Abstract
The Centers for Disease Control and Prevention currently recommends a 2-tier serologic approach to Lyme disease laboratory diagnosis, comprised of an initial serum enzyme immunoassay (EIA) for antibody to Borrelia burgdorferi followed by supplementary IgG and IgM Western blotting of EIA-positive or -equivocal samples. Western blot accuracy is limited by subjective interpretation of weakly positive bands, false-positive IgM immunoblots, and low sensitivity for detection of early disease. We developed an objective alternative second-tier immunoassay using a multiplex microsphere system that measures VlsE1-IgG and pepC10-IgM antibodies simultaneously in the same sample. Our study population comprised 79 patients with early acute Lyme disease, 82 patients with early-convalescent-phase disease, 47 patients with stage II and III disease, 34 patients post-antibiotic treatment, and 794 controls. A bioinformatic technique called partial receiver-operator characteristic (ROC) regression was used to combine individual antibody levels into a single diagnostic score with a single cutoff; this technique enhances test performance when a high specificity is required (e.g., ≥95%). Compared to Western blotting, the multiplex assay was equally specific (95.6%) but 20.7% more sensitive for early-convalescent-phase disease (89.0% versus 68.3%, respectively; 95% confidence interval [95% CI] for difference, 12.1% to 30.9%) and 12.5% more sensitive overall (75.0% versus 62.5%, respectively; 95% CI for difference, 8.1% to 17.1%). As a second-tier test, a multiplex assay for VlsE1-IgG and pepC10-IgM antibodies performed as well as or better than Western blotting for Lyme disease diagnosis. Prospective validation studies appear to be warranted.
INTRODUCTION
Lyme disease (LD) is the most common vector-borne disease in the United States, with a reported incidence of nearly 35,000 new cases annually (10, 21). There are three disease stages: stage I is the early acute phase, characterized by a rash (erythema migrans [EM]) that occurs in at least 70% of patients; stage II represents early disseminated infection, including lymphocytic meningitis, cranial neuropathy, radiculopathy, and Lyme carditis; and stage III represents late disseminated infection, such as Lyme arthritis, axonal peripheral neuropathy, and encephalomyelitis (39). Diagnosis of stage I disease is based on clinical, not serological, criteria, while stages II and III typically require serologic confirmation (37). Despite the predominance of stage I disease, more than 3.4 million tests for LD were ordered in 2008 in the United States (A. Hinckley, Centers for Disease Control and Prevention [CDC], personal communication). Overuse of serology has led to significant problems with false-positive results and misdiagnosis (38).
When first introduced for LD diagnosis, whole-cell enzyme immunoassays (EIAs) and indirect immunofluorescence assays (IFAs) for serum antibodies to Borrelia burgdorferi suffered from a lack of standardization, poor reproducibility, and high false-positive rates (11, 25). Following the Second National Conference on the Serologic Diagnosis of Lyme Disease (27 to 29 October 1994; Dearborn, MI), a 2-tier serologic approach was recommended, comprised of an initial serum EIA or IFA for antibody to B. burgdorferi followed by supplementary IgG and IgM Western blotting of positive or indeterminate samples (9). Furthermore, only IgG blots were recommended for serologic diagnosis more than 30 days after disease onset. Although Western blotting is very sensitive for stage II and III disease, multiple limitations to blot accuracy have been identified: a low sensitivity for stage I disease, false-positive IgM immunoblots, and subjective interpretation of weakly positive bands (1, 5). Western blotting is also labor-intensive and expensive. The goal of the current study was to develop an objective alternative to Western blotting as a second-tier assay.
Diagnostic serology has evolved and now utilizes recombinant and synthetic peptide antigens, such as C6, the 26-mer invariant portion of VlsE1 (variable major protein-like sequence 1); recombinant VlsE1 itself; and pepC10, a 10-mer conserved portion of OspC (2). These surface antigens are expressed by Borrelia burgdorferi during the early phase of mammalian infection (39). The predominant immune responses to C6 and VlsE1 are IgG mediated, even in early disease, while pepC10 generates an early and sometimes lasting IgM response (2, 5, 28). While more specific than whole-cell EIAs, these new assays might not be as specific as Western blotting (5, 40). Although the results are preliminary, microarrays for serologic detection of products of expressed open reading frames represent a promising new technology (3). Diagnostic alternatives to serology remain limited. Cultures of blood and body fluids for B. burgdorferi demonstrate low sensitivity (1). PCR assays for B. burgdorferi DNA from synovial fluid and skin are often positive prior to antibiotic therapy but require invasive procedures to obtain suitable samples and are prone to false-positive results if contamination risk is not rigorously controlled (1). At present, no assays for direct detection of B. burgdorferi have been approved by the Food and Drug Administration (19).
Given the complex nature of the host immune response to B. burgdorferi infection, the use of multiple serologic assays has been proposed to enhance either test sensitivity or specificity (2, 6, 12, 17, 34, 35). Tests derived from continuous data generate binary (positive or negative) results when a cutoff value is chosen to achieve a desired specificity (e.g., 99%). Combining binary test results by using Boolean “OR” logic, such as detecting either IgG antibody to VlsE1 or IgM antibody to pepC10 by kinetic EIA, can generate a more sensitive but less specific assay than the individual test components (2). In contrast, combining tests by using Boolean “AND” logic may produce a more specific but less sensitive assay (35).
Some potentially useful information about specific antibody levels is lost in creating a binary test. For some antibody combinations, multivariate regression models can outperform standard binary assays: they can identify the most important diagnostic tests among multiple options and weight their individual contributions when calculating an overall diagnostic score (31). Regression models can be used for either disease classification (i.e., disease or no disease) or prediction (i.e., probability of disease). Our analyses focus on using receiver-operator characteristic (ROC) regression models to improve disease classification. ROC curves plot the tradeoff between sensitivity and specificity as the test cutoff is varied; in general, the greater the area under the ROC curve (AUC), the better the test. Recent publications have explored the use of ROC curves to compare and optimize the performance of diagnostic tests (30, 32). Full ROC regression analysis optimizes classifier performance by maximizing the area under the entire ROC curve (29). Partial ROC regression is a relatively new bioinformatic technique that can augment test performance within clinically significant portions of the ROC curve (e.g., 95% specificity or higher) (14). We chose to evaluate partial ROC regression models for LD diagnosis because of the need for high test specificity.
The multiplex is a device that can perform multiple antibody assays simultaneously with the same serum sample (20), making this platform attractive for LD diagnosis. We developed a new multiplex immunoassay for VlsE1-IgG and pepC10-IgM antibodies, interpreted using partial ROC regression techniques, as an objective alternative to Western blotting.
MATERIALS AND METHODS
Study population.
Data set A consisted of the following: (i) 79 prospectively collected sera from patients with culture-proven, early-acute-phase LD (stage I; EM) and 78 early-convalescent-phase sera from the same patients; (ii) 4 retrospectively collected convalescent-phase samples from patients with culture-proven EM; (iii) 47 prospectively collected sera from patients with stage II and III LD (16 with early neurological disease, 2 with myocarditis, and 29 with Lyme arthritis); and (iv) 34 retrospectively collected sera obtained following treatment for extracutaneous disease (n = 16) and erythema migrans (n = 18) (A. Steere, Boston, MA, and the CDC, Fort Collins, CO). Of the 16 patients with early neurological disease, most had more than one disease manifestation, including facial palsy (n = 11), meningitis (n = 7), radiculopathy (n = 4), and optic neuritis (n = 2). All prospectively collected sera were obtained during prior investigations and evaluated retrospectively for this study. All patients from data set A had histories consistent with exposure to North American B. burgdorferi and met the CDC case definition for Lyme disease (7, 8).
Data set B consisted of 446 consecutive uncharacterized samples submitted to the New York State Department of Health (NYSDOH) for routine LD serology between 2006 and 2007 (S. J. Wong, Albany, NY); no clinical data were available for the patients. Of the latter samples, 164 were standard 2-tier serology positive.
Samples were collected with informed consent during previous studies (set A) or for nonresearch purposes (set B). This research was approved by the institutional review boards of Saint Francis Medical Center, Trenton, NJ, and the NYSDOH, Albany, NY; samples were deidentified prior to testing, and requirements for additional informed consent were waived. Laboratory personnel were blind to the multiplex diagnostic score when performing other assays.
Controls.
Uninfected controls included 300 healthy blood donors from New Mexico (where LD is not endemic), 300 healthy blood donors from New England (where LD is endemic), 99 patients from New Mexico undergoing routine screening examinations, and 95 patients with potentially cross-reacting conditions from an area where LD is endemic. The latter conditions included Epstein-Barr virus infection (20), toxoplasmosis (10), rheumatoid arthritis (10), anti-nuclear antibody-positive status (10), leptospirosis (10), syphilis (10), rubella (10), and other conditions (15).
Stratification of immune responses.
Initial samples collected less than 6 months after the start of treatment for stage II or III disease were considered representative of the maximal immune response (group 1), while samples collected 6 or more months after the beginning of treatment were considered representative of a waning immune response (group 2) (33). Similarly, early-convalescent-phase samples collected less than 30 days after the start of antibiotic treatment were considered representative of the maximal immune response (group 1), while samples collected 30 or more days after the beginning of treatment were considered representative of a waning immune response (group 2) (18). The immune responses of those with untreated early acute disease were considered separately.
Recombinant VlsE1 and pepC10 antigens.
Recombinant VlsE1 protein was produced using Escherichia coli Sure2 with a pVlsE1-His3 fusion protein plasmid construct (supplied by S. Norris, University of Texas Medical School, Houston, TX) and was purified using His and heparin affinity columns (27). Synthetic pepC10 (PVVAESPKKP-OH) was obtained from NeoMPS, Inc. (San Diego, CA).
Multiplex microsphere assay.
The AtheNA Multi-Lyte test system (Zeus Scientific, Inc., Branchburg, NJ) is a sandwich immunoassay based on flow cytometric separation of fluorescent microparticles by use of Luminex xMAP technology (Luminex Corp., Austin, TX) (20). Briefly, multiple sets of 5.6-μm polystyrene beads are each impregnated with fluorescent dyes that give them distinct spectral signatures (20). For this study, VlsE1 and pepC10 antigens were covalently bound to the surfaces of separate sets of beads. All patient samples and assay controls were diluted 1:21 by combining 10 μl of specimen with 200 μl of the specimen diluent and mixing them for 30 s on a shaker plate at 800 rpm. A 50-μl mixture of bead sets containing VlsE1-conjugated microspheres, 4 calibrators, and a bead set to detect nonspecific binding was added to each filtration well, resuspended by vortexing and sonication for 30 s each, and then incubated with 10 μl of diluted specimen for 30 min. All incubation steps required mixing on a shaker plate as described above, followed by vacuum washing with 200 μl of phosphate-buffered saline (PBS) three times. The bead sets were incubated with 150 μl of phycoerythrin (PE)-labeled goat anti-human IgG gamma antibody (Moss Inc., Pasadena, MD) for 30 min and then vacuum washed. A 50-μl mixture containing 2 additional bead sets was added to each filtration well: 1 set was conjugated with VlsE1, and the other was conjugated with pepC10. The bead sets were resuspended as described above, and another 10 μl of diluted specimen was added to each well and incubated for 30 min. The bead sets were vacuum washed, and 150 μl of PE-labeled goat anti-human IgM mu (Moss, Inc.) was added to each well and incubated for 30 min. After being vacuum washed, the bead sets were resuspended in 150 μl of PBS and the results were read by a flow cytometer. Using a proprietary method, IgG and IgM levels were measured simultaneously, using a single excitation laser and a single reporter molecule (PE). Antibody levels were measured in AtheNA units (AU), but the test combination (AtheNA bioinformatic score [BIS]) was computed using the algorithm described below and rescaled such that a score of ≥1.0 was considered positive; interpretive software is available through the corresponding author. All patient specimens were processed by S. J. Wong at the NYSDOH; control specimens were processed by both Zeus Scientific, Inc., and the NYSDOH.
Routine tests.
All specimens were tested with the following assays: (i) Zeus whole-cell EIA, (ii) IgG and IgM Western blotting for whole-cell EIA-positive and -equivocal specimens (MarDx Diagnostics, Inc., Carlsbad, CA), and (iii) C6 IgG/IgM EIA (Immunetics, Inc., Boston, MA). All assays were performed in accordance with the manufacturers' instructions. Western blots were interpreted in accordance with current CDC guidelines for 2-tier testing (9).
Multiplex assay precision.
For standardization purposes, individual cutoffs for VlsE1-IgG and pepC10-IgM antibodies were determined in AU by Zeus Scientific, Inc. (Branchburg, NJ). The cutoff for VlsE1-IgG, 31 AU/ml, was 8 standard deviations above the mean for 25 healthy controls from an area where LD is not endemic, while the cutoff for pepC10-IgM, 24 AU/ml, was 4 standard deviations above the mean. There were 3 testing sites and 2 technicians at each site. Five serum standards were prepared, with antibody concentrations ranging from negative and near the cutoff to highly positive. Each standard was run in triplicate twice each day for 5 consecutive days by each technician at each location.
Statistical methods.
A broad selection of second-tier classifiers was created to provide alternatives to Western blotting. Each multivariate classifier, including partial ROC regression models, generated a composite score for each sample by using a weighted linear combination of pepC10-IgM and VlsE1-IgG antibody levels measured by the multiplex assay. Examples of second-tier classifiers included VlsE1-IgG alone, pepC10-IgM alone, binary combinations of pepC10-IgM and VlsE1-IgG using individual antibody cutoffs, and combinations of antibody levels by logistic likelihood regression, full ROC regression, partial ROC regression trained using the 95% to 100% specificity portion of the ROC curve (95% pROC), and partial ROC regression trained using the 60% to 100% specificity portion of the ROC curve (60% pROC). See Appendix A for a detailed description of the partial ROC regression technique. The 95% pROC analysis focused on achieving high specificity, while the 60% pROC analysis emphasized high sensitivity.
To determine the optimal training set, each classifier was trained on each disease stage from data set A, using all samples from a given stage. A classifier trained on one disease stage from data set A was then tested against the other disease stages in data set A and against data set B. Training controls consisted of 249 sera from an area where LD is not endemic, and testing controls consisted of 545 samples from blood donors from both areas where LD is endemic and those where it is not endemic, as well as samples from patients with potential cross-reacting conditions. Partial ROC areas were used to identify the optimal disease stage for training purposes; otherwise, classifier sensitivity and specificity were used to compare performances. A median bootstrap method was used to generate 95% confidence intervals for the differences between classifier sensitivities, specificities, and partial ROC areas (16).
Classifier sensitivities were compared at both Western blot specificity (95.6%) and 99.0% specificity among 545 testing controls, corresponding to 69.2% specificity and 93.6% specificity, respectively, among the 78 EIA-reactive testing controls (the target population for second-tier assays). Classifier performance was compared at 99% specificity because the latter value generates a higher test accuracy in low-incidence settings (2, 15). Our primary end point was to determine if the overall sensitivity of the multiplex assay used as a second-tier test was noninferior to Western blotting (margin, ≤10%; two-tailed α = 0.05) at 95.6% specificity. As a secondary end point, the overall specificity of the multiplex assay was compared to that of Western blotting by bootstrapping at an equivalent sensitivity. Post-antibiotic-treatment sera (group 2) were excluded from the analysis of primary and secondary end points because their significance was uncertain.
Within-site variance was determined using one-way analysis of variance (ANOVA). Because only three test sites were utilized, calculating between-site variance using 2 degrees of freedom might significantly overestimate its true value. Therefore, between-site variance was estimated by dividing the sums of squares between sites by 60, the number of test repetitions at each site.
In addition to the bootstrap technique, differences in classifier performance were assessed using the Wilcoxon rank sum test where appropriate. All reported P values were calculated using the latter test unless stated otherwise. MATLAB software was used to estimate regression models, perform the ANOVA, and make statistical comparisons (MathWorks, Natick, MA).
RESULTS
Choice of training set among samples from patients with disease.
Table 1 evaluates the impact of using training data from different disease stages to maximize the AUC between 95% and 100% specificities and between 60% and 100% specificities. We observed no significant advantage to using training data from one disease stage to maximize the AUC for any other disease stage for a given partial ROC classifier; the same observation held true for logistic likelihood regression and full ROC regression classifiers (data not shown). Therefore, we were not able to identify disease stage-specific classifiers. By training the 95% pROC classifier using stage II and III data and the 60% pROC classifier using early-acute-phase data, we were able to validate both classifiers against early-convalescent-phase data with little risk of overfitting (i.e., overestimating the true performance).
Table 1.
Partial ROC regression model performance by disease stage, as measured by the partial AUCa
| Specificity quantile and disease stage (no. of cases) | Classifier training set |
|||
|---|---|---|---|---|
| EA | EC | II/III | EC+II/IIIb | |
| 95% to 100% specificity training quantile | ||||
| EA (79) | 0.4036 | 0.4087 | 0.4144 | 0.4133 |
| EC (82) | 0.7296 | 0.7341 | 0.7645 | 0.7436 |
| II/III (47) | 0.9252 | 0.9352 | 0.9640 | 0.9580 |
| EC+II/IIIb (129) | 0.8010 | 0.8067 | 0.8373 | 0.8216 |
| 60% to 100% specificity training quantile | ||||
| EA (79) | 0.5973 | 0.5976 | 0.5875 | 0.5948 |
| EC (82) | 0.8558 | 0.8558 | 0.8434 | 0.8558 |
| II/III (47) | 0.9815 | 0.9814 | 0.9836 | 0.9824 |
| EC+II/IIIb (129) | 0.9016 | 0.9016 | 0.8948 | 0.9017 |
Each partial AUC was rescaled by dividing the measured partial AUC by the maximum possible AUC for a given specificity quantile. Abbreviations: EA, stage I, early acute phase; EC, stage I, early convalescent phase; II/III, stages II and III; EC+II/III, combined data sets.
Represents all group 1 sera.
Choice of training quantile for controls.
We evaluated the following specificity quantiles to train partial ROC regression classifiers: 60% to 100%, 80% to 100%, 90% to 100%, and 95% to 100% specificities. The overall sensitivity at Western blot specificity (95.6%) was the same using either the 60% to 100% or 80% to 100% specificity quantile but fell as the specificity quantile narrowed to between 90% and 100% (data not shown). A more detailed comparison of the 60% pROC and 95% pROC classifiers is described below.
Classifier scatterplot.
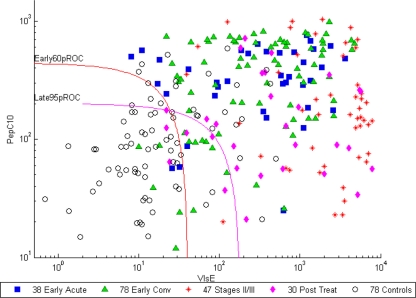
The log-log scatterplot in Fig. 1 demonstrates VlsE1-IgG and pepC10-IgM antibody levels by disease stage and illustrates the ability of the 95% pROC and 60% pROC regression classifiers to distinguish EIA-reactive case-patients from controls; these samples represent the target population of a second-tier assay.
Fig. 1.
Log-log scatterplot of VlsE1-IgG and pepC10-IgM antibody levels in EIA-reactive sera from case patients (n = 193) and controls (n = 78) from data set A. All antibody levels are reported in AtheNA units. Specimens above and to the right of each classifier curve are positive by that classifier. Abbreviations: Early60pROC, 60% pROC trained on early-acute-phase sera; Late95pROC, 95% pROC trained on disease stage II and III sera; Early Conv, early convalescent phase; stages II/III, stage II and III sera; Post Treat, post-antibiotic-treatment (group 2) sera.
Heuristic comparison of classifier performances.
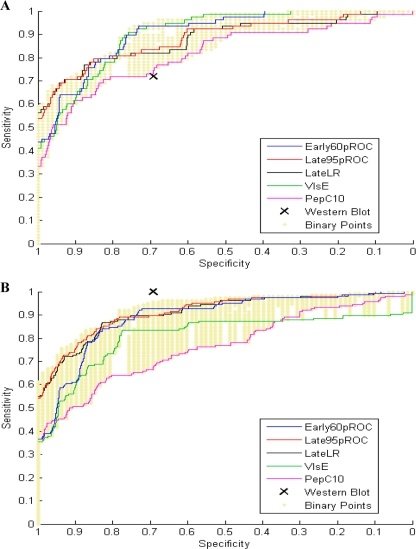
The ROC curves in Fig. 2 A provide a heuristic means by which to compare classifier performances among EIA-reactive early-convalescent-phase sera (76 EIA-positive and 2 EIA-equivocal sera) and controls (57 EIA-positive and 21 EIA-equivocal sera). Between 80% and 100% specificities, the 95% pROC and logistic regression models outperformed other classifiers, including single-antibody assays. Between 60% and 80% specificities, the 60% pROC and VlsE1-IgG classifiers demonstrated greater sensitivity than the other models, including Western blotting. Potential binary combinations of VlsE1-IgG and pepC10-IgM antibodies are represented by golden dots in each panel of Fig. 2; by varying the cutoff for each antibody separately, we could generate a range of sensitivities while maintaining the same specificity (or vice versa). The partial ROC regression models displayed in Fig. 2A appear more sensitive than most possible binary combinations at any given specificity.
Fig. 2.
(A) Classifier performances among EIA-reactive sera from patients with early-convalescent-phase disease for data set A (n = 78) and among EIA-reactive controls (n = 78). (B) Classifier performances among standard 2-tier test-positive samples from data set B (n = 164) and among EIA-reactive controls (n = 78). Abbreviations: Early60pROC, 60% pROC regression classifier trained using early-acute-phase sera; Late95pROC, 95% pROC regression classifier trained using stage II/III sera; LateLR, logistic regression model trained using stage II/III sera; VlsE, VlsE1-IgG; PepC10, pepC10-IgM. See the text for an explanation of the binary points.
Figure 2B compares classifier performances among 2-tier test-positive samples from data set B. Because no classifier for data set B could generate a sensitivity that exceeded that of Western blotting, we could determine only the relative sensitivities of alternative classifiers for that data set; real differences between classifiers may be muted by this sensitivity ceiling. Data set B (Fig. 2B) demonstrated the same relative sensitivities among classifiers between 80% and 100% specificities as those with data set A (Fig. 2A). The partial ROC regression classifiers in Fig. 2B also appear more sensitive than most possible binary combinations at any given specificity.
Statistical comparison of classifier performances.
In order to identify the best model(s), we compared classifier sensitivities for two data sets at two different specificities (Table 2). Because the proposed multiplex assay is part of a 2-tiered approach, it was necessary to consider all early-convalescent-phase sera from data set A in calculating overall test performance. Among early-convalescent-phase sera, the 95% pROC and logistic regression classifiers provided the best sensitivity at 99% specificity; combining antibody levels using these regression techniques generated 65.9% sensitivity, compared to 53.7% for VlsE-IgG alone and 48.8% for pepC10-IgM alone (P < 0.05 by bootstrapping for each antibody).
Table 2.
Sensitivities of classifiers used as second-tier assays with 2 data sets at 2 different specificitiesa,b
| Sample group and classifier | Sensitivity at specificity of: |
|
|---|---|---|
| 99% | 95.6% | |
| All early convalescent-phase sera from data set A (n = 82) | ||
| Logistic regressionc | 0.6585 | 0.7805 |
| 95% pROCc | 0.6585 | 0.7927 |
| 60% pROCd | 0.6098 | 0.8902 |
| Full ROC regressiond | 0.5976 | 0.8902 |
| VlsE1-IgG | 0.5366 | 0.8902 |
| Western blotting (IgG and IgM) | NAe | 0.6829 |
| Two-tier test-positive sera from data set B (n = 164)f | ||
| Logistic regressionc | 0.6951 | 0.8963 |
| 95% pROCc | 0.7134 | 0.8963 |
| 60% pROCd | 0.5854 | 0.9268 |
| Full ROC regressiond | 0.5793 | 0.9268 |
| VlsE1-IgG | 0.5305 | 0.8354 |
| Western blotting (IgG and IgM) | NAe | 1.00 |
All 545 testing controls were used for analysis.
See the text for pepC10-IgM data.
Partial ROC regression (95% to 100% specificity quantile) and logistic classifiers were trained using all stage II/III sera from data set A.
Partial ROC regression (60% to 100% specificity quantile) and full ROC regression classifiers were trained using all early-acute-phase sera from data set A.
NA, not applicable.
All sensitivities for data set B are reported relative to that of Western blotting.
At Western blot specificity (95.6%), the 60% pROC and full ROC classifiers provided optimal sensitivity among early-convalescent-phase sera and were statistically superior to (i) Western blotting (difference in sensitivity, 20.7%; 95% confidence interval [95% CI], 12.1% to 30.9%), (ii) the 95% pROC model (difference in sensitivity, 9.7%; 95% CI, 4.0% to 16.2%), and (iii) the logistic model (difference in sensitivity, 11.0%; 95% CI, 4.8% to 17.7%). The performance of VlsE1-IgG assay alone at 95.6% specificity was inconsistent between data sets: it was equal to that of the 60% pROC model with data set A but significantly less sensitive than this model with data set B (P < 0.05 by bootstrapping); in contrast, the performance of the 60% pROC model appeared robust for the choice of data sets. IgM antibody to pepC10 was the least sensitive assay with both data sets at 95.6% specificity.
Differences in classifier performance can also be expressed in terms of specificity at a fixed sensitivity. Comparing the specificity of regression classifiers to that of binary combinations is difficult because there is a range of separate cutoffs for VlsE1-IgG and pepC10-IgM antibodies that together can generate the same sensitivity but produce different specificities. To aid in comparisons, we identified a single cutoff value in AtheNA units for both antibodies, such that the overall sensitivity of the binary combination was equal to that of the 95% pROC classifier among samples from data set A; although the test sensitivity was 70.2% for both classifiers, the specificity of the 95% pROC model was 1.3% higher than that of the binary combination (95.6% versus 94.3%; 95% CI for the difference, 0.6% to 2.2%). The latter difference translated to a 9% improvement in specificity among EIA-reactive controls. When the sensitivity of the 95% pROC classifier was statistically equivalent to that of Western blotting (124/208 samples [59.6%] versus 130/208 samples [62.5%]; 95% CI for the difference, −8.3% to +2.4% among stages I through III combined), the regression model was 3.5% more specific than Western blotting (99.1% versus 95.6%; 95% CI for the difference, 1.9% to 5.1%); this difference translated to a 24.4% improvement in specificity among EIA-reactive controls.
Assay precision.
For VlsE-IgG, the within-site coefficient of variation ranged from 19.2% for negative samples to 5.8% for highly positive samples; the between-site coefficient of variation ranged from 13.4% for moderately positive samples to 11.6% for negative samples and 5.4% for highly positive samples. For pepC10-IgM, the within-site coefficient of variation ranged from 15.3% for negative samples to 9.8% for highly positive samples; the between-site coefficient of variation ranged from 10.6% for low-positive samples to 7.5% for negative samples and 1.3% for highly positive samples. The dynamic range for each antibody was approximately 4 log AtheNA units (data not shown).
Comparison of multiplex assay to Western blotting.
The standard 2-tier model enjoys a specificity advantage over single-tier assays: because only EIA-reactive samples are evaluated further, sera positive by Western blotting but negative by EIA are eliminated from consideration. Some studies suggest that Western blot specificity is improved 8% to 9% by including a first-tier evaluation (11, 18, 25). The same phenomenon was observed when the multiplex assay utilized a 60% pROC classifier as the second tier of a 2-tiered approach: an initial EIA improved the overall specificity of the multiplex assay by 10.3% (i.e., from 85.3% to 95.6%).
Employing the 60% pROC classifier as the second tier of a 2-tiered model, the multiplex assay was 20.7% more sensitive than Western blotting for early-convalescent-phase disease (Table 3) and 12.5% more sensitive for stages I through III combined (156/208 samples [75%] versus 130/208 samples [62.5%]; 95% CI for the difference, 8.1% to 17.1%; P = 0.008). Because the early-acute-phase and convalescent-phase sera in our study were not from independent groups, we also evaluated classifier performance by using only early-convalescent-phase and stage II/III sera (constituting all group 1 samples). The multiplex assay was 16.3% more sensitive than Western blotting with the latter group (120/129 samples [93.0%] versus 99/129 samples [76.7%]; 95% CI for the difference, 10.0% to 23.4%; P = 0.014).
Table 3.
Sensitivity of Western blotting versus multiplex assay by disease stage
| Stage (no. of cases) | Sensitivity (no. [%] of positive samples) |
||||
|---|---|---|---|---|---|
| IgG blotting | IgM blotting | Either IgG or IgM blotting | 2-Tier blottinga | Multiplex assayb | |
| Early acute phase (79) | 6 (7.6) | 29 (36.7) | 31 (39.2) | 31 (39.2) | 36 (45.7) |
| Early convalescent phase (82) | 17 (20.7) | 60 (73.2) | 63 (76.8) | 56 (68.3) | 73 (89.0)c |
| Stages II and III combinedd (47) | 39 (83.0) | 32 (68.1) | 45 (95.7) | 43 (91.5) | 47 (100) |
| Neuro/carditise (stage II) (18) | 11 (61.1) | 13 (72.2) | 16 (88.8) | 15 (83.3) | 18 (100) |
| Arthritis (stage III) (29) | 28 (96.6) | 19 (65.5) | 29 (100) | 28 (96.6) | 29 (100) |
| Posttreatmentf stages II and III (16) | 13 (81.25) | 4 (25) | 13 (81.3) | 13 (81.3) | 16 (100) |
| Posttreatmentf stage I (18) | 4 (22.2) | 9 (50.0) | 10 (55.6) | 4 (22.2) | 11 (61.1)g |
Standard 2-tier criteria were used for blot interpretation (9). Only IgG blots were used for diagnosis more than 30 days after disease onset.
Employed as a second-tier test, using the 60% pROC classifier trained on early acute disease; the cutoff was set at 95.6% specificity.
Difference in sensitivity versus 2-tier blot results, 20.7% (95% CI, 12.1 to 30.9%; P = 0.0009).
All samples were group 1 sera. The difference in sensitivity of the multiplex assay versus 2-tier blotting was 8.5% (95% CI, 1.8% to 17.8%; P = 0.042).
Early neurological disease or myocarditis.
Post-antibiotic-treatment samples were group 2 sera.
Difference in sensitivity versus 2-tier blot results, 38.9% (95% CI, 6.7% to 61.7%; P = 0.041).
The false-positive rates of both assays were identical (Table 4). Although positive assays among healthy blood donors from areas of endemicity might be related to past B. burgdorferi infection, the majority of false-positive results in our study were due to IgM rather than IgG blots; prior infection might have resulted in more positive IgG blots than we observed (41).
Table 4.
False-positive rates of Western blotting versus multiplex assaya
| Control group (no. of patients) | No. (%) of false-positive results |
|
|---|---|---|
| Western blot | Multiplex assay | |
| Blood donors from area of endemicity (300) | 15 (5.0) | 18 (6.0) |
| Blood donors from area where LD is not endemic (150) | 7 (4.7) | 2 (1.3) |
| Donors with cross-reacting conditions (95) | 2 (2.1) | 4 (4.2) |
| Total (545) | 24 (4.4) | 24 (4.4) |
The cutoff for the 60% partial ROC classifier was set to generate the same false-positive rate as that for Western blotting.
Four multiplex-positive samples from patients with stage II/III disease were EIA positive in our laboratory but Western blot negative by standard 2-tier criteria (MarDx, Carlsbad, CA) (9). There was enough serum remaining to retest 3 of the 4 samples at a second reference laboratory (A. Steere, Massachusetts General Hospital, Boston, MA), using a Western blot with a VlsE stripe from a different manufacturer (Viralab, Oceanside, CA). On retesting, sera from 2 patients with early neurological disease were positive by EIA and IgM blotting but failed to meet standard 2-tier criteria because they were collected 45 and 64 days after disease onset (37); it is likely that both patients had Lyme disease because (i) recent studies suggest that IgM blots may be useful for diagnosis of neurological disease within 6 weeks of onset (40) and (ii) the first patient demonstrated an IgG-VlsE band and the second patient had EM. A third patient, seen in 1982, had EM and flu-like symptoms followed 1 month later by the onset of facial palsy and meningitis; serologic testing was not available at that time. Serology in the current study was positive only by EIA, but additional serum was not available for retesting by the second reference laboratory. The fourth sample came from a patient with arthritis and was positive by both EIA and IgG blotting on repeat testing. It is possible that differences in Western blot reagents could have contributed to the discordant results between reference laboratories. On the whole, our results suggest no significant difference in test performance between the multiplex assay and Western blotting for the 47 group 1 sera from patients with stage II/III disease (Table 3). Although the multiplex assay was marginally more sensitive than Western blotting for post-antibiotic-treatment sera (group 2), persistently positive serology by either method was not indicative of treatment failure.
The multiplex assay was slightly more sensitive than the C6 IgG/IgM EIA among Western blot-positive sera from data set B (93% versus 86%) and among early-convalescent-phase sera from data set A (89% versus 85%), although neither difference was statistically significant. The multiplex assay was otherwise equivalent to C6 IgG/IgM EIA and was equally specific (96%).
DISCUSSION
The current study evaluated a multiplex microsphere assay for LD diagnosis using VlsE1-IgG and pepC10-IgM antibodies. Because multiplex systems can perform multiple tests simultaneously in the same sample well, this technology lends itself particularly to the study of LD, an illness with a complex multiantibody host immune response (39). We explored the use of regression classifiers to generate a single diagnostic score from two separate antibody levels; given the importance of high specificity for diagnostic tests for Lyme disease (39), partial ROC regression models were utilized to maximize multiplex performance at specificities of ≥95%. The multiplex assay used in this study performed as well as or better than Western blotting as a second-tier test.
When the sensitivities of the 95% pROC regression model and Western blotting were equivalent, the 95% pROC model was 3.5% more specific than Western blotting (95% CI, 1.9% to 5.1%). When the specificities of the 60% pROC regression model and Western blotting were equivalent, the 60% pROC regression model was significantly more sensitive than Western blotting, being 20.7% more sensitive for early-convalescent-phase disease (95% CI, 12.1% to 30.9%) and 12.5% more sensitive overall (95% CI, 8.1% to 17.1%); about 2/3 of the improvement in overall sensitivity was related to better detection of early-convalescent-phase disease.
No one classifier was superior under all conditions. If the objective of testing is to rule out Lyme disease in a low-risk setting, then the 95% pROC model is a reasonable choice because of its high specificity; in some instances, the pretest risk of LD may be low enough to justify deferring testing altogether (Appendix B). If the clinical picture suggests stage II or III Lyme disease, then the 60% pROC model may be preferred because of its high sensitivity.
Logistic likelihood regression analysis is one of the most commonly used statistical methods for both disease classification and prediction; it has been used to interpret the antibody response to B. burgdorferi by Western blotting (24), kinetic EIA (34), and flagellin-based EIA (13). Unlike logistic models, which maximize the likelihood of disease at a given specificity, ROC regression methods maximize the AUC (29). Pepe et al. (31) demonstrated that regression models that optimize the AUC can offer advantages over logistic models when selected biomarkers are combined. Data from set A demonstrated that full ROC regression and 60% partial ROC regression models were significantly more sensitive than the logistic model at Western blot specificity (95.6%), reinforcing the value of AUC optimization methods for classifier development.
Western blotting was only 69.2% specific among our EIA-reactive control sera, reducing its overall specificity to 95.6%; this specificity is lower than that reported by other investigators (5, 40). Of all false-positive Western blots in the current study, 79% were due solely to IgM antibody, illustrating the limitations associated with that assay (36). If achieving 99% specificity among the healthy population is an important benchmark from a public health perspective (2), then specificity among our EIA-reactive controls would need to be improved to at least 93.6%. Second-tier approaches have been proposed that eliminate IgM blotting by using IgG Western blots in conjunction with a VlsE band (5). The multiplex assay described above offers another alternative.
There are multiple limitations to the current study. The number of patients with stage II and III disease was insufficient to detect significant differences in assay performance. Some samples from data set A and all samples from data set B were collected retrospectively, potentially biasing the study population (30). We were unable to produce stage-specific classifiers, but it is possible that expanding the number of antibodies assayed might help to achieve that goal (e.g., IgGs to DbpA and BmpA). Although we detected benefits from utilizing VlsE1-IgG and pepC10-IgM antibodies together, we cannot extrapolate our results to other antibody combinations. Each test panel requires careful evaluation of classifier performance over a range of acceptable specificities. Because there was no clinical information accompanying the sera in data set B, we cannot be certain how many 2-tier test-positive patients actually had Lyme disease. We did not assess the role of paired acute- and convalescent-phase serology; demonstrating expansion of the IgG immune response by Western blotting may be helpful in diagnosing recent disease (40).
A full decision-analytic evaluation comparing the multiplex assay to Western blotting is warranted but is beyond the scope of this study. We did not calculate costs or benefits related to different clinical outcomes, nor did we provide a means to determine the pretest probability of LD. Formal decision analysis requires these elements, along with knowledge of intrinsic test performance, to help guide test utilization (Appendix B). Integrating pretest risk assessment into clinical workflow is a goal that has not yet been realized; computer-based decision support systems may soon assist with that assessment (23).
A prospective study using the current multiplex assay, particularly for patients with stage II or III disease, would address the above methodological issues and provide guidance to help integrate clinical with laboratory information. The score that we create through our regression model can be expressed as a likelihood ratio and utilized in a Bayesian context (i.e., pretest and posttest probabilities). We believe that the multiplex platform with employment of ROC regression techniques offers substantial promise for improving Lyme disease diagnosis.
ACKNOWLEDGMENTS
We thank Yingying Fan of the Marshall School of Business, University of Southern California, Los Angeles, CA, for her statistical contributions to this work; Hoshang Batliwala of Zeus Scientific, Branchburg, NJ, and Karen Hughes of the Massachusetts General Hospital, Boston, MA, for their laboratory support; and Brad Biggerstaff of the Centers for Disease Control and Prevention, Fort Collins, CO, for his statistical advice.
The findings and conclusions in this article are those of the authors and do not necessarily represent the views of the Centers for Disease Control and Prevention.
Financial support was provided by SBIR-AT-NIAID grants 1R43AI069564-01 and -01S1 to Infectious Disease Consultants, PC, from the National Institute of Allergy and Infectious Diseases, National Institutes of Health, and by a grant from Zeus Scientific, Inc., to Health Research, Inc., Menands, NY, a nonprofit organization which supported the work performed by S.J.W. and K.K. at the NYSDOH, Albany, NY, for this study.
R.B.P. holds 2 patents on bioinformatic methods for Lyme disease diagnosis and has affiliations with Zeus Scientific, Inc., and Infectious Disease Consultants, PC. C.G.H., L.L., and J.F. were consultants to Infectious Disease Consultants, PC. M.K. is an employee of Zeus Scientific, Inc. B.J.B.J., A.C.S., K.K., and S.J.W. report no conflicts of interest.
APPENDIX A
Partial ROC regression classifier. The score function is derived from a linear combination of test results, βTY, where D is the disease, Y1,..., Yk is a set of k diagnostic tests for D, Y is a vector of diagnostic test results y1,..., yk, D′ is not D, β is a vector of coefficients β1,..., βk for Y, and βT is the transpose of β. For a given cutoff value, c, a test is positive if βTY ≥ c.
With ROC regression, the test panel and β coefficients are chosen simultaneously to maximize the AUC of the empirical ROC, as approximated by the following equation:
where I is the indicator function, N is the total number of study subjects, nD is the number of patients with disease D, nH is the number of healthy controls, nD + nH = N, i = 1,..., nD, i ∈ D are patients with disease, j = 1,..., nH, and j ∈ H are healthy controls. The ROC curve is smoothed using the sigmoid function as follows:
wherein bias related to values of x close to zero is reduced by introducing a series of positive numbers, σn, such that Sn(x) = S(x/σn) and σn approaches zero as n approaches infinity (29). An optimal set of β coefficients is determined by an iterative gradient descent algorithm using the sigmoid maximum rank correlation estimator (SMRC) described by Ma and Huang (22, 29), as follows:
Raw test results are transformed into a likelihood ratio, and a logistic likelihood model is used to select the initial β coefficients and anchor marker. If feature selection is desired, then a gradient LASSO is applied to the SMRC; for tuning, an L1 constraint of ≤u is chosen using a V-fold cross-validation technique (26, 29). If the regression features are already known, as in the current study, then the SMRC alone is used to optimize β.
If t0 is the maximum false-positive rate permitted by a physician interpreting the tests and is a multiple of 1/nH, then the β coefficients and test panel are chosen simultaneously through partial ROC regression in order to generate the largest area below the partial ROC curve for the (1 − t0) quantile of individuals without disease; the score cutoff, c, is chosen such that SH(c) = t0 (the survival function of patients without disease with a score of c) when the score function βTYj ≥ c. The features are fitted to a truncated set of controls by using the above sigmoid maximum rank correlation estimator and gradient LASSO (14, 26, 29, 34). If the features are already known, then the SMRC estimator alone is used to optimize β; we observed estimator convergence within 100 iterations.
APPENDIX B
Test/no-test cutoff = [1 + B(sensitivity)/C(1 − specificity)]−1;
where B is the regret associated with failing to treat disease, C is the regret associated with treating someone without disease, and the sensitivity and specificity of a given test are known (4).
Footnotes
Published ahead of print on 2 March 2011.
REFERENCES
- 1. Aguero-Rosenfeld M. E., Wang G., Schwartz I., Wormser G. P. 2005. Diagnosis of lyme borreliosis. Clin. Microbiol. Rev. 18:484–509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bacon R. M., et al. 2003. Serodiagnosis of Lyme disease by kinetic enzyme-linked immunosorbent assay using recombinant VlsE1 or peptide antigens of Borrelia burgdorferi compared with 2-tiered testing using whole-cell lysates. J. Infect. Dis. 187:1187–1199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Barbour A. G., et al. 2008. A genome-wide proteome array reveals a limited set of immunogens in natural infections of humans and white-footed mice with Borrelia burgdorferi. Infect. Immun. 76:3374–3389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Biggerstaff B. J. 2000. Comparing diagnostic tests: a simple graphic using likelihood ratios. Stat. Med. 19:649–663 [DOI] [PubMed] [Google Scholar]
- 5. Branda J. A., et al. 2010. 2-Tiered antibody testing for early and late Lyme disease using only an immunoglobulin G blot with the addition of a VlsE band as the second-tier test. Clin. Infect. Dis. 50:20–26 [DOI] [PubMed] [Google Scholar]
- 6. Burbelo P. D., et al. 2010. Rapid, simple, quantitative, and highly sensitive antibody detection for Lyme disease. Clin. Vaccine Immunol. 17:904–909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. CDC 1997. Case definitions for infectious conditions under public health surveillance. MMWR Morb. Mortal. Wkly. Rep. 46(RR-10):14–15 [PubMed] [Google Scholar]
- 8. CDC 1990. Case definitions for public health surveillance. MMWR Morb. Mortal. Wkly. Rep. 39(RR-13):19–21 [PubMed] [Google Scholar]
- 9. CDC 1995. Recommendations for test performance and interpretation from the second National Conference on Serologic Diagnosis of Lyme Disease. MMWR Morb. Mortal. Wkly. Rep. 44:590–591 [PubMed] [Google Scholar]
- 10. CDC 2010. Summary of notifiable diseases—United States, 2008. MMWR Morb. Mortal. Wkly. Rep. 57:36. [PubMed] [Google Scholar]
- 11.CDC/ASTPHLD. Proceedings of the Second National Conference on Serologic Diagnosis of Lyme Disease; CDC/ASTPHLD, Dearborn, MI. 1994. [Google Scholar]
- 12. Coleman A. S., et al. 22 December 2010. BBK07 immunodominant peptides as serodiagnostic markers of Lyme disease. Clin. Vaccine Immunol. 18:406–413 doi:10.1128/CVI.00461-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Dessau R. B., Ejlertsen T., Hilden J. 2010. Simultaneous use of serum IgG and IgM for risk scoring of suspected early Lyme borreliosis: graphical and bivariate analyses. APMIS 118:313–323 [DOI] [PubMed] [Google Scholar]
- 14. Dodd L. E., Pepe M. S. 2003. Partial AUC estimation and regression. Biometrics 59:614–623 [DOI] [PubMed] [Google Scholar]
- 15. Dressler F., Whalen J. A., Reinhardt B. N., Steere A. C. 1993. Western blotting in the serodiagnosis of Lyme disease. J. Infect. Dis. 167:392–400 [DOI] [PubMed] [Google Scholar]
- 16. Efron B., Tibshirani R. 1995. An introduction to the bootstrap. Chapman and Hall, New York, NY [Google Scholar]
- 17. Embers M. E., Jacobs M. B., Johnson B. J., Philipp M. T. 2007. Dominant epitopes of the C6 diagnostic peptide of Borrelia burgdorferi are largely inaccessible to antibody on the parent VlsE molecule. Clin. Vaccine Immunol. 14:931–936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Engstrom S. M., Shoop E., Johnson R. C. 1995. Immunoblot interpretation criteria for serodiagnosis of early Lyme disease. J. Clin. Microbiol. 33:419–427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Food and Drug Administration 2010. In vitro diagnostics. Food and Drug Administration, Silver Spring, MD: http://www.accessdata.fda.gov/scripts/cdrh/cfdocs/cfivd/index.cfm Accessed 10 August 2010 [Google Scholar]
- 20. Fulton R. J., McDade R. L., Smith P. L., Kienker L. J., Kettman J. R., Jr 1997. Advanced multiplexed analysis with the FlowMetrix system. Clin. Chem. 43:1749–1756 [PubMed] [Google Scholar]
- 21. Goodman J. L., Dennis D. T., Sonenshine D. E. (ed.). 2005. Tick-borne diseases of humans. ASM Press, Washington, DC [Google Scholar]
- 22. Han A. K. 1987. Non-parametric analysis of a generalized regression model. J. Econom. 35:303–316 [Google Scholar]
- 23. Hejlesen O. K., Olesen K. G., Dessau R., Beltoft I., Trangeled M. 2005. Decision support for diagnosis of Lyme disease. Stud. Health Technol. Inform. 116:205–210 [PubMed] [Google Scholar]
- 24. Honegr K., et al. 2001. Criteria for evaluation of immunoblots using Borrelia afzelii, Borrelia garinii and Borrelia burgdorferi sensu stricto for diagnosis of Lyme borreliosis. Epidemiol. Mikrobiol. Imunol. 50:147–156 [PubMed] [Google Scholar]
- 25. Johnson B. J., et al. 1996. Serodiagnosis of Lyme disease: accuracy of a two-step approach using a flagella-based ELISA and immunoblotting. J. Infect. Dis. 174:346–353 [DOI] [PubMed] [Google Scholar]
- 26. Kim Y., Kim J. 10 August 2010, accession date Gradient LASSO for feature selection. Proceedings of the 21st International Conference on Machine Learning, Banff, Canada. www.machinelearning.org/icml.html [Google Scholar]
- 27. Lawrenz M. B., et al. 1999. Human antibody responses to VlsE antigenic variation protein of Borrelia burgdorferi. J. Clin. Microbiol. 37:3997–4004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Liang F. T., et al. 1999. Sensitive and specific serodiagnosis of Lyme disease by enzyme-linked immunosorbent assay with a peptide based on an immunodominant conserved region of Borrelia burgdorferi VlsE. J. Clin. Microbiol. 37:3990–3996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Ma S., Huang J. 2005. Regularized ROC method for disease classification and biomarker selection with microarray data. Bioinformatics 21:4356–4362 [DOI] [PubMed] [Google Scholar]
- 30. Pepe M. S. 2003. The statistical evaluation of medical tests for classification and prediction. Oxford University Press, Oxford, United Kingdom [Google Scholar]
- 31. Pepe M. S., Cai T., Longton G. 2006. Combining predictors for classification using the area under the receiver operating characteristic curve. Biometrics 62:221–229 [DOI] [PubMed] [Google Scholar]
- 32. Pepe M. S., Feng Z., Janes H., Bossuyt P. M., Potter J. D. 2008. Pivotal evaluation of the accuracy of a biomarker used for classification or prediction: standards for study design. J. Natl. Cancer Inst. 100:1432–1438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Philipp M. T., Marques A. R., Fawcett P. T., Dally L. G., Martin D. S. 2003. C6 test as an indicator of therapy outcome for patients with localized or disseminated Lyme borreliosis. J. Clin. Microbiol. 41:4955–4960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Porwancher R. 13 March 2008. Bioinformatic approach to disease diagnosis. Publication no. US20080064118. United States Patent and Trademark Office, Alexandria, VA [Google Scholar]
- 35. Porwancher R. 2003. Improving the specificity of recombinant immunoassays for Lyme disease. J. Clin. Microbiol. 41:2791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Porwancher R. 1999. A reanalysis of IgM Western blot criteria for the diagnosis of early Lyme disease. J. Infect. Dis. 179:1021–1024 [DOI] [PubMed] [Google Scholar]
- 37. Rahn D. W., Evans J. (ed.). 1998. Lyme disease. American College of Physicians, Philadelphia, PA [Google Scholar]
- 38. Reid M. C., Schoen R. T., Evans J., Rosenberg J. C., Horwitz R. I. 1998. The consequences of overdiagnosis and overtreatment of Lyme disease: an observational study. Ann. Intern. Med. 128:354–362 [DOI] [PubMed] [Google Scholar]
- 39. Steere A. 2010. Borrelia burgdorferi (Lyme disease, Lyme borreliosis), p. 3071–3081 In Mandell G., Bennett J., Dolin R., Mandell D. (ed.), Bennett's principles and practice of infectious diseases, 7th ed., vol. 2. Churchill Livingstone, Philadelphia, PA [Google Scholar]
- 40. Steere A. C., McHugh G., Damle N., Sikand V. K. 2008. Prospective study of serologic tests for Lyme disease. Clin. Infect. Dis. 47:188–195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Steere A. C., Sikand V. K., Schoen R. T., Nowakowski J. 2003. Asymptomatic infection with Borrelia burgdorferi. Clin. Infect. Dis. 37:528–532 [DOI] [PubMed] [Google Scholar]