Abstract
VIM-27 metallo-β-lactamase, an Ala57 → Ser variant of VIM-1, was identified in three Klebsiella pneumoniae isolates belonging to sequence type 147. blaVIM-27 was part of a class 1 integron carried by non-self-transferable plasmids. Kinetic parameters and MIC determinations indicated that VIM-27 hydrolyzed most β-lactams, especially imipenem and cefoxitin, less effectively than VIM-1.
TEXT
VIM-type enzymes comprise a group of zinc-dependent β-lactamases (metallo-β-lactamases [MβLs]) of unknown origin that exhibit wide hydrolysis spectra, including carbapenems, and that are not inactivated by the clinically available β-lactamase inhibitors. The respective genes commonly occur as cassettes of class 1 integrons carried by a variety of Gram-negative species, including Klebsiella pneumoniae (1). Of the 27 VIM variants (www.lahey.org/studies), at least 3 differ by only one (VIM-4 [12] and VIM-5 [4]) or two (VIM-19 [13, 14]) amino acid residues from the prototype VIM-1 (8), constituting a distinct cluster within the VIM group. We describe here VIM-27, an A57S mutant of VIM-1 produced by K. pneumoniae.
Identification of blaVIM-27 and its genetic environment.
K. pneumoniae 254B, an MβL-positive isolate that exhibited resistance to multiple drugs, including all β-lactams but aztreonam (Table 1), was from a patient treated in the university hospital of Larissa in 2010. A PCR assay using the primers INT-F (10) and 3′-CS (9) resulted in a product of 3,548 bp whose sequence matched that of a segment of the VIM-1-encoding integron In-e541 (positions 8,341 to 11,888; GenBank accession no. AY339625) (11), except a G-to-T transversion within the blaVIM-1 coding sequence (position 9,107). This difference, confirmed by sequencing of products of two independent PCR assays, resulted in an A57S variant of VIM-1 (numbering as proposed in reference 5), designated VIM-27. Isoelectric focusing of crude cell extracts and staining with nitrocefin showed that K. pneumoniae 254B produced two β-lactamases, with apparent isoelectric points of 5.1 (corresponding to a VIM MβL) and 7.6 (most likely the inherent penicillinase of the species).
TABLE 1.
Etest MICs of β-lactams against a VIM-27-producing K. pneumoniae clinical isolate and E. coli DH5α clones producing VIM MβLs under isogenic conditions
| Antibiotic | Etest MIC (μg/ml) |
|||
|---|---|---|---|---|
| K. pneumoniae 254B | E. coli (pB-vim27) | E. coli (pB-vim1) | E. coli DH5α | |
| Ticarcillin | NTa | ≥512 | ≥5,122 | 2 |
| Ticarcillin-CLAb | ≥512 | ≥512 | ≥512 | 2 |
| Piperacillin | ≥512 | ≥512 | ≥512 | 1 |
| Piperacillin-TAZc | 256 | ≥512 | ≥512 | 0.5 |
| Cefoxitin | ≥512 | 96 | ≥512 | 2 |
| Cefotaxime | 256 | 128 | 256 | 0.12 |
| Ceftazidime | ≥512 | ≥512 | ≥512 | 0.25 |
| Cefepime | 96 | 12 | 32 | 0.06 |
| Aztreonam | 0.5 | 0.047 | 0.047 | 0.032 |
| Imipenem | 16 | 1.5 | 4 | 0.12 |
| Meropenem | 16 | 0.38 | 0.38 | <0.06 |
Not tested.
Clavulanate (CLA) was present at a fixed concentration of 2 μg/ml.
Tazobactam (TAZ) was present at a fixed concentration of 4 μg/ml.
For a preliminary estimation of the frequency of blaVIM-27 carriers, we examined 20 blaVIM-positive K. pneumoniae isolates referred to the National School of Public Health (NSPH) during July 2010 throughout the country. By PCR and sequencing, we identified two additional isolates from two hospitals in Athens also carrying blaVIM-27 in an In-e541-like integron. All three VIM-27-positive isolates exhibited similar levels of resistance to β-lactams, including carbapenems, as well as similar resistance phenotypes for non-β-lactam drugs (data not shown).
None of the blaVIM-27 carriers was capable of transferring the gene to Escherichia coli by conjugation performed in mixed broth cultures (16). Plasmid DNA from the clinical isolates was extracted using a Qiagen Midi kit (Qiagen, Hilden, Germany) and used to transform E. coli DH5α. β-Lactam-resistant transformants containing blaVIM-27 were obtained from all three preparations. They carried plasmids of approximately 50 kb that hybridized strongly with a blaVIM-specific probe (not shown). Sequence typing, performed as described previously (2), showed that the VIM-27 isolates belonged to the sequence type 147 (ST147). Isolates of ST147 comprise a minor cluster of VIM producers isolated in this setting since 2004 (unpublished data).
Construction of VIM-encoding recombinant plasmids.
A PCR using total DNA from K. pneumoniae 254B as a template and the primers INT-F and aacA7-R (5′-CGCTGTTGGTAAGTTGAGTG-3′) yielded a product of 1,874 bp that included the P1 “strong” promoter of the integron, blaVIM-27, and a fragment of the aacA7 gene cassette (corresponding to nucleotides 8,341 to 10,214 in In-e541). The product was cloned into the Topo TA vector (Invitrogen, Carlsbad, CA). An EcoRI fragment of 1,852 bp was then introduced into the polycloning site of pBCSK(+) (Stratagene, La Jolla, CA), resulting in the plasmid pB-vim27. A similar plasmid encoding VIM-1 (pB-vim1) was constructed following the same procedure and using the plasmid p541 as a template (11). Plasmids pB-vim27 and pB-vim1 were introduced into E. coli DH5α, and the resulting clones were used for expression of MβLs and comparative MIC determinations.
Purification and hydrolysis spectra of VIM MβLs.
β-Lactamases were released by mild ultrasonic treatment of bacterial cells suspended in Tris-HCl (20 mM, pH 7.5) (buffer A). Proteins were fractionated by ammonium sulfate precipitation as described previously (3). Fractions with β-lactamase activity were dialyzed against buffer A and loaded on Sepharose Q columns (Bio-Rad Laboratories, Hercules, CA) preequilibrated with buffer A, and the VIM enzymes were eluted with a 0 to 1 M NaCl gradient. The final purification step was carried out by size exclusion chromatography using Sephadex G-50 Superfine columns (GE Healthcare Biosciences AB, Uppsala, Sweden). The purity of the final preparations was >90%, as determined by sodium dodecyl sulfate-polyacrylamide gel electrophoresis. Protein concentrations were determined by the Bradford method.
Hydrolysis of β-lactams at various concentrations was monitored using a Hitachi U-2001 spectrophotometer. Wavelengths and extinction coefficients have been reported elsewhere (6). Kinetic parameters of VIM-27 were typical of this group of MβLs (Table 2). The most preferable substrate of those tested was cephalothin. The enzyme exhibited high rates of hydrolysis against cefotaxime and cefepime. However, apparent Km values for the latter substrates were high. Hydrolytic efficiency against cefoxitin was relatively low. A lower kcat value was observed with imipenem; however, this was compensated by a relatively high affinity between VIM-27 and the drug, resulting in a meaningful “imipenemase” activity. VIM-27 was inactive against aztreonam (data not shown). Comparison of the kinetic parameters of VIM-27 with those of VIM-1, determined under the same conditions, showed that the former enzyme was a less efficient β-lactamase exhibiting lower kcat/Km for all substrates tested. The most marked decrease in efficiency was against cefoxitin and imipenem (4.7- and 3.2-fold, respectively). The respective decreases for cefotaxime and cefepime were marginal (1.6-fold) though reproducible. Inhibition by EDTA was studied using imipenem as the reporter substrate, and results were expressed as 50% inhibitory concentrations (IC50). VIM-27 and VIM-1 were similarly sensitive to EDTA (IC50s of 5.6 ± 0.6 and 6.4 ± 0.7 μM, respectively).
TABLE 2.
Kinetic parameters of VIM-27 for various β-lactams and comparison with VIM-1a
| β-Lactam | VIM-27 (Ser57) | VIM-1 (Ala57) | ||||
|---|---|---|---|---|---|---|
| kcat (s−1) | Km (μM) | kcat/Km (μM−1·s−1) | kcat (s−1) | Km (μM) | kcat/Km (μM−1·s−1) | |
| Cephalothin | 357 ± 32 | 43 ± 5.1 | 8.3 | 476 ± 42 | 43 ± 4.2 | 11 |
| Cefoxitin | 8.7 ± 1.0 | 273 ± 18 | 0.03 | 12 ± 0.8 | 88 ± 10 | 0.14 |
| Cefotaxime | 292 ± 11 | 355 ± 25 | 0.82 | 278 ± 19 | 209 ± 15 | 1.3 |
| Cefepime | 130 ± 7.9 | 258 ± 18 | 0.50 | 175 ± 18 | 220 ± 20 | 0.80 |
| Imipenem | 3.0 ± 0.5 | 11.4 ± 1.5 | 0.26 | 3.7 ± 0.2 | 4.4 ± 0.8 | 0.84 |
Values are the means of data from four independent measurements.
Resistance conferred by VIM-27 and VIM-1 under isogenic conditions.
MICs of β-lactam antibiotics tested against E. coli (pBvim-27) and E. coli(pBvim-1) were determined by the Etest method (AB bioMérieux, Solna, Sweden). Results were in line with the kinetic data, the most significant differences in the resistance levels being those for cefoxitin and imipenem (Table 1).
Conclusions.
The small number of K. pneumoniae isolates screened for blaVIM-27 does not allow for an estimation of the spread of this novel MβL variant. And yet, detection of three blaVIM-27-positive isolates in different locations in a limited time period may indicate establishment of these microorganisms in Greek hospitals. blaVIM-27 genes were found in similar integrons carried by nonconjugative plasmids that were similar in size and harbored by genetically related isolates. It can thus be hypothesized that at present, dissemination of blaVIM-27 is probably clonal.
Assuming that VIM-27 is a direct descendant of VIM-1, the prevalent MβL among clinical enterobacteria in Greece (15), a positive antibiotic selection pressure driving its emergence and/or spread cannot be supported. It should be underlined, however, that the Ala57Ser change results in a minor loss of activity against β-lactams that could be compensated by other resistance mechanisms common in clinical isolates. Indeed, MICs of carbapenems and expanded-spectrum cephalosporins for the VIM-27 producers were between the respective MIC50 and MIC90 values observed for the population of VIM-positive K. pneumoniae isolates during 2009 and 2010 (unpublished records of the NSPH).
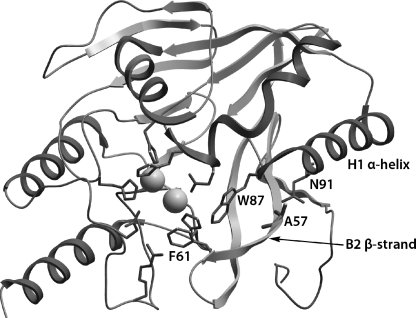
Residue 57 in VIM β-lactamases is included in the B2 β-strand. Based on the crystal structure of VIM-4 (7), its distance from the active site precludes a direct interaction with amino acids important for catalysis (Fig. 1). Nevertheless, the replacement of the aliphatic Ala by the polar Ser may change the interaction of B2 with the H1 α-helix through a hydrogen bond with the adjacent Asn91. Such an interaction could affect positioning of active site residues, such as Phe61 in B2 and Trp87 in H1.
FIG. 1.
Drawing of the three-dimensional structure of VIM-4 (PDB identifier 2WHG) showing the position of amino acid 57. The side chain of Ala57 lies at the vicinity of the side chain of Asn91. The B2 β-strand and the H1 α-helix, which contain the active site residues Phe61 and Trp87, respectively, are also shown. Side chains of the above amino acids and of other active site residues are indicated as sticks. Spheres represent the two Zn2+ ions.
Nucleotide sequence accession number.
The nucleotide sequence of blaVIM-27 has been assigned GenBank accession number HQ858608.
Acknowledgments
This work was supported by funding from the Hellenic Pasteur Institute and the European Commission (TROCAR contract HEALTH-F3-2008-223031).
Footnotes
Published ahead of print on 25 April 2011.
REFERENCES
- 1. Cornaglia G., et al. 2007. Metallo-beta-lactamases as emerging resistance determinants in Gram-negative pathogens: open issues. Int. J. Antimicrob. Agents 29:380–388 [DOI] [PubMed] [Google Scholar]
- 2. Diancourt L., Passet V., Verhoef J., Grimont P. A., Brisse S. 2005. Multilocus sequence typing of Klebsiella pneumoniae nosocomial isolates. J. Clin. Microbiol. 43:4178–4182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Docquier J.-D., et al. 2003. On functional and structural heterogeneity of VIM-type metallo-β-lactamases. J. Antimicrob. Chemother. 51:257–266 [DOI] [PubMed] [Google Scholar]
- 4. Gacar G. G., et al. 2005. Genetic and enzymatic properties of metallo-β-lactamase VIM-5 from a clinical isolate of Enterobacter cloacae. Antimicrob. Agents Chemother. 49:4400–4403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Garau G., et al. 2004. Update of the standard numbering scheme for class B β-lactamases. Antimicrob. Agents Chemother. 48:2347–2349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kotsakis S. D., Miriagou V., Tzelepi E., Tzouvelekis L. S. 2010. Comparative biochemical and computational study of the role of naturally occurring mutations at Ambler positions 104 and 170 in GES β-lactamases. Antimicrob. Agents Chemother. 54:4864–4871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Lassaux P., et al. 2011. Biochemical and structural characterization of the subclass B1 metallo-β-lactamase VIM-4. Antimicrob. Agents Chemother. 55:1248–1255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Lauretti L., et al. 1999. Cloning and characterization of blaVIM, a new integron-borne metallo-β-lactamase gene from a Pseudomonas aeruginosa clinical isolate. Antimicrob. Agents Chemother. 43:1584–1590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Levesque C., Piche L., Larose C., Roy P. H. 1995. PCR mapping of integrons reveals several novel combinations of resistance genes. Antimicrob. Agents Chemother. 39:185–191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Mavroidi A., et al. 2001. An integron-associated β-lactamase (IBC-2) from Pseudomonas aeruginosa is a variant of the extended-spectrum β-lactamase IBC-1. J. Antimicrob. Chemother. 48:627–630 [DOI] [PubMed] [Google Scholar]
- 11. Miriagou V., Tzelepi E., Gianneli D., Tzouvelekis L. S. 2003. Escherichia coli with a self-transferable, multiresistant plasmid coding for metallo-β-lactamase VIM-1. Antimicrob. Agents Chemother. 47:395–397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Pournaras S., Tsakris A., Maniati M., Tzouvelekis L. S., Maniatis A. N. 2002. Novel variant (blaVIM-4) of the metallo-β-lactamase gene blaVIM-1 in a clinical strain of Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 46:4026–4028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Robin F., Aggoune-Khinache N., Delmas J., Naim M., Bonnet R. 2010. Novel VIM metallo-β-lactamase variant from clinical isolates of Enterobacteriaceae from Algeria. Antimicrob. Agents Chemother. 54:466–470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Rodriguez-Martinez J.-M., Nordmann P., Fortineau N., Poirel L. 2010. VIM-19, a metallo-β-lactamase with increased carbapenemase activity from Escherichia coli and Klebsiella pneumoniae. Antimicrob. Agents Chemother. 54:471–476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Vatopoulos A. 2008. High rates of metallo-β-lactamase-producing Klebsiella pneumoniae in Greece—a review of the current evidence. Euro Surveill. 13:8023. [PubMed] [Google Scholar]
- 16. Vatopoulos A. C., Philippon A., Tzouvelekis L. S., Komninou Z. Z., Legakis N. J. 1990. Prevalence of a transferable SHV-5 type β-lactamase in clinical isolates of Klebsiella pneumoniae and Escherichia coli in Greece. J. Antimicrob. Chemother. 26:635–648 [DOI] [PubMed] [Google Scholar]