Abstract
Open reading frame 2 (ORF2) of porcine circovirus type 2 (PCV2) codes for the 233-amino-acid capsid protein (CP). Baculovirus-based vaccines that express only ORF2 are protective against clinical disease following experimental challenge or natural infection. The goal of this study was to identify regions in CP preferentially recognized by sera from experimentally infected and vaccinated pigs and to compare these responses to those of pigs diagnosed with porcine circovirus-associated disease (PCVAD), including porcine multisystemic wasting syndrome (PMWS) and porcine dermatitis and nephropathy syndrome (PDNS). The approach was to react porcine sera with CP polypeptide fragments followed by finer mapping studies using overlapping oligopeptides that covered amino acids 141 to 200. The results showed that vaccinated pigs preferentially recognized only the largest polypeptide fragment, CP(43-233). A subset of experimentally infected pigs and pigs with PDNS showed strong reactivity against a CP oligopeptide, 169-STIDYFQPNNKR-180. Alanine scanning identified Y-173, F-174, Q-175, and K-179 as important for antibody recognition. The results from this study support the notion of PCV2 modulation of immunity, including antibody responses that may represent a precursor for disease. The recognition of CP(169-180) and other polypeptides provides opportunities to devise diagnostic tests for monitoring the immunological effectiveness of vaccination.
INTRODUCTION
First described in Canada in the early 1990s, porcine circovirus-associated disease (PCVAD) has emerged as an economically important disease worldwide (1, 48). A central feature of PCVAD is the involvement of porcine circovirus type 2 (PCV2). PCVAD encompasses a group of complex, multifactorial diseases, of which porcine multisystemic wasting syndrome (PMWS) and porcine dermatitis and nephropathy syndrome (PDNS) are common syndromes (7, 49, 52, 53). Even though Krakowka et al. (25) reported the appearance of PDNS in gnotobiotic pigs in the absence of PCV2, pigs with clinical PDNS possess high levels of PCV2-specific antibodies, which are implicated in disease progression (53). In 2008, poor growth performance in herds without overt clinical signs was reported as another manifestation of PCVAD (20). Factors such as host genetics, other infectious agents, and the pathogenic potential of the PCV2 isolate contribute to the disease (2, 3, 14, 23, 24, 40, 41, 43).
The majority of PCV2 isolates can be divided into one of two genotypes, known as PCV2a and PCV2b (11, 19, 39). A third genotype, designated PCV2c, describes a small group of historical isolates found in Denmark (48). The genotypic classification of PCV2 is complicated by field isolates composed of PCV2a and PCV2b sequences (18, 32). The PCV2 genome is dominated by three open reading frames (ORFs). The 233-amino-acid capsid protein (CP), coded for by ORF2, forms a homopolymer that surrounds the single-stranded ambisense 1.7-kb DNA genome (38). The principal sequence differences between PCV2a and PCV2b genotypes localize to ORF2, where the nucleotide and peptide sequence identities are approximately 93%. Baculovirus-based vaccines that express PCV2 ORF2 are sufficient to offer protection from disease (4, 15, 19, 31, 42, 51). ORF1 is 945 nucleotides (nt) in length and codes for two replicase proteins, Rep and Rep′ (12, 34). A third gene, ORF3, is in a different reading frame embedded within ORF1 and codes for a protein associated with apoptosis (28). The significance of ORF3 with regard to the onset and severity of PCVAD is still unknown (9).
The current model for the virion capsid structure locates the arginine-rich N-terminal end of CP projecting inward, where it interacts with the viral genome. Based on studies of another circovirus, psittacine beak and feather disease virus, the arginine-rich domain functions as a nuclear localization signal sequence, which shuttles the viral DNA across the nuclear pore complex and into the nucleus, the site of PCV2 replication (10, 17, 29, 37).
Previous studies describing the humoral response following PCV2 infection indicate that seroconversion occurs between 10 and 28 days postinfection or after vaccination (1, 35, 45). There are reports that pigs with PMWS seroconvert later than or produce a reduced antibody titer compared to subclinically infected animals (5, 35). Another difference between subclinical infection and PMWS is reduced neutralizing antibody (NA) titers in PMWS pigs compared to those in subclinally infected or vaccinated pigs (16, 45). There is no PCV2-based model system that can reproduce PDNS. However, clinically moribund pigs show a hyperimmune response leading to significant antibody production, which may contribute to immune complex formation and PDNS (56).
For the purpose of mapping antibody epitopes in CP, Lekcharoensuk et al. (26) used a panel of seven anti-CP monoclonal antibodies (MAbs), which were reacted with cells transfected with infectious PCV chimeric DNA clones composed of different combinations of PCV1 and PCV2a ORF2 sequences. Immunoreactive regions were reported between residues 47 and 85, 165 and 200, and 200 and 233. Using overlapping oligopeptides and sera from infected and immunized pigs, Mahé et al. (33) described antibody-reactive regions between amino acids 23 and 43, 71 and 85, 117 and 131, and 171 and 202 in CP. In a follow-up study, Truong et al. (55) identified amino acids 117 to 131 as a dominant antibody recognition region. Shang et al. (50) reported four regions in CP associated with MAb recognition: amino acids 156 to 162, 175 to 192, 195 to 202, and 228 to 233.
The purpose of this study was to test the hypothesis that sera from pigs experimentally infected or vaccinated or with clinically diagnosed PCVAD produce different responses to epitopes within PCV2 CP. The original intent was to identify specific epitopes that could offer protection versus those epitopes involved in immunopathogenesis. The epitope mapping methodology incorporated in this study is an extension of previous work demonstrating that bacterially expressed CP antigens react with antibody from PCV2-infected pigs (30, 38, 54). We extended this approach by evaluating the reactivities of individual polypeptide fragments to different combinations of epitopes followed by finer epitope mapping using overlapping oligopeptides. The results describe several different recognition patterns within the different groups of pigs, including the identification of a single epitope, 169-STIDYFQPNNKR, which was preferentially recognized by pigs diagnosed with PDNS and a subset of pigs experimentally infected with PCV2. Alanine substitutions within CP(169-180) showed that Y-173, F-174, Q-175, and K-179 amino acid residues contribute to antibody recognition. The results from this study support the notion of immune dysregulation, characterized by a hyperimmune response during PDNS and a diminished response during PMWS. Furthermore, the methods incorporated in this study provide a means for characterizing the immune response upon vaccination, natural infection, and disease.
MATERIALS AND METHODS
Experimental PCV2 infection.
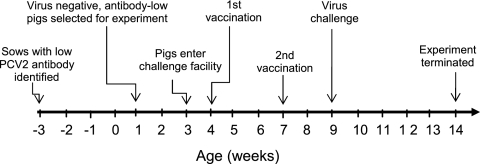
All animal experiments were performed after review and approval by Kansas State University Institutional Animal Use and Biosafety committees. PCV2-negative pigs were derived from sows with low PCV2 antibody titers (44). Upon entry into the challenge facility, pigs were confirmed as negative for PCV2 DNA by PCR. At the time of virus challenge, pigs were confirmed as negative for PCV2 DNA and PCV2 antibody as measured by PCR and indirect fluorescent-antibody (IFA) assay, respectively. Pigs were randomly assigned to one of seven groups, summarized in Table 1. The timeline for vaccination and infection is diagrammed in Fig. 1. PCV2 vaccination was performed using a two-dose commercial baculovirus-expressed PCV2 ORF2 product (Intervet) administered at 4 and 7 weeks of age according to label instructions. Two weeks after the last vaccine dose, pigs were challenged with PCV2 or PCV2 plus porcine reproductive and respiratory syndrome virus (PRRSV). The PCV2 challenge inoculum consisted of a lymph node homogenate from a pig with PMWS. Virus from the homogenate was titrated on swine testicle (ST) cells in quadruplicate on a 96-well plate. Three days after infection, the cells were fixed and stained with fluorescein isothiocyanate (FITC)-labeled anti-PCV (Veterinary Medical Research and Development, Inc. [VMRD]). The 50% tissue culture infectious dose (TCID50) per ml was calculated according to the Reed-Muench method (47). The concentration of virus in the challenge homogenate was determined to be approximately 108 TCID50/ml. The inoculum was negative for other common pathogens, such as porcine parvovirus and swine influenza virus, but was positive for PRRSV. The homogenate was filtered to remove bacteria. The rationale for using a homogenate was based on the inability to obtain significant quantities of PCV2b in culture. PRRSV and other heat-labile infectious agents were removed by heat inactivation at 60°C for 30 min. Pigs were challenged intranasally with approximately 105 TCID50 of PCV2. For dual challenge, 105 TCID50 of PRRSV was added back to the PCV2 inoculum. All pigs were monitored for clinical signs and bled weekly. Sera were assayed for PCV2 and PRRSV nucleic acid and virus-specific antibodies by the Kansas State Veterinary Diagnostic Laboratory (KSVDL) using standard molecular and serological diagnostic techniques. The experiment was terminated at 6 weeks after virus challenge. Serum samples used for this study were obtained at the conclusion of the experiment or 5 weeks after infection and 7 weeks after vaccination.
Table 1.
Experimental infection/vaccination treatment groups
| Group | Description | n | Treatment |
||
|---|---|---|---|---|---|
| Vaccine | PCV2 | PRRSV | |||
| 1 | Control | 7 | − | − | − |
| 2 | Vaccine | 7 | + | − | − |
| 3 | PRRSV | 7 | − | − | + |
| 4 | PCV2 | 7 | − | + | − |
| 5 | PCV2/vaccine | 7 | + | + | − |
| 6 | PCV2/PRRSV | 7a | − | + | + |
| 7 | PCV2/PRRSV/vaccine | 7 | + | + | + |
Three of the seven pigs died following infection and were not subjected to further study.
Fig. 1.
Timeline for experimental PCV2 vaccination and infection.
Selection of PCVAD pigs.
PMWS and PDNS pigs were submitted as diagnostic cases to the KSVDL. The diagnosis of PDNS or PMWS was made as previously described (20). The PMWS pigs presented as emaciated with greatly enlarged superficial inguinal lymph nodes. Histologically, secondary lymphoid organs were almost completely depleted of lymphocytes. PCV2 antigen, as determined by diagnostic immunohistochemistry (IHC) and PCR, was present in histological lesions. PDNS pigs were identified by the presence of multifocal erythematous lesions on the hindquarters. At the anatomical level, the kidneys were greatly enlarged with cortical petechiae covering the kidney surface. The glomeruli were swollen and fibrinous with necrosis of glomerular tufts. All pigs were confirmed to be infected with a PCV2b virus. For the purposes of this study, 10 PDNS and 10 PMWS serum samples were selected.
PCV2 IFA and measurement of virus-neutralizing activity.
Total PCV2 antibodies were measured by indirect fluorescent-antibody (IFA) staining of infected cells. Rapidly dividing ST cells, maintained in minimum essential medium (MEM) with 10% fetal bovine serum (FBS) and antibiotics, were infected with a laboratory isolate of PCV2 on 96-well plates. After 3 days, the plates were fixed for 10 min in 80% acetone. An initial dilution of 1:40 was followed by 1:2 dilutions of each serum sample. Samples were diluted in phosphate-buffered saline (PBS) with 5% FBS (PBS-FBS) and incubated with antigen for 2 h at room temperature. After being washed in PBS, bound antibody was detected with FITC-labeled anti-pig antibody (Jackson Labs) according to the manufacturer's recommendations. Plates were read on an inverted fluorescence microscope, and titers were calculated as the reciprocal of the last serum dilution that showed fluorescence staining. For measurement of virus-neutralizing activity, serial dilutions of serum in 100 μl of cell culture medium were mixed with a constant quantity of PCV2 (50 to 300 TCID50), incubated for 1 h at 37°C, and placed onto four replicate wells of 1-day-old ST cells in 96-well plates. Plates were incubated for 3 days at 37°C and then fixed and stained with FITC-labeled anti-PCV2 (VMRD). The log2 50% neutralizing activity (NA50) endpoint was calculated by the method of Spearman and Karber (14a).
Cloning, expression, and purification of recombinant PCV2 polypeptides.
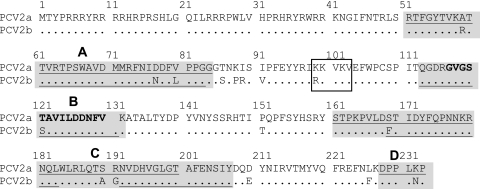
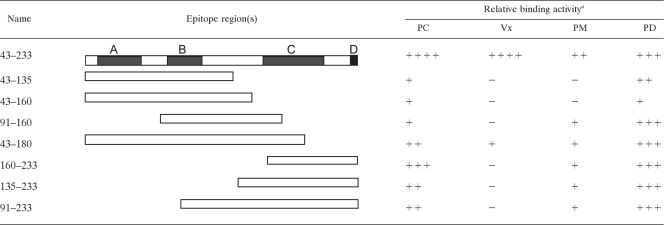
Previous immunological studies identified several antibody recognition sites in CP, including at least one immunorelevant epitope (26, 33, 55). The epitope recognition regions from these previous studies were combined and summarized in Fig. 2. From these studies, we located four general antibody recognition regions, labeled A (51 to 84), B (113 to 131), C (161 to 207), and D (228 to 233). The CP amino acid sequence of the PCV2 used to infect pigs is the same PCV2b genotype as the CP used to prepare polypeptide fragments (GenBank accession no. HQ713495). CP was 99% identical to the CP of the PCV2b used for experimental infection. There were two amino acid mutations: phenylalanine to asparagine at position 46 and phenylalanine to aspartic acid at position 115. To prepare recombinant CP polypeptides, primers were designed to amplify regions of CP containing one or more of the antibody recognition regions. The 5′ ends of the forward and reverse primers contained additional SacII and HindIII restriction sites, respectively. The primers used for amplification and cloning of the individual ORF2 cDNA fragments are listed in Table 2. PCR products were cloned into PCR2.1 Topo (Promega), and plasmids were propagated in Escherichia coli. Plasmids were cut with SacII and HindIII, and the inserted DNA was cloned in frame into the SacII and HindIII sites in the E. coli expression vector pHUE (6). The pHUE vector is designed to yield a 6×His ubiquitin fusion protein. For recombinant protein expression, E. coli was grown in LB plus ampicillin at 37°C until bacteria reached an optical density at 600 nm (OD600) of 0.4 to 0.6. Protein expression was induced by the addition of IPTG (isopropyl-β-d-thiogalactopyranoside; 1.0-μg/ml final concentration), and cultures were grown for an additional 4 h. Bacteria were harvested, and proteins were purified using a PrepEase His-tagged protein purification kit (USB) according to the manufacturer's protocol. A modification of the procedure included the addition of urea at a final concentration of 8 M to the bacterial disruption and Ni column elution buffers. After elution from the nickel column, the protein-containing fractions were concentrated on a 5,000-molecular-weight (MW)-cutoff spin column (Millipore), and total protein was measured using a protein assay (Bio-Rad) according to the manufacturer's protocol. The purified proteins were visualized by SDS-PAGE.
Fig. 2.
Locations of immunoreactive regions in PCV2 CP. Underlined regions in the PCV2a genotype peptide sequence show the locations of the immunoreactive regions described by Lekcharoensuk et al. (26) using MAbs prepared against whole purified virus (strain ISU 31, GenBank accession number AJ223185). The underlined regions in the PCV2b genotype sequence (GenBank accession number AF201311) summarize the results obtained from the pepscan analysis by Mahé et al. (33). The epitope maps are combined to yield the shaded peptide sequences, identified as epitope regions A, B, C, and D. Amino acids in bold show the location of an immunodominant region described by Truong et al. (55). The box identifies the putative receptor binding region described in the work of Misinzo et al. (36).
Table 2.
Primer sequences used for preparing CP polypeptidesa
| CP region (amino acids) | Forward primer | Reverse primer |
|---|---|---|
| 43–135 | 5′CCGCGGTGGTAATGGCATCTTCAACA | 5′AAGCTTTTAGGCTGTGGCCTTTGATA |
| 91–233 | 5′CCGCGGTGGAGTGCCCTTTGAATACT | 5′GCGCAAGCTTTTAAGGGTTAAGTGGC |
| 136–233 | 5′CCGCGGTGGACTCACCTATGACCCCT | 5′GCGCAAGCTTTTAAGGGTTAAGTGGC |
| 91–160 | 5′CCGCGGTGGAGTGCCCTTTGAATACT | 5′AAGCTTTTAGTAGCGGGTGTGGTAGC |
| 160–233 | 5′CTCCGCGGTGGATACTTTACCCCCAA | 5′GCGCAAGCTTTTAAGGGTTAAGTGGC |
| 43–160 | 5′CCGCGGTGGTAATGGCATCTTCAACA | 5′AAGCTTTTAGTAGCGGGTGTGGTAGC |
| 43–180 | 5′CCGCGGTGGTAATGGCATCTTCAACA | 5′GCGCAAGCTTTTAATCTTTTGTTGTT |
| 43–233 | 5′CCGCGGTGGTAATGGCATCTTCAACA | 5′GCGCAAGCTTTTAAGGGTTAAGTGGC |
Additional SacII and HindIII restriction sites are underlined.
CP polypeptide and oligopeptide ELISA.
Commercially prepared 20-mer oligopeptides (21st Century Biochemicals) were conjugated to bovine serum albumin (BSA) by the manufacturer. For the purpose of conjugation to BSA, a cysteine was added to the N- or C-terminal end of the oligopeptide. Each CP polypeptide or BSA-conjugated oligopeptide was diluted to 4 μg/ml in 0.05 M carbonate binding buffer (pH 9.6), and 100 μl was added to each well of an enzyme-linked immunosorbent assay (ELISA) plate (Costar). Antigen-coated ELISA plates were incubated at 4°C for approximately 15 h. Wells were washed three times with PBS-0.01% Tween 20 (PBST) and then blocked for at least 1 h at room temperature in PBS with 10% goat serum (PBS-GS). Next, 100 μl of 1:100 dilutions of pig sera in PBS-GS was added to the plates and then incubated for 2 h at room temperature. After being washed with PBST, 100 μl of peroxidase-labeled goat anti-swine antibody (Accurate Chemical & Scientific Corp.) diluted 1:2,000 in PBS-GS was added to each well, and incubation continued for 1 h at room temperature. After extensive washing with PBST, peroxidase activity was detected by addition of 100 μl of the chromogenic substrate ABTS [2,2′-azinobis(3-ethylbenzthiazolinesulfonic acid); KPL]. Following a 20-min incubation away from light, absorbance values were read at 405 nm using a Maxline microplate reader (Molecular Devices Corporation).
Binding ratio calculation.
To compare results across ELISA plates, each ELISA plate included an internal positive control, which consisted of wells coated with the largest CP polypeptide [CP(43-233)] incubated with serum from a PDNS pig with a high PCV2 antibody titer. The antibody binding ratio was calculated as the A405 value of the test sample minus background divided by the A405 value of the internal positive control minus background. Antibody binding ratios for samples and control were derived from a 1:100 dilution.
Field vaccination study.
For the purpose of determining antibody responses to CP(43-233) and CP(169-180) before and after vaccination, samples were collected from a 150-head sow source herd that was being vaccinated for the first time. The source of the vaccine was a baculovirus-expressed ORF2 PCV2 vaccine, the same vaccine that was used to experimentally vaccinate pigs in the experimental study. Pigs were vaccinated at 3 and 6 weeks of age according to the vaccine label instructions. Blood was collected from 33 randomly selected 3-week-old pigs derived from 10 different dams. The same pigs were bled a second time at 16 weeks of age, or 10 weeks after administration of the last vaccine dose. Sera were reacted with CP(43-233) and CP(169-180) in an ELISA as previously described.
RESULTS
Expression and immunoreactivity of CP polypeptides.
The largest CP fragment contained the full-length protein minus the first 42 amino acid residues. The removal of the first 42 amino acids, which contain the arginine-rich domain, increases polypeptide expression in bacteria (54). Analysis of CP(43-233) by SDS-PAGE showed a single protein band that migrated at the predicted molecular weight (data not shown). The remaining ORF2 constructs, CP(43-135), CP(43-160), CP(43-180), CP(91-160), CP(160-233), CP (135-233), and CP(91-233), were cloned into pHUE, expressed, and affinity purified. Unlike CP(43-233), the smaller polypeptides were retained within the bacteria as insoluble inclusion bodies. Therefore, 8 M urea was added to the extraction and purification buffers. SDS-PAGE confirmed that all recombinant polypeptides were expressed and possessed the predicted size (data not shown). CP polypeptides were applied as a coating directly to ELISA plates as His-ubiquitin fusion proteins. As mentioned above, urea was required to solubilize the polypeptides, with the exception of the largest polypeptide, CP(43-233). The synthesis of a truncated protein combined with the presence of urea likely disrupted conformational epitopes within CP; however, all polypeptides demonstrated the ability to react with sera from diseased and infected pigs (see below).
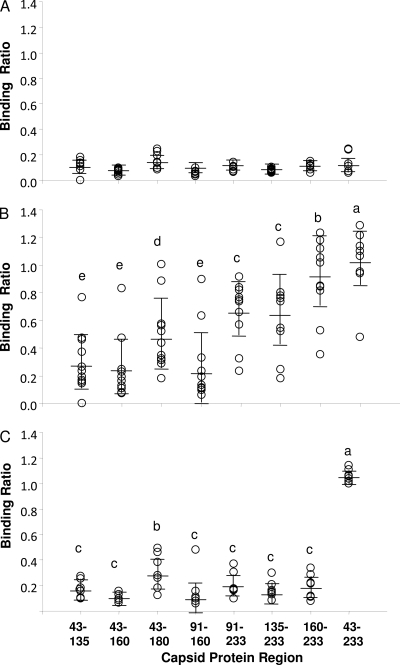
The first antibody measurements were performed on sera from experimentally infected pigs (described in Table 1). To reduce variation in results across ELISA plates, each ELISA plate included an internal positive-control serum reacted with CP(43-233). The results in Fig. 3A showed only low binding ratios for the uninfected control pigs. The same serum samples were also negative for PCV2 antibody as measured by IFA assay and negative for PCV2 DNA by PCR (data not shown). The results confirmed that the control pigs remained negative for PCV2 infection during the experiment. The antibody binding ratios for pigs infected with PCV2 and PCV2-PRRSV are shown in Fig. 3B. Since there was no significant difference in the antibody responses between the pigs infected with PCV2 and those dually infected with PCV2-PRRSV, the data for the two groups were combined. All PCV2-challenged pigs were positive for PCV2 infection by PCR and for PCV2 antibody by IFA assay, which confirmed that all pigs were productively infected. PRRSV reverse transcription-PCR (RT-PCR) and serology confirmed that all dual-infected pigs were productively infected with PRRSV. Seven of the 11 pigs in this group showed no clinical evidence of PDNS or PMWS. Three pigs in the dual-challenged group died and were not included in the study. The remaining pigs showed some disease signs, including reduced weight gain and accumulation of PCV2 antigen in lymph nodes (data not shown). As shown in Fig. 3B, the PCV2 group showed measurable antibody activity against all polypeptides by at least one pig; however, the highest mean binding ratio was obtained for the full-length CP(43-233) polypeptide and the lowest for CP(43-135), CP(43-160), and CP(91-160). The remaining polypeptides, CP(91-233), CP(135-233), CP(160-233), and CP(43-180), possess mean ratios that were intermediate between those of CP(43-233) and the low-responding polypeptides. The responses for the seven pigs receiving only the PCV2 vaccine are shown in Fig. 3C. All vaccinated pigs were positive for PCV2 antibody by IFA assay and were confirmed to be negative for PCV2 by PCR (data not shown). The highest binding ratios were observed for the CP(43-233) polypeptide, with only background levels of binding against the smaller CP fragments. The only exception was CP(43-180), which showed a small, but significant, increase in binding relative to the other small polypeptide fragments. In order to demonstrate that vaccinated pigs were protected, 14 vaccinated pigs were challenged with PCV2 (seven pigs) or PCV2 plus PRRSV (seven pigs). All challenged pigs remained clinically normal with no mortality and were negative for PCV2 DNA in serum (data not shown).
Fig. 3.
Immunoreactivities of bacterium-expressed polypeptides with sera from experimentally PCV2-infected and vaccinated pigs. The ELISA results show the responses of sera from PCV2-negative (A), PCV2-infected (B), and CP-vaccinated (C) pigs obtained at 5 weeks after experimental infection with PCV2 or 7 weeks after vaccination as described in the timeline shown in Fig. 1. The antibody binding ratio was calculated as described in Materials and Methods. The open circles show the responses of individual pigs. The means and standard deviations are represented by horizontal lines. Means with the same lowercase letter do not significantly differ as determined by the Student-Newman-Keuls method (P < 0.05).
Serological responses of pigs with PCVAD.
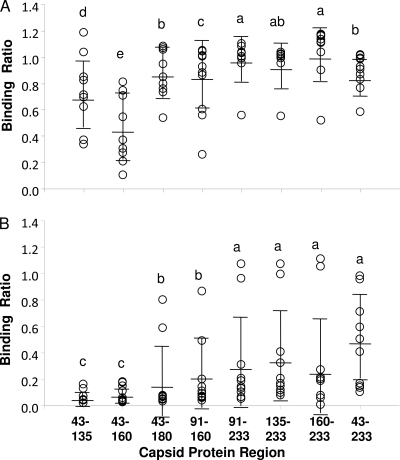
Currently, there are no PCV2 experimental infection models that consistently reproduce all of the elements of PMWS or PDNS. Therefore, sera from pigs submitted to the KSVDL with a diagnosis of PCVAD were evaluated for recognition of CP polypeptides. Even though PCV2 may not be the proximal cause of PDNS (25), pigs with PDNS typically possess high levels of PCV2 antibodies (56). As shown in Fig. 4A, 9 of 10 PDNS pigs possessed high binding activity against CP(43-233). Overall, relatively high levels of antibody binding were observed for seven of the eight polypeptides, with the highest mean antibody binding ratio being that against the smallest polypeptide, CP(160-233). Significantly lower binding ratios were observed for CP(43-135) and CP(43-160). The results for sera from 10 pigs diagnosed with PMWS are shown in Fig. 4B. Unlike the PDNS pigs, overall binding ratios against the CP fragments were relatively low for a majority of the pigs. Two of the 10 pigs showed elevated binding ratios against CP(43-233), while four pigs exhibited only background activity against CP(43-233), and the remaining four showed intermediate activities. The overall low binding ratios against CP(43-233) likely reflect decreased antibodies as a result of the overall depletion of lymphocytes, which occurs during end-stage PMWS. Responses against the other oligopeptides were also variable, except for CP(43-135) and CP(43-160), which were negative for binding.
Fig. 4.
Immunoreactivities of PCV2 CP polypeptides with sera from pigs with PDNS and PMWS. The same ELISA method described in the Fig. 3 legend was used to assess antibody activity in sera from pigs diagnosed with PDNS (A) or PMWS (B). The open circles show the responses of individual pigs. The horizontal lines show means and standard deviations. Means with the same lowercase letter do not significantly differ as determined by the Student-Newman-Keuls method (P < 0.05).
A summary of the antibody binding activities of the different groups of pigs against the different CP fragments is presented in Table 3. For PCV2-infected, PMWS, and PDNS pigs, the highest levels of antibody binding were obtained primarily for CP polypeptides that contained epitope C (Fig. 2).
Table 3.
Summary of antibody responses to PCV2 capsid polypeptides
PC, PCV2 infected; Vx, vaccinated; PM, PMWS; PD, PDNS; −, no measurable binding activity; +, low binding activity; ++, intermediate binding activity; +++, high binding activity; ++++, very high binding activity.
Pepscan mapping of the C-terminal region of CP.
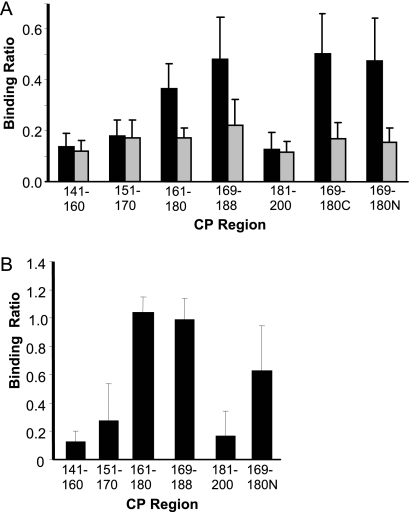
Further studies were performed to determine the smallest oligopeptide recognized by PCV2-infected pigs. Based on the results of Table 3, 20-mer oligopeptides, with 10 overlapping residues, spanning epitope C and the flanking region (residues 141 to 200), were prepared and reacted with sera from PCV2-infected and PDNS pigs. The results for the experimentally infected PCV2 pigs showed a large variation in binding activity. Closer inspection of the data revealed that pigs could be divided into two groups. For instance, as presented in Fig. 5A, 4 of the 11 serum samples showed minimal binding activity against all oligopeptides (gray bars), similar to the responses of vaccinated and uninfected control pigs (data not shown). The remaining PCV2-infected pigs exhibited a pattern shown by the black bars, with relatively high activity against the CP(161–180) and CP(169–188) oligopeptides and lower binding activity against the flanking oligopeptides. The combined region CP(161–188) is within the epitope C region. Two 12-mer oligopeptides covering the region overlapped by CP(161–180) and CP(169–188) were prepared and tested for antibody binding. The BSA-conjugated oligopeptides, CP(169–180C) and CP(169–180N), were constructed with an aminohexonic acid (Ahx) spacer added to the C- or N-terminal end, respectively. The spacer fragment was designed to increase antibody accessibility by extending the oligopeptide beyond the surface of the BSA molecule. Both oligopeptides showed binding activities similar to those of the CP(161–180) and CP(169–188) parent oligopeptides. Results for sera from PDNS pigs are shown in Fig. 5B. Similar to the responses shown by black bars in Fig. 5A, the highest binding activities for the PDNS pigs were against CP(161–180), CP(169–188), and CP(169–180).
Fig. 5.
Immunoreactivities of pig sera with CP oligopeptides. (A) Antibody responses of experimentally infected pigs against a series of 20-mer overlapping oligopeptides that cover residues 141 to 200 of the PCV2 CP. The gray bars show the means and standard deviations for a subset of four pigs that possessed low antibody levels of binding against all oligopeptides. The black bars show the means and standard deviations for the remaining six pigs. (B) Responses of sera from PDNS pigs. The results show the mean and 1 standard deviation of the antibody binding ratio for each group of pigs.
Alanine scanning mutagenesis in CP(169–180).
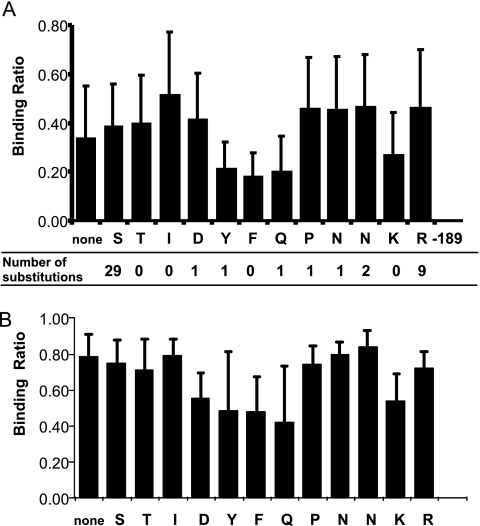
To identify the individual amino acids that contributed to antibody recognition within CP(169–180), single alanine substitutions were made along the CP(169–180) peptide sequence, 169-STIDYFQPNNKR-180. The results for the subset of experimentally infected PCV2 pigs that showed high binding against CP(169–180) in Fig. 5A are presented in Fig. 6A. The results showed reduced binding for oligopeptides with alanine substitutions at tyrosine-173, phenylalanine-174, glutamine-175, and lysine-179. Similar alanine scanning results were obtained for sera from PDNS pigs (Fig. 6B).
Fig. 6.
Alanine scanning mutagenesis. (A) Mean and standard deviation results for the same six pigs as those shown in Fig. 5A (black bars) that recognized CP(169–180). BSA-conjugated oligopeptides that possessed single alanine substitutions were tested. The numbers below panel A show the numbers of amino acid substitutions found in a comparison of 462 peptide sequences obtained from GenBank and diagnostic lab submissions. (B) Alanine scanning results for the same PDNS pigs as those shown in Fig. 5B.
Four hundred sixty-two CP(169–180) peptide sequences, obtained from GenBank and diagnostic lab submissions, were analyzed for amino acid differences. The results, presented below Fig. 6A, showed that 45 of the 462 sequences possessed changes within CP(169–180): all were single amino acid substitutions. For those residues that contributed to binding, tyrosine-173, phenylalanine-174, glutamine-175, and lysine-179, there were only two amino acid substitutions. Therefore, the core peptide region that forms the epitope within CP(169–180) is highly conserved.
Virus-neutralizing activity in PCV2-infected and vaccinated pigs.
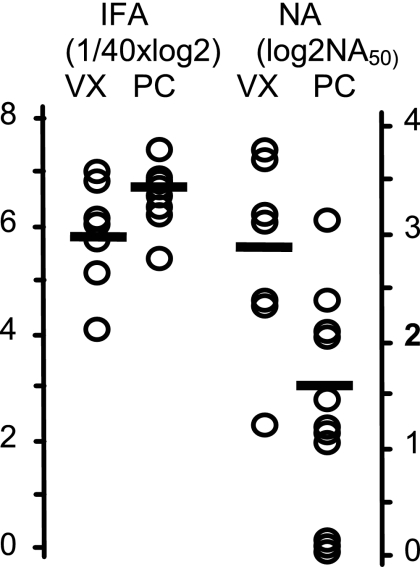
Virus-neutralizing activity in serum samples from the same PCV2 pigs in Fig. 3B and vaccinated pigs in Fig. 3C is shown in Fig. 7. The mean total antibody level for PCV2-infected pigs (IFA titer = 6.7), as measured by IFA assay, was almost twice the level of the mean value for the vaccinated pigs (IFA titer = 5.8). In contrast, the mean NA level for vaccinated pigs (log2 NA50 = 2.8) was approximately four times higher than the mean level obtained from sera of PCV2-infected pigs (log2 NA50 = 1.6).
Fig. 7.
Total and neutralizing antibody responses following vaccination and infection. Serum samples from PCV2-infected (PC) pigs (Fig. 3B) and PCV2-vaccinated (VX) pigs (Fig. 3C) were tested for the presence of total antibody (IFA) and neutralizing activity (NA). Open circles represent values for individual pigs, and the horizontal bars represent the mean value for each group.
Serological response to CP(169–180) following PCV2 vaccination in the field.
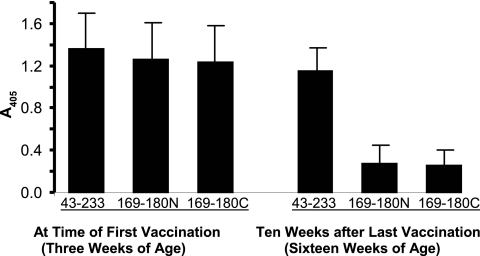
We determined the antibody responses to CP(43–233) and CP(169–180) before and after vaccination of a small pig herd that had a history of clinically apparent PCV2 infection, which was confirmed by serology and PCR (data not shown). The results, summarized in Fig. 8, showed that the antibody responses of the 3-week-old pigs were directed against both CP(43–233) and CP(169–180) oligopeptides. The presence of CP-specific antibody in prevaccination pigs is primarily the result of maternally derived antibodies from dams that had been naturally infected with PCV2 (19). At 16 weeks of age (10 weeks after the second vaccine dose), the pattern of antigen recognition changed, with the response against CP(169–180) greatly diminished relative to that against CP(43–233). The effectiveness of vaccination was confirmed by the disappearance of clinical disease from the herd and by the decreased prevalence of PCR-positive serum samples. Together, the results illustrate a pattern showing the initial presence of antibodies derived from natural infection, which were replaced with CP(43–233)-specific antibodies generated in response to vaccination with CP.
Fig. 8.
Recognition of CP(43–233) and CP(169–180) in a herd before and after PCV2 vaccination. Results are from a herd vaccinated for PCV2 for the first time. Samples were tested when pigs received the first dose of vaccine (3 weeks of age), and the same pigs were tested at 16 weeks of age, or 10 weeks after the second vaccine dose. The results are shown as the means and standard deviations for 33 pigs reacted with CP(43–233) and CP(169–180) oligopeptides. All samples were assayed on a single plate, and results (vertical axis) are presented as A405 values. The oligopeptides CP(169–180N) and CP(169–180C) were linked to BSA via the N- or C-terminal end of the oligopeptide, respectively.
DISCUSSION
Previous studies of humoral immunity during PCV2 infection have primarily focused on mapping antigenic regions of CP, including the characterization of immunorelevant epitopes (26, 33, 55). The present study describes differences in the antibody responses toward PCV2 CP immunoreactive regions following vaccination and experimental infection and during severe disease. The results illustrate the complexity of the immune response during PCV2 infection while providing information on the immunological basis for protection during vaccination and the initiation of immunopathogenesis. Analysis of antibody reactivities of infected, vaccinated, and PCVAD pigs against individual CP polypeptide fragments and oligopeptides identified at least four unique antibody recognition patterns, which are summarized in Table 3. The first pattern is illustrated by pigs with severe PMWS. Overall, there was a decreased reactivity to all CP polypeptide antigens (Fig. 4B). This outcome is consistent with the immune suppression associated with PMWS, a disease syndrome characterized by an almost complete depletion of lymphocytes with a corresponding loss or severe dysregulation of immune function (13). Immunohistochemical staining of PMWS lymph nodes typically shows large accumulations of PCV2 antigen (8). Presumably, dividing lymphocytes, activated in response to infection or other immune stimuli, become a primary target for PCV2 replication. Cytopathogenesis in lymphocytes is attributed to the function of PCV2 ORF3, which is not required for virus replication in culture but has been linked to apoptosis (21, 22, 28). In sharp contrast, pigs with clinically apparent PDNS showed high reactivity to all CP polypeptide fragments (Fig. 4), including the oligopeptide CP(169-180) (Fig. 5B). This outcome is consistent with hyperactive humoral response and immune complex formation. Pathogenesis is linked to the deposition of antigen-antibody complexes in the kidney and other organs followed by the activation of complement (56). The role that PCV2 plays in PDNS remains unclear. Krakowka et al. (25) reported the induction of PDNS in gnotobiotic pigs after infection with a group 1 torque teno virus (TTV) and PRRSV. To date, there are no models of PCV2 infection that reproduce PDNS. One likely possibility is that PCV2 infection may not be the proximal cause of PDNS but may play a role in the evolution of the disease process and expression of full-blown disease.
A third pattern of antibody recognition was found in the responses of pigs experimentally infected with PCV2. The results showed the highest antibody response to CP(43-233) followed by reactivity with polypeptides that contained epitope C. A dichotomy in the response to the epitope C region was evident from the oligopeptide mapping results, in which PCV2 pigs could be divided into two groups: those that produced a response similar to that of PDNS pigs and recognized CP(169-180) and those that responded similarly to vaccinates. Finally, a fourth antibody response pattern was found in pigs vaccinated with a baculovirus-expressed CP antigen. Vaccinated pigs almost exclusively recognized the largest polypeptide, CP(43-233), and showed much lower responses to smaller polypeptides, including those polypeptide fragments that contained epitope C (Fig. 3C). This pattern of antibody recognition suggests that vaccination produces antibodies that primarily recognize a single large conformational epitope. Evidence for the protective nature of this response was found in the complete protection of vaccinated pigs challenged with PCV2 or with PCV2 and PRRSV and suggests that vaccination with only CP is sufficient to deliver sterilizing immunity.
The principal difference between PCVAD and vaccinated pigs localized to CP(169-180), within epitope C. PDNS pigs and a subset of PCV2 pigs preferentially recognized CP(169-180). To further demonstrate the specific nature of the recognition, alanine scanning mutagenesis identified specific residues as important for antibody recognition. Furthermore, the key amino acid residues involved in antibody binding are highly conserved among PCV2 isolates. The significance of this epitope in disease progression is not entirely clear. However, the results suggest that antibodies directed against this epitope are not involved in immune protection. Protection from infection and disease is likely dependent on antibodies directed against a single, conformational epitope. The results support the hypothesis that antibodies preferentially directed against epitope C are nonprotective and signal the initial immune defect that leads to disease. One possibility is that epitope C functions as a decoy epitope, allowing PCV2 to evade humoral immunity by focusing the antibody response toward a nonprotective epitope. Evidence for this possibility is found in the total and neutralizing antibody responses of PCV2-infected versus CP-vaccinated pigs (Fig. 7). Even though the total PCV2 antibody response of infected pigs was almost twice the response of vaccinated pigs, the mean neutralizing activity for the vaccinated group was almost four times that of the infected group. Therefore, the apparent paradox of decreased neutralizing activity in the face of an overall robust humoral response can be resolved if the response is directed toward nonneutralizing epitopes. The diversion of humoral immunity away from the larger neutralizing epitope is a strategy that has been proposed for HIV and PRRSV (27, 46).
One interesting aspect of PCV2 infection is that only a subpopulation of infected pigs go on to develop full-blown disease, while other infected pigs remain unaffected. The mixed antibody response of PCV2-infected pigs provides insight into a possible mechanism for differences in disease susceptibility within the same population of PCV2-infected pigs. For example, those pigs that respond to PCV2 in a manner similar to that of pigs following vaccination produce an effective antibody response that results in virus clearance and protection. In contrast, those pigs that produce a response similar to PDNS pigs, i.e., against nonprotective epitopes, are more susceptible to prolonged virus replication and disease.
A practical application of the results from this study is illustrated by the data obtained for herd vaccination, shown in Fig. 8. Prior to vaccination, maternally derived antibody (MDA) acquired during natural infection showed reactivity to both CP(43-233) and CP(169-180), a pattern similar to that in experimental PCV2 infection. After vaccination and decay of MDA, the antibody recognition pattern switched to recognition of only CP(43-233). Therefore, differential recognition of CP(43-233) and CP(169-180) provides the basis for diagnostic approaches that can assess the level of protective antibody in a vaccinated herd, as well as methods that can differentiate infected from vaccinated animals (DIVA).
ACKNOWLEDGMENTS
This work was supported by National Pork Board grant 06-073 and USDA NRI grant 2009-35204-05290.
Footnotes
Published ahead of print on 23 March 2011.
The authors have paid a fee to allow immediate free access to this article.
REFERENCES
- 1. Allan G. M., Ellis J. A. 2000. Porcine circoviruses: a review. J. Vet. Diagn. Invest. 12:3–14 [DOI] [PubMed] [Google Scholar]
- 2. Allan G. M., et al. 1999. Experimental reproduction of severe wasting disease by co-infection of pigs with porcine circovirus and porcine parvovirus. J. Comp. Pathol. 121:1–11 [DOI] [PubMed] [Google Scholar]
- 3. Allan G., et al. 2003. Reproduction of postweaning multisystemic wasting syndrome in pigs experimentally inoculated with a Swedish porcine circovirus 2 isolate. J. Vet. Diagn. Invest. 15:553–560 [DOI] [PubMed] [Google Scholar]
- 4. Blanchard P., et al. 2003. Protection of swine against post-weaning multisystemic wasting syndrome (PMWS) by porcine circovirus type 2 (PCV2) proteins. Vaccine 21:4565–4575 [DOI] [PubMed] [Google Scholar]
- 5. Bolin S. R., Stoffregen W. C., Nayar G. P., Hamel A. L. 2001. Postweaning multisystemic wasting syndrome induced after experimental inoculation of cesarean-derived, colostrum-deprived piglets with type 2 porcine circovirus. J. Vet. Diagn. Invest. 13:185–194 [DOI] [PubMed] [Google Scholar]
- 6. Catanzariti A., Soboleva T. A., Jans D. A., Board P. G., Baker R. T. 2004. An efficient system for high-level expression and easy purification of authentic recombinant proteins. Protein Sci. 13:1331–1339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Chae C. 2005. A review of porcine circovirus 2-associated syndromes and diseases. Vet. J. 169:326–336 [DOI] [PubMed] [Google Scholar]
- 8. Chae C. 2004. Postweaning multisystemic wasting syndrome: a review of aetiology, diagnosis and pathology. Vet. J. 168:41–49 [DOI] [PubMed] [Google Scholar]
- 9. Chaiyakul M., Hsu K., Dardari R., Marshall F., Czub M. 2010. Cytotoxicity of ORF3 proteins from a nonpathogenic and a pathogenic porcine circovirus. J. Virol. 84:11440–11447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Cheung A. K., Bolin S. R. 2002. Kinetics of porcine circovirus type 2 replication. Arch. Virol. 147:43–58 [DOI] [PubMed] [Google Scholar]
- 11. Cheung A. K. 2003. Comparative analysis of the transcriptional patterns of pathogenic and nonpathogenic porcine circoviruses. Virology 310:41–49 [DOI] [PubMed] [Google Scholar]
- 12. Cheung A. K. 2003. Transcriptional analysis of porcine circovirus type 2. Virology 305:168–180 [DOI] [PubMed] [Google Scholar]
- 13. Darwich L., Segalés J., Mateu E. 2004. Pathogenesis of postweaning multisystemic wasting syndrome caused by porcine circovirus 2: an immune riddle. Arch. Virol. 149:857–874 [DOI] [PubMed] [Google Scholar]
- 14. Dorr P. M., Almond G. W., Wayne S. R., Gebreyes W. A. 2007. Epidemiologic assessment of porcine circovirus type 2 coinfection with other pathogens in swine. J. Am. Vet. Med. Assoc. 230:244–250 [DOI] [PubMed] [Google Scholar]
- 14a. Finney D. J. 1964. The Spearman-Karber method, p. 524–530 In Finney D. J. (ed.), Statistical method in biological assay, 2nd ed. Charles Griffin, London, United Kingdom [Google Scholar]
- 15. Fort M., et al. 2008. Porcine circovirus type 2 (PCV2) vaccination of conventional pigs prevents viremia against PCV2 isolates of different genotypes and geographic origins. Vaccine 26:1063–1071 [DOI] [PubMed] [Google Scholar]
- 16. Fort M., Olvera A., Sibila M., Segalés J., Mateu E. 2007. Detection of neutralizing antibodies in postweaning multisystemic wasting syndrome (PMWS)-affected and non-PMWS-affected pigs. Vet. Microbiol. 125:244–255 [DOI] [PubMed] [Google Scholar]
- 17. Heath L., Williamson A., Rybicki E. P. 2006. The capsid protein of beak and feather disease virus binds to the viral DNA and is responsible for transporting the replication-associated protein into the nucleus. J. Virol. 80:7219–7225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hesse R., Kerrigan M., Rowland R. R. R. 2008. Evidence for recombination between PCV2a and PCV2b in the field. Virus Res. 132:201–207 [DOI] [PubMed] [Google Scholar]
- 19. Horlen K. P., et al. 2008. A field evaluation of mortality rate and growth performance in pigs vaccinated against porcine circovirus type 2. J. Am. Vet. Med. Assoc. 232:906–912 [DOI] [PubMed] [Google Scholar]
- 20. Horlen K. P., et al. 2007. A cluster of farms experiencing severe porcine circovirus associated disease: clinical features and association with the PCV2b genotype. J. Swine Health Prod. 15:270–278 [Google Scholar]
- 21. Juhan N. M., LeRoith T., Opriessnig T., Meng X. J. 2010. The open reading frame 3 (ORF3) of porcine circovirus type 2 (PCV2) is dispensable for virus infection but evidence of reduced pathogenicity is limited in pigs infected by an ORF3-null PCV2 mutant. Virus Res. 147:60–66 [DOI] [PubMed] [Google Scholar]
- 22. Karuppannan A. K., et al. 2009. Attenuation of porcine circovirus 2 in SPF piglets by abrogation of ORF3 function. Virology 383:338–347 [DOI] [PubMed] [Google Scholar]
- 23. Krakowka S., et al. 2001. Activation of the immune system is the pivotal event in the production of wasting disease in pigs infected with porcine circovirus-2 (PCV-2). Vet. Pathol. 38:31–42 [DOI] [PubMed] [Google Scholar]
- 24. Krakowka S., et al. 2000. Viral wasting syndrome of swine: experimental reproduction of postweaning multisystemic wasting syndrome in gnotobiotic swine by coinfection with porcine circovirus 2 and porcine parvovirus. Vet. Pathol. 37:254–263 [DOI] [PubMed] [Google Scholar]
- 25. Krakowka S., et al. 2008. Evaluation of induction of porcine dermatitis and nephropathy syndrome in gnotobiotic pigs with negative results for porcine circovirus type 2. Am. J. Vet. Res. 69:1615–1622 [DOI] [PubMed] [Google Scholar]
- 26. Lekcharoensuk P., et al. 2004. Epitope mapping of the major capsid protein of type 2 porcine circovirus (PCV2) by using chimeric PCV1 and PCV2. J. Virol. 78:8135–8145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Lin G., Nara P. L. 2007. Designing immunogens to elicit broadly neutralizing antibodies to the HIV-1 envelope glycoprotein. Curr. HIV Res. 5:514–541 [DOI] [PubMed] [Google Scholar]
- 28. Liu J., Chen I., Kwang J. 2005. Characterization of a previously unidentified viral protein in porcine circovirus type 2-infected cells and its role in virus-induced apoptosis. J. Virol. 79:8262–8274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Liu Q., Tikoo S. K., Babiuk L. A. 2001. Nuclear localization of the ORF2 protein encoded by porcine circovirus type 2. Virology 285:91–99 [DOI] [PubMed] [Google Scholar]
- 30. Liu Q., Willson P., Attoh-Poku S., Babiuk L. A. 2001. Bacterial expression of an immunologically reactive PCV2 ORF2 fusion protein. Protein Expr. Purif. 21:115–120 [DOI] [PubMed] [Google Scholar]
- 31. Lyoo K., et al. 4 August 2010. Comparative efficacy of three commercial PCV2 vaccines in conventionally reared pigs. Vet. J. [Epub ahead of print.] doi:10.1016/j.tvjl.2010.06.015 [DOI] [PubMed] [Google Scholar]
- 32. Ma C., et al. 2007. Evidence for recombination in natural populations of porcine circovirus type 2 in Hong Kong and mainland China. J. Gen. Virol. 88:1733–1737 [DOI] [PubMed] [Google Scholar]
- 33. Mahé D., et al. 2000. Differential recognition of ORF2 protein from type 1 and type 2 porcine circoviruses and identification of immunorelevant epitopes. J. Gen. Virol. 81:1815–1824 [DOI] [PubMed] [Google Scholar]
- 34. Mankertz A., Hillenbrand B. 2001. Replication of porcine circovirus type 1 requires two proteins encoded by the viral rep gene. Virology 279:429–438 [DOI] [PubMed] [Google Scholar]
- 35. Meerts P., et al. 2006. Correlation between the presence of neutralizing antibodies against porcine circovirus 2 (PCV2) and protection against replication of the virus and development of PCV2-associated disease. BMC Vet. Res. 2:6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Misinzo G., Delputte P. L., Meerts P., Lefebvre D. J., Nauwynck H. J. 2006. Porcine circovirus 2 uses heparan sulfate and chondroitin sulfate B glycosaminoglycans as receptors for its attachment to host cells. J. Virol. 80:3487–3494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Nakai K., Kanehisa M. 1992. A knowledge base for predicting protein localization sites in eukaryotic cells. Genomics 14:897–911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Nawagitgul P., et al. 2000. Open reading frame 2 of porcine circovirus type 2 encodes a major capsid protein. J. Gen. Virol. 81:2281–2287 [DOI] [PubMed] [Google Scholar]
- 39. Olvera A., Cortey M., Segalés J. 2007. Molecular evolution of porcine circovirus type 2 genomes: phylogeny and clonality. Virology 357:175–185 [DOI] [PubMed] [Google Scholar]
- 40. Opriessnig T., et al. 2006. Evidence of breed-dependent differences in susceptibility to porcine circovirus type-2-associated disease and lesions. Vet. Pathol. 43:281–293 [DOI] [PubMed] [Google Scholar]
- 41. Opriessnig T., McKeown N. E., Zhou E., Meng X., Halbur P. G. 2006. Genetic and experimental comparison of porcine circovirus type 2 (PCV2) isolates from cases with and without PCV2-associated lesions provides evidence for differences in virulence. J. Gen. Virol. 87:2923–2932 [DOI] [PubMed] [Google Scholar]
- 42. Opriessnig T., Patterson A. R., Madson D. M., Pal N., Halbur P. G. 2009. Comparison of efficacy of commercial one dose and two dose PCV2 vaccines using a mixed PRRSV-PCV2-SIV clinical infection model 2-3-months post vaccination. Vaccine 27:1002–1007 [DOI] [PubMed] [Google Scholar]
- 43. Opriessnig T., et al. 2004. Experimental reproduction of postweaning multisystemic wasting syndrome in pigs by dual infection with Mycoplasma hyopneumoniae and porcine circovirus type 2. Vet. Pathol. 41:624–640 [DOI] [PubMed] [Google Scholar]
- 44. Opriessnig T., Yu S., Thacker E. L. 2004. Derivation of porcine circovirus type 2-negative pigs from positive breeding herds. J. Swine Health Prod. 12:186–191 [Google Scholar]
- 45. Opriessnig T., Meng X., Halbur P. G. 2007. Porcine circovirus type 2 associated disease: update on current terminology, clinical manifestations, pathogenesis, diagnosis, and intervention strategies. J. Vet. Diagn. Invest. 19:591–615 [DOI] [PubMed] [Google Scholar]
- 46. Ostrowski M., et al. 2002. Identification of neutralizing and nonneutralizing epitopes in the porcine reproductive and respiratory syndrome virus GP5 ectodomain. J. Virol. 76:4241–4250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Reed J., Muench H. 1938. A simple method of estimating fifty percent endpoints. Am. J. Hyg. 27:493–497 [Google Scholar]
- 48. Segalés J., et al. 2008. PCV-2 genotype definition and nomenclature. Vet. Rec. 162:867–868 [DOI] [PubMed] [Google Scholar]
- 49. Segalés J., Allan G. M., Domingo M. 2005. Porcine circovirus diseases. Anim. Health Res. Rev. 6:119–142 [DOI] [PubMed] [Google Scholar]
- 50. Shang S., et al. 2009. Fine mapping of antigenic epitopes on capsid proteins of porcine circovirus, and antigenic phenotype of porcine circovirus type 2. Mol. Immunol. 46:327–334 [DOI] [PubMed] [Google Scholar]
- 51. Shen H. G., et al. 2010. Comparison of commercial and experimental porcine circovirus type 2 (PCV2) vaccines using a triple challenge with PCV2, porcine reproductive and respiratory syndrome virus (PRRSV), and porcine parvovirus (PPV). Vaccine 28:5960–5966 [DOI] [PubMed] [Google Scholar]
- 52. Sorden S. D. 2000. Update on porcine circovirus and postweaning multisystemic wasting syndrom (PMWS). Swine Health Prod. 8:133–138 [Google Scholar]
- 53. Thomson J. R., Higgins R. J., Smith W. J., Done S. H. 2002. Porcine dermatitis and nephropathy syndrome. Clinical and pathological features of cases in the United Kingdom (1993-1998). J. Vet. Med. A Physiol. Pathol. Clin. Med. 49:430–437 [DOI] [PubMed] [Google Scholar]
- 54. Trundova M., Celer V. 2007. Expression of porcine circovirus 2 ORF2 gene requires codon optimized E. coli cells. Virus Genes 34:199–204 [DOI] [PubMed] [Google Scholar]
- 55. Truong C., et al. 2001. Identification of an immunorelevant ORF2 epitope from porcine circovirus type 2 as a serological marker for experimental and natural infection. Arch. Virol. 146:1197–1211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Wellenberg G. J., Stockhofe-Zurwieden N., de Jong M. F., Boersma W. J. A., Elbers A. R. W. 2004. Excessive porcine circovirus type 2 antibody titres may trigger the development of porcine dermatitis and nephropathy syndrome: a case-control study. Vet. Microbiol. 99:203–214 [DOI] [PubMed] [Google Scholar]