), T (5.2), H (1.6), Q (1.5), S (1.3) |
SP1 sequences were retrieved from the Los Alamos database.
Amino acid residues present with a prevalence of <1% are not listed.
It should be noted that the SP1 polymorphism residues identified in this study and elsewhere are located at the very C terminus of a CA-SP1 junction region exhibiting an α-helical secondary structure (1, 9). We previously proposed that BVM might bind to this putative helical structure of the Gag CA-SP1 protein and therefore disrupt CA-SP1 processing without interfering with the function of the putative structural element involved in virus assembly and budding (8). A possible explanation for the observed SP1 polymorphism-associated drug resistance involves a mechanism in which mutations at positions 7 and 8 in SP1 may alter the helical structure and consequently affect BVM interaction with the helix, leading to drug resistance. A more complete understanding of the structure and function of this helical structure of Gag CA-SP1 in HIV-1 biology and the mechanism of BVM action will be required before more definitive conclusions can be drawn.
The results presented here continue to support the theme that amino acid variation in the SP1 polymorphism motif inhibits the ability of the HIV-1 maturation inhibitor bevirimat to block CA-SP1 cleavage (3, 14, 18). Our results extend previous findings by demonstrating that SP1 residue 7 is a primary determinant of SP1 polymorphism-associated drug resistance, and an additional mutation at SP1 residue 8 seems to potentiate a threshold effect defined by SP1 residue 7. The results also extend our previous characterization of the molecular determinants of BVM susceptibility to include Gag residues that are located in the middle region of SP1, distal from the CA-SP1 cleavage site. Future studies to address the mechanisms by which SP1 polymorphisms confer drug resistance should allow for a better understanding of the molecular target for the HIV-1 maturation inhibitor bevirimat and should aid in the discovery and design of additional active maturation inhibitors effective against both WT HIV-1 and resistant viruses.
ACKNOWLEDGMENTS
We thank Ruth M. Ruprecht for providing the 1084i clone and Elizabeth Kolb for editing the manuscript. The HIV-1 NL4-3 clone, NL4-3-ΔENV-EGFP clone, TZM-bl indicator cell line, and HIV-Ig were obtained from the NIH AIDS Research and Reference Reagent Program.
This research was supported by the SDSU AES Fund (grant 3AH203 to F.L.) and by Public Health Service grants (AI071788 and AI076125) to F.L.
Footnotes
Published ahead of print on 18 April 2011.
REFERENCES
- 1. Accola M. A., Hoglund S., Gottlinger H. G. 1998. A putative alpha-helical structure which overlaps the capsid-p2 boundary in the human immunodeficiency virus type 1 Gag precursor is crucial for viral particle assembly. J. Virol. 72:2072–2078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Adamson C. S., et al. 2006. In vitro resistance to the human immunodeficiency virus type 1 maturation inhibitor BVM (bevirimat). J. Virol. 80:10957–10971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Adamson C. S., Sakalian M., Salzwedel K., Freed E. O. 2010. Polymorphisms in Gag spacer peptide 1 confer varying levels of resistance to the HIV-1 maturation inhibitor bevirimat. Retrovirology 7:36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Adamson C. S., Waki K., Ablan S. D., Salzwedel K., Freed E. O. 2009. Impact of human immunodeficiency virus type 1 resistance to protease inhibitors on evolution of resistance to the maturation inhibitor bevirimat (BVM). J. Virol. 83:4884–4894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Grisson R. D., et al. 2004. Infectious molecular clone of a recently transmitted pediatric human immunodeficiency virus clade C isolate from Africa: evidence of intraclade recombination. J. Virol. 78:14066–14069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kantor R., et al. 2005. Impact of HIV-1 subtype and antiretroviral therapy on protease and reverse transcriptase genotype: results of a global collaboration. PLoS Med. 2:e112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Li F., et al. 2003. BVM: a potent HIV inhibitor that disrupts core condensation by targeting a late step in Gag processing. Proc. Natl. Acad. Sci. U. S. A. 100:13555–13560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Li F., et al. 2006. Determinants of activity of the HIV-1 maturation inhibitor BVM. Virology 356:217–224 [DOI] [PubMed] [Google Scholar]
- 9. Liang C., et al. 2002. Characterization of a putative alpha-helix across the capsid-SP1 boundary that is critical for the multimerization of human immunodeficiency virus type 1 Gag. J. Virol. 76:11729–11737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Margot N. A., Gibbs C. S., Miller M. D. 2010. Phenotypic susceptibility to bevirimat in isolates from HIV-1-infected patients without prior exposure to bevirimat. Antimicrob. Agents Chemother. 54:2345–2353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Martin D. E., Blum R., Doto J., Galbraith H., Ballow C. 2007. Multiple-dose pharmacokinetics and safety of bevirimat, a novel inhibitor of HIV maturation, in healthy volunteers. Clin. Pharmacokinet. 46:589–598 [DOI] [PubMed] [Google Scholar]
- 12. Martin D. E., et al. 2007. Safety and pharmacokinetics of bevirimat (BVM), a novel inhibitor of human immunodeficiency virus maturation, in healthy volunteers. Antimicrob. Agents Chemother. 51:3063–3066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Martin D. E., Salzwedel K., Allaway G. P. 2008. Bevirimat: a novel maturation inhibitor for the treatment of HIV-1 infection. Antivir. Chem. Chemother. 19:107–113 [DOI] [PubMed] [Google Scholar]
- 14. McCallister S., et al. 2008. HIV-1 Gag polymorphisms determine treatment response to bevirimat (BVM). Antivir. Ther. 2008:A10 [Google Scholar]
- 15. Ndung'u T., Renjifo B., Essex M. 2001. Construction and analysis of an infectious human immunodeficiency virus type 1 subtype C molecular clone. J. Virol. 75:4964–4972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Seclen E., et al. 2010. High prevalence of natural polymorphisms in Gag (CA-SP1) associated with reduced response to bevirimat, an HIV-1 maturation inhibitor. AIDS 24:467–469 [DOI] [PubMed] [Google Scholar]
- 17. Smith P. F., et al. 2007. Phase I and II study of the safety, virologic effect, and pharmacokinetics/pharmacodynamics of single-dose 3-o-(3′,3′-dimethylsuccinyl)betulinic acid (bevirimat) against human immunodeficiency virus infection. Antimicrob. Agents Chemother. 51:3574–3581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Van Baelen K., et al. 2009. Susceptibility of human immunodeficiency virus type 1 to the maturation inhibitor bevirimat is modulated by baseline polymorphisms in Gag spacer peptide 1. Antimicrob. Agents Chemother. 53:2185–2188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Velazquez-Campoy A., et al. 2003. Protease inhibition in African subtypes of HIV-1. AIDS Rev. 5:165–171 [PubMed] [Google Scholar]
- 20. Zhou J., Chen C. H., Aiken C. 2004. The sequence of the CA-SP1 junction accounts for the differential sensitivity of HIV-1 and SIV to the small molecule maturation inhibitor 3-O-{3′,3′-dimethylsuccinyl}-betulinic acid. Retrovirology 1:15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Zhou J., et al. 2004. Small-molecule inhibition of human immunodeficiency virus type 1 replication by specific targeting of the final step of virion maturation. J. Virol. 78:922–929 [DOI] [PMC free article] [PubMed] [Google Scholar]