Abstract
Hepatitis C virus (HCV) infection is a major burden to public health worldwide, affecting approximately 3% of the human population. Although HCV detection is currently based on reliable tests, the field of medical diagnostics has a growing need for inexpensive, accurate, and quick high-throughput assays. By using the recombinant HCV antigens NS3, NS4, NS5, and Combined, we describe a new bead-based multiplex test capable of detecting HCV infection in human serum samples. The first analysis, made in a singleplex format, showed that each antigen coupled to an individual bead set presented high-level responses for anti-HCV-positive reference serum pools and lower-level responses for the HCV-negative pools. Our next approach was to determine the sensitivity and specificity of each antigen by testing 93 HCV-positive and 93 HCV-negative sera. When assayed in the singleplex format, the NS3, NS4, and NS5 antigens presented lower sensitivity values (50.5%, 51.6%, and 55.9%, respectively) than did the Combined antigen, which presented a sensitivity of 93.5%. All antigens presented 100% specificity. These antigens were then multiplexed in a 4-plex assay, which resulted in increased sensitivity and specificity values, performing with 100% sensitivity and 100% specificity. The positive and negative predictive values for the 4-plex assay were 100%. Although preliminary, this 4-plex assay showed robust results that, aligned with its small-sample-volume requirements and also its cost- and time-effectiveness, make it a reasonable alternative to tests currently used for HCV screening of potentially infected individuals.
INTRODUCTION
According to the World Health Organization, hepatitis C virus (HCV) affects approximately 200 million people worldwide, almost 3% of the world's population. HCV infection is characterized by a great propensity to progress to persistent infection, leading to chronic liver disease, which, in certain patients, may evolve into cirrhosis and hepatocellular carcinoma (10). International studies have estimated that because the risk of HCV-related chronic liver disease is associated with the duration of infection, it is likely that the incidence of HCV-related complications will increase in the upcoming decades, being quadrupled in 2015 (5). To curb this trend, health services need to improve the screening of infected individuals in order to treat them when liver disease is asymptomatic and not life-threatening.
Currently, the routine detection of HCV is based on the detection of anti-HCV IgG antibodies in serum or plasma by an enzyme immunoassay (EIA). Cloning of the HCV genome and sequence analysis have led to the development of a variety of antigens and synthetic peptides that have been successfully used in these immunoassays, improving the reliability of the test and increasing the detection of anti-HCV earlier in the course of infection (1, 2, 6). In spite of this, false-positive results with EIAs are still prevalent, especially among low-risk subjects, such as blood donors, or populations without liver-related diseases (4). This requires supplemental or confirmatory tests, potentially increasing the volume of sample needed as well as the associated technologist and instrument time required for testing, most of the time leading to unnecessary health care costs and difficulties in diagnosis (3). These tests also have important impairments: low processing speed, long labor time, low-throughput capacity, limited multiplex capability, and high cost (12, 19).
In the past decade, several technologies have emerged as diagnostic tools capable of improving detection by using multiplex principles. The diagnostic process becomes faster and less expensive and the hands-on time in laboratories decreases substantially since these platforms can be fully automated (18). One of the most promising multiplex techniques uses digital signal processing to classify small polystyrene beads. The beads are internally dyed with distinct proportions of red and near-infrared fluorophores, and these proportions define an intrinsic fluorescence or spectral address for each bead population (13, 16). Each group of beads can be coupled to a specific capture molecule, including protein antigens, acting as solid supports for the detection of their respective antibodies. Since the beads can be distinguished by their spectral addresses, they can be combined to produce multiplex assays, thereby allowing the rapid screening of multiple antibodies using a small volume of plasma. The captured antibodies are detected and quantified following the addition of a fluorescently labeled reporter antibody whose emission is measured by a flow-based detector (13, 16).
Bead-based immunoassays allow a quantitative and qualitative analysis of multiple targets with a unique combination of features, including rapid data acquisition, excellent sensitivity and specificity, and multiplexed analysis capabilities (20). This system is an open platform that allows the detection of several molecules, with applications for the screening of serum antibodies against a plethora of infectious agents (8, 9, 14, 15) and also for vaccine trials (22).
In this study, we developed a bead-based high-throughput immunoassay for the determination of antibodies against HCV in patient serum samples using the antigenic properties of four recombinant proteins: NS3, NS4, NS5, and Combined. These proteins have been widely used in screening tests for HCV detection and represent the most relevant epitopes for HCV diagnosis, associated with both acute and chronic phases of the disease (21, 23). We describe its application as a rapid, less-time-consuming, and less-serum-demanding assay, demonstrating that this platform is suitable for epidemiologic and also diagnostic applications for HCV management.
MATERIALS AND METHODS
Human sera.
A total of 186 serum samples were analyzed in the study, 93 sera positive and 93 negative for HCV, obtained from the Technology Institute for Immunobiologicals (Bio-Manguinhos)-Fiocruz. All serum samples were previously analyzed by using conventional serologic tests (enzyme-linked immunosorbent assay [ELISA]) and assigned a definitive serostatus (positive or negative for anti-HCV antibodies). Samples were tested with two commercially available HCV ELISA kits from different manufacturers and were classified as being serologically “positive” or “negative” according to the instructions provided by each manufacturer. Of these 186 samples, 53 (23 positive and 30 negative) were obtained from the National Panel for Blood Screening Quality Control (AEQ), which was elaborated and tested by Bio-Manguinhos-Fiocruz and the National Institute for Health Quality Control (INCQS)-Fiocruz. Six serum pools were constructed from the AEQ samples: three “negative” pools consisting of 10 sera each, which had negative results in all assays, and three “positive” pools containing 10, 8, and 5 sera each, which were unequivocally positive with all tests. These samples were also individually used throughout the experiments.
Antigens.
The recombinant antigens NS3, NS4, NS5, and Combined were purchased from ProSpec-Tany TechnoGene Ltd. (Rehovot, Israel). The Combined antigen is a fusion protein comprised of nucleocapsid, NS3, NS4, and NS5 immunodominant epitopes.
Bead coupling to HCV antigens.
The coupling of recombinant antigens to paramagnetic carboxylated beads (Luminex Corp., TX) was performed according to the manufacturer's instructions. Briefly, 106 beads were vortexed and sonicated to ensure a homogeneous distribution. The bead suspension was then washed twice with double-distilled water (ddH2O) and suspended in 80 μl of activation buffer (100 mM sodium phosphate [pH 6.2]). Solutions (10 μl each) of N-hydroxysulfosuccinimide (sulfo-NHS; Pierce, IL) and 1-ethyl-3-(3-dimethylaminopropyl)-carbodiimide hydrochloride (EDC; Pierce), both diluted to 50 mg/ml in ddH2O, were added to stabilize the reaction and activate the beads. After mixing, the beads were incubated for 20 min in the dark at room temperature. The activated beads were subsequently washed with coupling buffer (0.1 M NaHCO3 [pH 8.0]), after which 100 μl of antigen solution was added and incubated with shaking for 2 h. Antigen concentrations for coupling were experimentally determined previously (data not shown). After incubation, the beads were washed (1× phosphate-buffered saline [PBS], 1% bovine serum albumin [BSA], 0.02% Tween 20, 0.05% sodium azide) and suspended in 200 μl of blocking/storage buffer (1× PBS, 1% BSA, 0.02% Tween 20, 0.05% sodium azide). The beads were counted with a hemocytometer, adjusted to a concentration of 106 beads/ml with storage buffer, and stored protected from light at 2°C to 8°C.
Bead-based immunoassay standard protocol.
Serum samples were diluted in assay buffer (1× PBS, 1% BSA, 0.02% Tween 20, 0.05% sodium azide), and the test was performed by using standard procedures according to instructions provided by the manufacturer (Luminex Corp.). A total of 50 μl, containing approximately 2,000 coupled beads, was added to each well of a flat-bottom 96-well plate. For the multiplex assays, the same number of coupled beads was added to each well for each bead set. Diluted serum (50 μl) and beads were mixed and incubated for 30 min in the dark. The beads were then washed twice with 100 μl of wash buffer (1× PBS, 1% BSA, 0.02% Tween 20, 0.05% sodium azide), and 100 μl goat anti-human IgG conjugated to phycoerythrin (Sigma-Aldrich, MO) was added and incubated for 30 min in the dark. The beads were washed twice with 100 μl of wash buffer, and the bead reporter fluorescence, expressed as the median fluorescence intensity (MFI), was determined with a LabScan 100 instrument (One Lambda, CA). All incubations were performed at 37°C with a microplate shaker (set at 600 rpm), and the wash steps were performed with a Hydroflex plate washer with a magnetic plate support (Tecan, NC).
Net MFI and cutoff determinations.
Samples were always assayed in duplicates, and the MFI values were considered valid when the bead count reached a minimum of 100 beads per bead set per well. The net MFI values were obtained by subtracting the mean MFI of the duplicates of each sample from the mean of the MFI obtained from the background wells (no serum added). The cutoff value for each antigen was determined by using receiver operating characteristic (ROC) analysis, defined as the MFI value that gave the best combination of sensitivity and specificity. Samples were then classified as “positive” or “negative” according to the cutoff values specific for each antigen. For the multiplex assay, a sample was considered “positive” if it had one or more positive results from the individual antigens.
Statistical analysis.
Test performance was assessed according to the cutoff values determined for each antigen. All samples (positive and negative) were initially assayed in duplicate. Upon repeat testing, specimens were classified as “positive” (reactive) or “negative” (nonreactive). The results of the second test confirmed the previous results in 100% of the cases. Sensitivity was defined as the correct identification of anti-HCV antibody-positive samples, according to the following formula: sensitivity = number of true-positive samples/(number of true-positive samples + number of false-negative samples) × 100%. Specificity was defined as the correct identification of anti-HCV antibody-negative samples, according to the following formula: specificity = number of true-negative samples/(number of true-negative samples + number of false-positive samples) × 100%. True positives and true negatives were defined as the numbers of anti-HCV-positive and -negative samples identified correctly by each test. False positives and false negatives were defined as the numbers of anti-HCV-negative or -positive samples identified incorrectly by each method. Other outcome measures were as follows: positive predictive value (PPV) [PPV = number of true-positive samples/(number of true positive samples + number of false-positive samples) × 100%] and negative predictive value (NPV) [NPV = number of true-negative samples/(number of true-negative samples + number of false-negative samples) × 100%].
RESULTS
Antigen performance in the singleplex format.
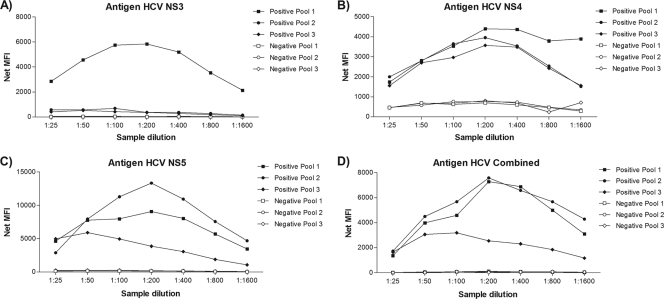
As the performance of each antigen influences the final outcome of the assay, we initially assessed the efficiency of antigen coupling to the beads and identified the best sample dilution to be used for the assay using six serum sample pools derived from the AEQ panel (three positive for anti-HCV antibody and three negative). Each HCV antigen was coupled to an individual bead set. Serial dilutions of pools ranging from 1:25 to 1:1,600 were prepared and assayed with the standard test protocol. The analyses of the performance of each antigen were made separately in order to assess the range of fluorescence generated by each individual bead set. All antigens presented high MFI values for the positive pools and lower values for the negative pools, demonstrating proper antigen coupling. In spite of the different behaviors of the three positive pools, a 1:200 dilution gave optimal signal results for all antigens (Fig. 1). This dilution proved to be the best compromise for the multiplexed format, where a common dilution is needed, and for that reason, it was used in all subsequent experiments. The test for the coupling efficiency showed that the highest coupling readings ranged from 2,000 to 12,000 MFI for the five different antigens, while background levels (no serum added) were lower than 100 MFI, proving that antigen coupling to the beads was successful (Fig. 1).
Fig. 1.
Efficiency of HCV antigens coupling to microspheres. The net MFIs of antigen detection for the antigens NS3 (A), NS4 (B), NS5 (C), and Combined (D) in serially diluted samples of HCV-positive pools 1, 2, and 3 and HCV-negative pools 1, 2, and 3 are shown. All samples were assayed in duplicate.
Singleplex specificity analysis.
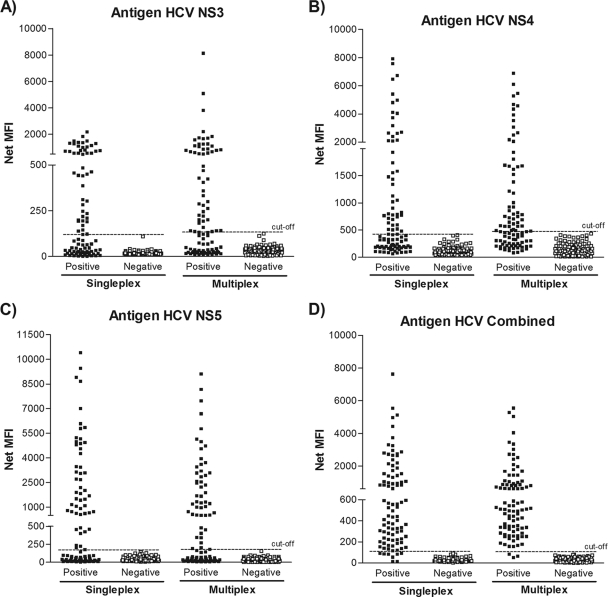
With the coupling efficiency confirmed and the sample dilution determined, our next step was to verify the specific anti-HCV responses of patient samples. For that purpose, 93 sera positive and 93 sera negative for anti-HCV antibody were individually assayed against all antigens. When the NS3, NS4, and NS5 antigens were assayed with individual samples in the singleplex format, they were not able to clearly differentiate the negative from the positive serum samples, whereas the Combined antigen succeeded in doing so (Fig. 2). This was not an unexpected result, since the Combined antigen is a fusion protein composed of different epitopes that include all three antigens mentioned above, reinforcing the importance of combining distinct antigens for better detection.
Fig. 2.
Comparison of the performances of the singleplex and multiplex assays. Shown are the net MFIs obtained by a 1:200 dilution of 93 HCV-positive serum samples and 93 HCV-negative sera evaluated in singleplex and multiplex formats against the antigens NS3, NS4, NS5, and Combined. For the multiplex assays, the same number of coupled microspheres was added to each well for each microsphere set. A sample was considered “positive” if it had one or more positive results from the individual antigens. All samples were assayed in duplicate.
Antigen performance in the multiplex format.
To determine if MFIs were equivalent when antigen-coupled microspheres were used in a singleplex format (i.e., a single antigen) or in a multiplex format (i.e., equal amounts of four antigen-coupled microspheres used simultaneously), the same positive and negative sera used previously were screened (Fig. 2). All antigens in the multiplex assay showed results for positive and negative samples similar to those of the singleplex assay (Fig. 2). Thus, multiplexing of the four HCV antigens did not alter the results obtained when antigens were tested individually.
Performance evaluation for the singleplex and multiplex tests.
To evaluate the accuracy parameters of the singleplex and multiplex tests, the samples were classified as “positive” or “negative” according to the cutoff values determined specifically for each antigen. Table 1 shows the sensitivities, specificities, positive predictive values (PPVs), and negative predictive values (NPVs) for the singleplex assays and for the multiplex test. All singleplex assays, with the exception of the Combined singleplex assay, presented unsatisfactory results, with poor sensitivity despite good specificity. Alternatively, the multiplex test showed excellent results, with 100% sensitivity, specificity, PPV, and NPV.
Table 1.
Accuracy parameters of the singleplex and multiplex test formats
| Antigen | % Sensitivity | % Specificity | PPV (%) | NPV (%) |
|---|---|---|---|---|
| NS3 | 50.5 | 100 | 100 | 66.9 |
| NS4 | 51.6 | 100 | 100 | 67.4 |
| NS5 | 55.9 | 100 | 100 | 69.4 |
| Combined | 93.5 | 100 | 100 | 93.9 |
| Multiplex | 100 | 100 | 100 | 100 |
DISCUSSION
The detection of anti-HCV antibodies is indispensable for the identification and screening of HCV-infected individuals. In this study, we have described the development of a multiplex assay for the simultaneous identification of human antibodies against five HCV antigens in a single serum dilution.
To confirm antigen coupling to the beads, our first approach was to determine the antibody responses to each antigen individually using serially diluted positive and negative serum pools. Although different patterns were observed for each dilution curve of the positive pools, these results were not surprising. Since antibody responses to HCV antigens can be influenced by HCV genotype, antigenic variation, or viremia, as well as major histocompatibility complex (MHC) or T-cell receptor (TCR) and immunoglobulin phenotypes, antibody sets may differ among infected individuals (7, 24). In fact, when the samples that comprise the pools were analyzed individually, we observed that they presented different responses to the four individual antigens evaluated and that their overall MFI signals were fully compatible with the heterogeneity in responsiveness observed among the pools (data not shown).
A major concern with multiplexing beads is antibody competition or blocking (11, 20). These occurrences are to be expected when antigens that share antibody epitopes are detected together in a multiplex format. As the Combined antigen has epitopes that are also present in the NS3, NS4, and NS5 antigens, one could predict a signal decrease caused by the competition of binding sites. However, the present work demonstrated for all antigens tested that multiplexing does not appear to alter the quality or sensitivity of the assays compared to those of the singleplex format. Another issue to be considered is the degrees of interference and cross-reactivity between the different bead sets. While multiplex immunoassays facilitate the analysis of various antibody responses simultaneously, the mixture of several antigens can lead to an unspecific binding of antibodies to the wrong antigen (12, 17). Nevertheless, in this study, no significant difference was found when antigen-coupled beads were used alone or in combination, indicating that the antigen-antibody complexes formed were specific and stable.
The main criteria for an HCV screening assay are to attain the highest sensitivity possible in combination with excellent specificity. In our study, the sensitivity, specificity, negative predictive value, and positive predictive value of the multiplex test were 100%, higher than those of the NS3, NS4, NS5, and Combined singleplex assays. Despite the small number of samples evaluated, the high values for specificity and sensitivity obtained are particularly important given the potential use of the HCV multiplex test for blood screening and/or epidemiological surveillance programs. It is important to mention that although the use of the Combined antigen was essential to achieve good sensitivity in the multiplex assay, the identification of the positive samples that were not reactive to the Combined antigen was possible only because these samples were responsive to the other three antigens used (NS3, NS4, and/or NS5), clearly demonstrating the benefit and importance of using and combining these distinct antigens to improve detection.
The multiplex test described herein has numerous advantages for the simultaneous measurement of antibodies to multiple HCV antigens. The total time required for performing this assay was 2 h, virtually the same time as those required for two commercially available kits (Abbott HCV EIA 3.0 and Ortho HCV ELISA 3.0). The estimated reaction cost of the multiplex assay was 2 to 4 times lower than that of available commercial methods in Brazil. In order to estimate the costs of the multiplex assay, we employed the approximate costs of beads, antigens, reagents, and disposable items used during the assay procedure. Test cost estimation was based on currently available commercial reagent prices in Brazil at the time when the study was conducted. Equipment, human resources, and other indirect costs were not considered for comparison and calculation. The cost reduction associated with this new assay would make its use possible for several health services in Brazil, allowing the establishment of the real prevalence of HCV in the country.
Further improvements to the assay could be gained by incorporating additional features to include new targets of interest, such as antigens/antibodies specific for other hepatitis viruses, like hepatitis B virus (HBV), for example. This would add further power to the assay by allowing the simultaneous detection of host antibodies and pathogen-specific antigens. In addition, the HCV multiplex assay has great potential for being used as both a screening test and a confirmatory test. As the reactivities of all four antigens can be individualized, it is possible that samples could be both screened and confirmed with this one test by establishing recombinant immunoblot assay (RIBA)-like criteria for the individual antigens. In such a situation, possibly a superoxide dismutase or another marker bead should be included to rule out interferences. This would decrease the costs associated with HCV diagnosis even more, improving treatment and surveillance strategies.
We believe that a more thorough study of the multiplex assay's performance including an analysis of samples from patients with a resolved infection versus samples from patients with an active infection and also comparing it to supplemental serological testing currently used for HCV detection would be important to establish the true potential of this multiplex test. Although further work will be required to establish the use of the HCV multiplex assay as a diagnostic tool, the test described herein is sensitive and rapid and shows excellent specificity. Therefore, the assay has the potential to become a viable alternative to standard tests and should simplify screening for HCV infection.
Footnotes
Published ahead of print on 23 February 2011.
REFERENCES
- 1. Abdel-Hamid M., et al. 2002. Comparison of second- and third-generation enzyme immunoassays for detecting antibodies to hepatitis C virus. J. Clin. Microbiol. 40:1656–1659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Alter H. J. 1992. New kit on the block: evaluation of second-generation assays for detection of antibody to the hepatitis C virus. Hepatology 15:350–353 [DOI] [PubMed] [Google Scholar]
- 3. Ansari M. H. K., Omrani M. D. 2006. Evaluation of diagnostic value of Elisa method (EIA) & PCR in diagnosis of hepatitis C virus in hemodialysis patients. Hepatitis Mon. 6:19–23 [Google Scholar]
- 4. Araújo E. S., et al. 2007. Consensus of the Brazilian Society of Infectious Diseases on the management and treatment of hepatitis C. Braz. J. Infect. Dis. 11:446–450 [DOI] [PubMed] [Google Scholar]
- 5. Armstrong G., Alter M. J., Mcquillan G. M., Margolis H. S. 2000. The past incidence of hepatitis C virus infection: implications for the future burden of chronic liver disease in the United States. Hepatology 31:777–782 [DOI] [PubMed] [Google Scholar]
- 6. Baath L., Widell A., Nordenfelt E. 1992. A comparison between one first generation and three second generation anti-HCV ELISAs: an investigation in high- and low-risk subjects in correlation with recombinant immunoblot assay and polymerase chain reaction. J. Virol. Methods 40:287–296 [DOI] [PubMed] [Google Scholar]
- 7. Beld M., et al. 1999. Quantitative antibody responses to structural (core) and nonstructural (NS3, NS4, and NS5) hepatitis C virus proteins among seroconverting injecting drug users: impact of epitope variation and relationship to detection of HCV RNA in blood. Hepatology 29:1288–1298 [DOI] [PubMed] [Google Scholar]
- 8. Bellisario R., Colinas R. J., Pass K. A. 2001. Simultaneous measurement of antibodies to three HIV-1 antigens in newborn dried blood-spot specimens using a multiplexed microsphere-based immunoassay. Early Hum. Dev. 64:21–25 [DOI] [PubMed] [Google Scholar]
- 9. Binnicker M. J., Jespersen D. J., Harring J. A., Rollins L. O., Beito E. M. 2008. Evaluation of a multiplex flow immunoassay for detection of Epstein-Barr virus-specific antibodies. Clin. Vaccine Immunol. 15:1410–1413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Boyer N., Marcellin P. 2000. Pathogenesis, diagnosis and management of hepatitis C. J. Hepatol. 32:S98–Sl12 [DOI] [PubMed] [Google Scholar]
- 11. Carson R. T., Vignali D. A. 1999. Simultaneous quantitation of 15 cytokines using a multiplexed flow cytometric assay. J. Immunol. Methods 227:41–52 [DOI] [PubMed] [Google Scholar]
- 12. de Voer R. M., et al. 2008. Development of a fluorescent-bead-based multiplex immunoassay to determine immunoglobulin G subclass responses to Neisseria meningitidis serogroup A and C polysaccharides. Clin. Vaccine Immunol. 15:1188–1193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Dunbar S. A. 2006. Applications of Luminex xMAP technology for rapid, high-throughput multiplexed nucleic acid detection. Clin. Chim. Acta 363:71–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Foti L., et al. 2009. Viability study of a multiplex diagnostic platform for Chagas disease. Mem. Inst. Oswaldo Cruz 104(Suppl. 1):136–141 [DOI] [PubMed] [Google Scholar]
- 15. Fouda G. G., et al. 2006. Multiplex assay for simultaneous measurement of antibodies to multiple Plasmodium falciparum antigens. Clin. Vaccine Immunol. 13:1307–1313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Fulton R. J., McDade R. L., Smith P. L., Kienker L. J., Kettman J. R., Jr 1997. Advanced multiplexed analysis with the FlowMetrix system. Clin. Chem. 43:1749–1756 [PubMed] [Google Scholar]
- 17. Gouri L., Balmer P., Joseph H., Dawson M., Borrow R. 2004. Development and evaluation of a Tetraplex flow cytometric assay for quantitation of serum antibodies to Neisseria meningitidis serogroups A, C, Y, and W-135. Clin. Diagn. Lab. Immunol. 11:272–279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Jani I. V., Janossy G., Brown D. W., Mandy F. 2002. Multiplexed immunoassays by flow cytometry for diagnosis and surveillance of infectious diseases in resource-poor settings. Lancet Infect. Dis. 2:243–250 [DOI] [PubMed] [Google Scholar]
- 19. Joos T. O., Stoll D., Templin M. F. 2002. Miniaturized multiplexed immunoassays. Curr. Opin. Chem. Biol. 6:76–80 [DOI] [PubMed] [Google Scholar]
- 20. Kellar K. L., Iannone M. A. 2002. Multiplexed microsphere-based flow cytometric assays. Exp. Hematol. 30:1227–1237 [DOI] [PubMed] [Google Scholar]
- 21. Pawlotsky J. M. 2002. Use and interpretation of virological tests for hepatitis C. Hepatology 36:S65–S73 [DOI] [PubMed] [Google Scholar]
- 22. Raedler M. D., et al. 2009. Serologic assay to quantify human immunoglobulin G antibodies to the Staphylococcus aureus iron surface determinant B antigen. Clin. Vaccine Immunol. 16:739–748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Sillanpää M., et al. 2009. Hepatitis C virus core, NS3, NS4B and NS5A are the major immunogenic proteins in humoral immunity in chronic HCV infection. Virol. J. 6:84–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Thimme R., Neumann-Haefelin C., Boettler T., Blum H. E. 2008. Adaptive immune responses to hepatitis C virus: from viral immunobiology to a vaccine. Biol. Chem. 389:457–467 [DOI] [PubMed] [Google Scholar]