Abstract
The magnitude of the immune responses elicited by plasmid DNA vaccines might be limited, in part, by the duration of vaccine antigen expression in vivo. To explore strategies for improving plasmid DNA vaccine efficacy, we studied the apoptotic process in myocytes of mice vaccinated intramuscularly. We found that after vaccination, the proapoptotic protein caspase 12 (Casp12) was upregulated in myocytes coincident with the loss of vaccine antigen expression. To harness this observation to improve plasmid DNA vaccine efficacy, we used RNA interference technology, coadministering plasmid DNA expressing a short hairpin RNA (shRNA) of Casp12 with plasmid DNA vaccine constructs. This treatment with shRNA Casp12, administered twice within the first 10 days following vaccine administration, increased antigen expression 7-fold, the antigen-specific CD8+ T cell immune response 6-fold, and antigen-specific antibody production 5-fold. This study demonstrates the critical role for Casp12 in plasmid DNA vaccine-induced immune responses and shows that increased antigen expression mediated by down-modulation of Casp12 can be used to potentiate vaccine efficacy.
INTRODUCTION
While there are many benefits to using plasmid DNA as a vaccine immunogen, the utility of this vaccine modality has been limited by its failure to elicit sufficiently potent immune responses (26, 40). The immunogenicity of viral or bacterial vectors can be limited in humans by anti-vector immunity, either preexisting or generated as a consequence of repeated vaccine administration (28, 33). In contrast, vector backbone immunity is not generated against plasmid DNA immunogens. Therefore, repeated administration of plasmid DNA immunogens can induce increasing immune responses. Nevertheless, the immune responses elicited by repeated administration of plasmid DNA vectors are limited in nonhuman primates and humans. This limited immunogenicity may be a consequence of the relative brief duration of vaccine antigen expression in vivo. Approximately 14 days after intramuscular (i.m.) administration of plasmid DNA, an adaptive immune response that mediates the apoptotic destruction of vaccine antigen-expressing myocytes is generated (10).
In the present study, we investigated the apoptotic process in the vaccine antigen-expressing myocytes of plasmid DNA-inoculated mice. We reasoned that a disruption of the apoptotic pathway through the use of RNA interference (RNAi) might prolong vaccine antigen expression, which might increase T cell and antibody responses. In fact, we found that RNAi targeting of caspase 12 (Casp12), a major proapoptotic protein activated 14 days after plasmid DNA vaccination, led to increased in vivo antigen expression and the augmentation of antigen-specific CD8+ T cell and antibody responses.
MATERIALS AND METHODS
Ethics statement.
All animals were housed and maintained in accordance with the Guide for the Care and Use of Laboratory Animals (22a, 24), and all studies and procedures were reviewed and approved by the Institutional Animal Care and Use Committee (IACUC) of the Beth Israel Deaconess Medical Center (BIDMC). BIDMC follows NIH guidelines for animal handling and has Animal Welfare Assurance number A3153-01 on file with the Office for Protection of Research Risks. This institution maintains full accreditation from the Association for Assessment and Accreditation of Laboratory Animal Care.
Mice.
Six- to eight-week-old, wild-type C57BL/6 mice were purchased from the Jackson Laboratory (36).
Vectors and immunization.
The plasmid DNA-luciferase (DNA-Luc) construct containing the cytotoxic T lymphocyte (CTL) epitope Gag AL11 tag was constructed as previously described (10). This vector contained the GL4.10 luciferase gene (Promega, Madison, WI) and the immunodominant H-2Db-restricted simian immunodeficiency virus (SIV)-Gag (AL11) epitope (AAVKNWMTQTL) flanked by triple-alanine spacers (plasmid DNA-Luc vaccine construct). The codon-optimized human immunodeficiency virus 1 (HIV-1) HXB2 env gene was cloned into the VRC vector (plasmid DNA-gp120 vaccine construct) as previously described (11). The VRC vector was provided by G. Nabel (Vaccine Research Center, National Institute of Allergy and Infectious Diseases, National Institutes of Health). Plasmid DNA-small hairpin RNA (shRNA) constructs were obtained from OriGene Technologies (Rockville, MD). Plasmid DNA was prepared using an endotoxin-free Qiagen Giga prep kit (Valencia, CA). The endotoxin concentration of the plasmid DNA preparations was below 0.1 U/μg plasmid DNA as determined with the E-Toxate kit (Sigma-Aldrich, St. Louis, MO). For immunizations, 50 μg of the plasmid DNA vaccine construct, with 200 μg of a plasmid DNA-shRNA construct, was suspended in 100 μl of sterile saline and administered at day 0 by intramuscular (i.m.) inoculation, divided between the quadriceps muscles. At day 10, 200 μg of a plasmid DNA-shRNA construct was administered.
Monoclonal antibodies.
Allophycocyanin (APC)- and phycoerythrin (PE)-labeled antibodies were used for the flow cytometric analyses. The dye-coupled antibody anti-CD8α-APC (53-6.7) was purchased from BD Bioscience (San Jose, CA). H-2Db/AL11 and H-2Dd/p18 tetramer-PE were prepared as previously described (2).
Immune assays.
Peripheral blood was collected and lysed with BD Pharm lyse buffer (BD Bioscience). Samples were stained using anti-CD8α-APC and H-2Db/AL11 or H-2Dd/p18 tetramer-PE and then analyzed on a fluorescence-activated cell sorting (FACS) array flow cytometer (BD Bioscience). Vaccine antigen-specific CD8+ T lymphocytes were identified by staining with the H-2Db/AL11 and H-2Dd/p18 (RGPGRAFVTI)-PE tetramer. CD8+ T lymphocytes from control mice immunized with the untagged plasmid DNA-Luc construct exhibited less than 0.1% tetramer staining.
For anti-luciferase IgG antibody titer measurements, recombinant luciferase protein (Promega, Madison, WI) was coated overnight at 5 μg/ml in 100 μl/well at 4°C onto Costar 96-well enzyme immunoassay (EIA)/radioimmunoassay (RIA) plates (Fisher Scientific, Pittsburgh, PA). Plates were washed and incubated with increasing dilutions of mouse serum. Luciferase protein standard curves were obtained by coating increasing dilutions of recombinant luciferase protein (Promega, Madison, WI) onto Costar 96-well EIA/RIA plates. Plates were blocked by bovine serum albumin (BSA) blocking solution, followed by monoclonal anti-luciferase antibody (Sigma-Aldrich, St. Louis, MO; catalog number 015K855). Labeling and assay development using a protein detector enzyme-linked immunosorbent assay (ELISA) kit (KPL) were performed according to the manufacturer's protocol. Serum binding anti-HIV gp120 Env antibody titers were determined by coating wells with 100-μl volumes of 1 μg/ml clade C gp140 trimer (CZA97.012) (23) overnight at 4°C onto Costar 96-well EIA/RIA plates. Plates were blocked by BSA blocking solution, followed by 1 h of room temperature (RT) incubation with a 1/4,000 dilution of a horseradish peroxidase (HRP)-conjugated goat anti-mouse secondary antibody (Jackson ImmunoResearch Laboratories, West Grove, PA). Assays were developed using the protein detector ELISA kit (KPL, Gaithersburg, MD) according to the manufacturer's protocol. Titers were analyzed at 405 nm on a Spectramax Plus ELISA plate reader (Molecular Devices, Sunnyvale, CA) using Softmax Pro version 4.7.1 software. ELISA endpoint titers were defined as the highest reciprocal serum dilution that yielded absorbance of >2-fold above that of the background.
Measurement of in vivo bioluminescence and antigen expression.
Animals were injected intraperitoneally (i.p.) with 100 μl of a 30-mg/ml solution of firefly luciferin (Xenogen, Alameda, CA) in phosphate-buffered saline (PBS) and 100 μl of a mixture containing 20 mg/ml ketamine and 1.72 μg/ml xylazine. After 20 min, imaging was performed using the IVIS series 100 imaging system (Xenogen) with an integration time of 1 min. Overlay images and luminescence measurements were obtained using Living Image software (version 2.50.1; Xenogen). To convert the in vivo bioluminescence of relative light units (RLU) of the plasmid DNA-Luc vaccine into the quantity of antigen expressed, we prepared a standard curve of emitted light per minute for different amounts of recombinant luciferase protein. The linear correlation between the amount of protein injected (10 ng to 50 μg) and the light emitted enabled the calculation of antigen expression using the following formula: concentration of antigen in nanograms = antilog [(log RLU − 4.2)/0.76].
Antiapoptotic plasmids.
Plasmid DNAs encoding the antiapoptotic proteins BCL-xl, BCL-2, X-linked inhibitor of apoptosis protein (XIAP), or dominant negative (dn) mutants of caspase 9 (dn-Casp9) and caspase 8 (dn-Casp8) were used as described previously (12–15). Two hundred micrograms of these plasmid DNA constructs was delivered with 50 μg of a plasmid DNA construct in 100-μl volumes. In vivo transfection of muscle tissue with plasmid DNA-short hairpin interference RNA constructs was done using continuously expressed 29-nucleotide shRNA constructs to silence target protein expression (OriGene). Effects of plasmid DNA-shRNA constructs on target protein expression were tested in vitro by Western blot analysis. The plasmid DNA-shRNA construct for Luc mRNA transcripts, plasmid DNA-shLuc (catalog number TR30002), showed greater than 90% inhibition of luciferase expression in vitro. The plasmid shRNA-GFP (catalog number TR30001) showed more than 90% inhibition of the green fluorescence protein (GFP) production in vitro. The plasmid-shCasp12 constructs (GGAGGACACATGAAAGAGATCCAATCTAC or CCTCTTTCATTTCCAAACTCGTTGACTGC) used showed a greater than 70% inhibition of their target genes in vitro. One hundred micrograms of each of the Casp12 constructs was injected simultaneously with the plasmid DNA vaccine constructs on the day of vaccination, followed by i.m. administration without the plasmid DNA vaccine on day 10.
Gene array for the analysis of apoptosis-associated genes.
To analyze the expression of apoptosis genes, the Oligo GEArray mouse apoptosis microarray (SA Biosciences, Frederick, MD; catalog number OMM-012) was used on snap-frozen quadricep muscles from vaccinated or unvaccinated mice 14 days after plasmid DNA vaccination according to the manufacturer's protocol. The Oligo GEArray mouse apoptosis microarray profiled the expression of 112 genes involved in apoptosis. Unvaccinated animals were used as controls. Gene expression was normalized against the housekeeping gene product GAPDH (glyceraldehyde-3-phosphate dehydrogenase).
Data analysis.
The statistical significance of differences between two experimental groups was determined using the Mann-Whitney test. A P value of <0.05 was considered to be significant. Statistical calculations were performed using the GraphPad Prism program (version 4.03). Error bars represent the standard errors of the means (SEM).
RESULTS
Expression of genes involved in apoptosis was associated with the loss of antigen expression in mice vaccinated with plasmid DNA. The reduction of vaccine antigen expression in vivo in mice vaccinated i.m. with plasmid DNA is temporally correlated with local accumulation of T cells in the muscle and involves an apoptotic Fas/FasL-dependent process (8, 10). Therefore, we coadministered plasmid DNA vaccine and plasmid DNAs encoding the antiapoptotic proteins BCL-xl, BCL-2, X-linked inhibitor of apoptosis protein (XIAP), or dominant negative (dn) mutants of caspase 9 (dn-Casp9) and caspase 8 (dn-Casp8) to evaluate their abilities to augment the immunogenicity of a plasmid DNA immunogen. The administration of these plasmid DNAs had no effect on the CD8+ T cell immune response in the vaccinated mice (day 14 P values of 0.17, 0.68, 0.36, 0.46 and 0.89) (data not shown).
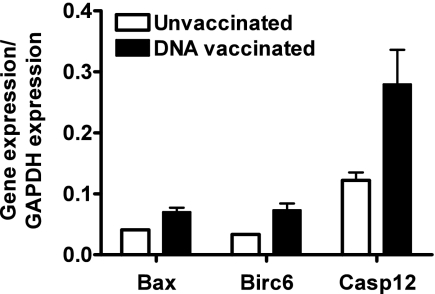
To explore further the apoptotic process in the vaccinated muscle, we extracted muscles from vaccinated and unvaccinated mice 14 days after vaccination. An antiapoptosis gene array allowed for gene expression profiling of 112 genes associated with apoptosis. We found that two proapoptotic genes, Casp12 and Bax, were activated in this tissue at this late time after vaccination. Expression of an antiapoptosis gene product, Birc6, was also increased (Fig. 1). These data suggested that Casp12 might be involved in the apoptosis of myocytes following plasmid DNA vaccine administration.
Fig. 1.
Apoptosis gene expression profile in muscle after plasmid DNA vaccination. Using an Oligo GEArray mouse apoptosis microarray, the expression levels of 112 genes in the quadricep were compared between vaccinated and unvaccinated mice. Gene expression was normalized using the GAPDH housekeeping gene. Genes with an expression ratio greater than 0.03 to GAPDH and significantly upregulated (Mann-Whitney test, P < 0.05, four animals per group) compared to the untreated controls at day 14 after plasmid DNA vaccine inoculation are shown: Bcl-2-associated X protein (Bax), baculoviral inhibitor of apoptosis repeat-containing protein 6 (Birc6) and caspase 12 (Casp12).
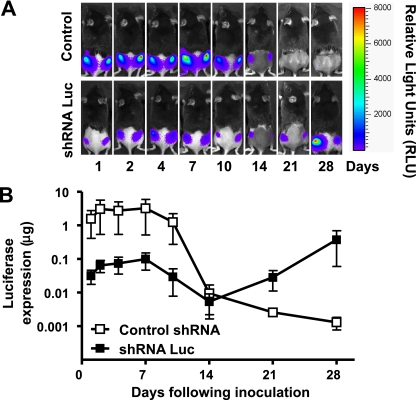
Coadministration of a plasmid DNA-shLuc construct with a plasmid DNA-Luc vaccine construct decreased the level of plasmid DNA luciferase expression. To explore the potential contribution of Casp12 in limiting plasmid DNA antigen expression in vivo, we sought to use RNAi technology (4, 18, 32) to silence the Casp12 gene. To determine the feasibility of this strategy, we codelivered DNA encoding luciferase and DNA encoding an shRNA that targets the luciferase gene and then characterized the expression of luciferase over time using a noninvasive bioluminescence imaging system. Results were compared to those in mice that received DNA-encoding luciferase and an shRNA control (shGFP). We found that luciferase expression was suppressed up to 1.5 logs for 10 days by shRNA directed against the luciferase gene. After 10 days, luciferase gene expression returned to normal levels. Control animals given shRNA directed against an irrelevant gene target showed normal levels of luciferase gene expression with the expected loss of antigen expression by day 14 following plasmid DNA vaccine administration (Fig. 2). This observation demonstrated that the administration of a plasmid DNA-shRNA construct can modulate the local expression of a plasmid DNA vaccine construct in mice.
Fig. 2.
Effect of codelivery of the plasmid DNA-Luc vaccine construct with the antigen-silencing plasmid DNA-shLuc construct on vaccine antigen expression. (A) Representative infrared signal images with overlaid photo images to compare luciferase expression following coinoculation of plasmid DNA-Luc vaccine and plasmid DNA-shLuc constructs to coinoculation of plasmid DNA-Luc vaccine and plasmid DNA-shRNA control constructs (shGFP). Scale shows measured luciferase activity as relative light units (RLU) for 1-min measurements. (B) Comparison of quantitated antigen expression after coinoculation of plasmid DNA-Luc vaccine and plasmid DNA-shLuc constructs to coinoculation of the plasmid DNA-Luc vaccine and plasmid DNA-shRNA control constructs (shGFP). In vivo expression of luciferase was measured by an in vivo imaging system (IVIS) and quantitated with a standard curve generated for quadricep muscle injections of the recombinant Luc protein. Means ± SEM for five animals per group. Experiments were done twice. Representative data are shown.
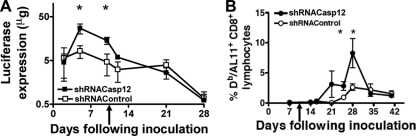
Coadministration of plasmid DNA-shCasp12 constructs with a plasmid DNA-Luc vaccine construct resulted in a delay in the loss of vaccine antigen expression and an increase in antigen-specific CD8+ T cell responses. Since the gene array experiment suggested that Casp12 might mediate the apoptosis associated with the loss of vaccine antigen expression, we explored whether reducing Casp12 expression using shRNA might modulate the loss of vaccine antigen expression and therefore enhance the vaccine's immunogenicity. Since the luciferase experiments suggested that the shRNA was effective for 10 days, we administered shRNA Casp12 twice, at day 0 and day 10. We then monitored antigen expression in vivo by measuring the bioluminescence of the luciferase antigen. We also determined the antigen-specific T cell immune response elicited by a DNA vaccine by measuring vaccine-elicited SIV-Gag AL11-specific CD8+ T cells.
We found that shCasp12 delivery had a significant effect on both vaccine antigen expression and vaccine-elicited antigen-specific CD8+ T cell immune responses. Coadministration of shCasp12 increased vaccine antigen expression up to 7-fold at day 4 and day 10 (Fig. 3A). Consistent with this finding, the antigen-specific CD8+ T cell immune response was increased up to 10-fold at days 24 and 28 following immunization (Fig. 3B). These data suggest that the delay in the loss of antigen expression associated with the administration of shCasp12 resulted in an increase in the plasmid DNA-induced CD8+ T cell immune response.
Fig. 3.
Effect of coexpression of the plasmid DNA-Luc vaccine construct with plasmid DNA-shCasp12 construct on luciferase expression and epitope-specific CD8+ T cell immune response. (A) Comparison of Luc antigen expression in mice coinoculated with the plasmid DNA-Luc vaccine construct and plasmid DNA-shCasp12 to mice coinoculated with the plasmid DNA-Luc vaccine and plasmid DNA-shRNA control constructs (shGFP). In vivo expression of luciferase was measured by IVIS imaging. (B) SIV-Gag AL11 epitope-specific CD8+ T cell responses in mice coinoculated with plasmid DNA-Luc vaccine and plasmid DNA-shCasp12 constructs or coinoculated with plasmid DNA-Luc vaccine and plasmid DNA-shRNA control constructs as described for panel A (shGFP). Mice were coinoculated on day 0 with the plasmid DNA-shCasp12 or plasmid DNA-shRNA control and plasmid DNA-Luc vaccine constructs and injected at day 10 with plasmid DNA-shCasp12 or plasmid DNA-shRNA control (arrow). Epitope-specific CD8+ T cell responses were measured by H-2Db/AL11 tetramer staining of CD8+ T cells at the indicated times following plasmid DNA inoculation. Mean ± SEM. *, significant difference (P < 0.05), Mann-Whitney test, five animals per group. Experiments were done twice. Representative data are shown.
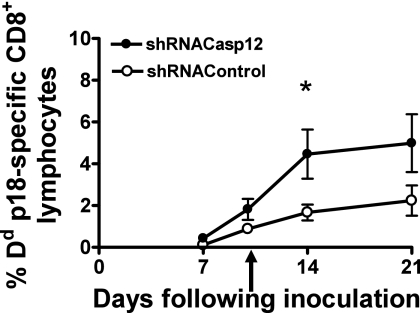
Coadministration of plasmid DNA-shCasp12 constructs with a plasmid DNA-HIV gp120 Env vaccine construct increased the Env-specific antigen-specific CD8+ T cell and antibody immune response. To explore further the utility of this strategy for enhancing plasmid DNA vaccine immunogenicity, we codelivered shCasp12 with a plasmid DNA vaccine construct expressing the HIV gp120 Env at day 0 (Fig. 4). We administered shRNA Casp12 or the shRNA control again at day 10. We observed up to a 5-fold increase in the HIV gp120 Env-specific CD8+ T cell immune response at day 14 following vaccine administration.
Fig. 4.
Effect of coexpression of plasmid DNA-gp120 vaccine with plasmid DNA-antiapoptosis shCasp12 constructs on epitope-specific CD8+ T cell response. Comparison of stained gp120-p18 epitope-specific CD8+ T cells in the peripheral blood of mice coinoculated with plasmid DNA-gp120 vaccine and plasmid DNA-shCasp12 constructs or plasmid DNA-gp120 vaccine and plasmid DNA-shRNA control (shGFP) constructs. Mice were coinoculated on day 0 with the plasmid DNA-shCasp12 or plasmid DNA-shRNA control constructs and the plasmid DNA-gp120 vaccine construct and injected at day 10 with plasmid DNA-shCasp12 or plasmid DNA-shRNA control (arrow). Epitope-specific CD8+ T cell responses were measured by gp120-p18 tetramer staining of CD8+ T cells at the indicated times following plasmid DNA inoculation. Mean ± SEM. *, significant difference (P < 0.05), Mann-Whitney test, five animals per group. Experiments were done twice. Representative data are shown.
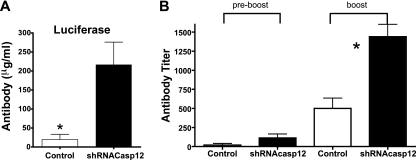
To determine if the plasmid DNA-shCasp12-induced prolongation of antigen expression had an effect on plasmid DNA vaccine-induced antibodies, we delivered the plasmid DNA-shRNA Casp12 construct with the plasmid DNA-Luc and the plasmid HIV gp120 Env vaccine constructs at day 0 and the shRNA Casp12 construct or the shRNA control at day 10. After 1 month, mice were boosted with plasmid DNA luciferase or plasmid HIV gp120 Env vaccine constructs without the shRNA Casp12. After 1 week, serum was collected and luciferase- or HIV gp120-specific antibody was measured by ELISA. We found increases in production of antigen-specific antibodies of 5-fold and 3-fold for the plasmid DNA-Luc and the HIV gp120 Env vaccine constructs, respectively, compared to that for control mice (Fig. 5). These data indicate that increasing antigen expression using shRNA can increase the production of antigen-specific antibodies induced by plasmid DNA vaccines.
Fig. 5.
Effect of coexpression of plasmid DNA-Luc vaccine or plasmid DNA-gp120 vaccine constructs with plasmid DNA-shCasp12 constructs on plasmid DNA vaccine-specific-induced antibody production. Mice were coinoculated on day 0 with the plasmid DNA-shCasp12 or plasmid DNA-shRNA control constructs and plasmid DNA-Luc vaccine or plasmid DNA-gp120 vaccine constructs and injected at day 10 with plasmid DNA-shCasp12 or plasmid DNA-shRNA control (arrow). Mice were boosted after 1 month with the plasmid DNA vaccine constructs alone. Serum was collected after 4 weeks (preboost) and 1 week after boosting and total Luc-specific (A) or gp120-specific (B) IgG antibody was measured by ELISA. Mean ± SEM. *, significant difference (P < 0.05), Mann-Whitney test, five animals per group. Experiments were done twice. Representative data are shown.
DISCUSSION
A variety of strategies for augmenting the immunogenicity of plasmid DNA vaccines have been explored (20). These include the use of a gene gun, electroporation, and the codelivery of DNA encoding antiapoptotic proteins. These proteins have included BCL-xl, BCL-2, XIAP, dn-Casp9, and dn-Casp8. The coadministration of DNA encoding these proteins led to an inhibition of dendritic cell (DC) apoptosis and an increase in the plasmid DNA-induced antigen-specific CD8+ T cell immune responses (13). However, this vaccine enhancement was seen for plasmid DNA vaccination administered by the intradermal (i.d.) but not the i.m. route (5, 17, 31). These differences in the effects seen following administration of DNA encoding antiapoptotic proteins might be a consequence of the difference in cell types that act as primary antigen-presenting cells following i.m. and i.d. plasmid DNA vaccine delivery. The primary source of vaccine antigen following i.m. administration seems to be the myocytes, while following i.d. administration it seems to be DCs (8, 9, 21, 35). We felt that a more detailed investigation into the mechanism of apoptosis was needed to determine the intracellular apoptosis-associated protein(s) responsible for the damping of the antigen expression in the myocytes after i.m. plasmid DNA vaccine administration.
The apoptosis gene array experiment carried out in the present study demonstrated that Casp12 was upregulated following DNA plasmid vaccine administration. It is certainly possible that other apoptosis-associated genes might be upregulated in the muscle of plasmid DNA-vaccinated mice earlier following vaccine administration. Nevertheless, we showed that coinjection of a plasmid DNA vaccine construct with a plasmid containing DNA that encodes an shCasp12 led to the prolongation of antigen expression and an increase in both antigen-specific CD8+ T cell and antibody immune responses. Casp12 is activated in response to endoplasmic reticulum (ER) stress (22), which might occur as a consequence of exhaustion of the translational machinery resulting from overexpression of the plasmid DNA antigen. Fourteen days after plasmid DNA vaccination, Casp12 activation might initiate the amplification of an apoptotic cascade downstream of the common sensors and/or effectors for cellular damage, Bax, Bak, and initiator Casp9 (29, 41), and therefore blockage of Casp12 expression might negate the apoptotic effect of all these factors.
This study provides an example of a successful harnessing of the RNAi technology. One obstacle in using RNAi in mammalian cells in vitro or in mammals in vivo has been that double-stranded RNAs (dsRNAs), especially those longer than 30 nucleotides, activate a cellular interferon response, leading to the nonspecific degradation of RNA transcripts and a general shutdown of all host cell protein translation (1, 34, 38). As a result, long dsRNA (>30 nucleotides), with a few exceptions (3, 7, 25), does not mediate long-lasting RNAi activity. The 10-day period of efficacy of the 29 nucleotide shRNAs seen in the present study is therefore likely a consequence of the balance between efficiently suppressing the expression of the gene of interest and at the same time lowering the cellular interferon response. We found that after 10 days the cells were no longer susceptible to the shRNA-mediated effects since target gene expression was restored (Fig. 2). We also observed a delay in the adaptive immune response due to the lowering of antigen expression (data not shown).
These findings differ from those of our other recent experiments in which the antigen is not the target of gene suppression but of the Casp12 gene. Our dose-finding experiments with shCasp12 showed that two injections with shCasp12, 10 days apart, were necessary to achieve an optimal effect on the CD8+ T cell immune response: one injection had only limited efficacy, and a third injection, 10 days after the second, resulted in no further increase of the antigen-specific CD8+ T cell immune response (data not shown). Thus, the shRNA-mediated effect was lost after the second injection, possibly due to the increasing strength of CD4+ T cell immune responses destroying antigen-producing myocytes after 14 days by a Fas/FasL mechanism (8). Our data demonstrate a 7-fold increase of antigen expression leading to a 6-fold increase of the antigen-specific CD8+ T cell immune response and a 5-fold increase in antigen-specific antibody production. Thus, an increase of antigen expression rather than a prolongation of antigen expression increased the adaptive immune responses.
CD4+ T cell-mediated apoptosis still occurred after 14 days. This was likely necessary for inducing CD8+ T cell immune responses, since the deletion of CD4+ T cells or major histocompatibility complex (MHC) class II molecules led to indefinite antigen expression without eliciting an antigen-specific CD8+ T cell immune response (8). A requirement for apoptosis for eliciting an adaptive immune response was also shown in earlier reports (5, 16, 27, 31). Nevertheless, the present experiments do not rule out the possibility that the effect on antigen-specific CD8+ T cell immune response is a consequence of the decrease of Casp12-mediated effects other than apoptosis (30, 39).
The generation of antigen-specific CD8+ T cells is increased by type I interferons (37). Short interference RNAs increase interferon production by the activation of a cellular interferon response (34). To rule out the possibility that these effects are the results of interferon production, we used the shRNA GFP in our control experiments.
It is not clear whether direct antigen presentation by myocytes to T cells, antigen presentation of myocyte-produced antigen by professional antigen-presenting cells to T cells, or cross-priming is the most important mechanism for generating a strong T cell immune response (6, 9, 19, 35). Thus, the apoptotic pathways that are activated may differ with the route of administration of the DNA immunogen and differences in the recruited antigen-presenting cells.
The setting of plasmid DNA vaccination is an ideal one for applying RNAi technology. Plasmid DNA-shRNA constructs can be administered with the same vector system as that used for the vaccine immunogens. Moreover, plasmid DNA-shRNA constructs can be administered directly to the target tissue by injection. The utility of this strategy in the present studies underscores the importance of apoptosis in plasmid DNA vaccination.
ACKNOWLEDGMENTS
We thank Michelle Lifton (BIDMC and Harvard Medical School) for advice, support, and reagents. We thank Maytal Bivas-Benita (BIDMC and Harvard Medical School) for the help with mouse i.m. injections. We thank Maytal Bivas-Benita and Christa Osuna-Gutierrez for the critical reading of the manuscript. The antiapoptotic plasmid DNA constructs used were a gift from J. Marie Hardwick and T.-C. Wu (Johns Hopkins University). The clade C gp140 trimer (CZA97.012) was a gift from Dan Barouch (BIDMC and Harvard Medical School).
This work was supported by the National Institute of Allergy and Infectious Diseases Center for HIV/AIDS Vaccine Immunology, grant AI067854.
R.G.-L. designed and performed the research, analyzed data, and wrote the paper. K.F.-B. performed the research and analyzed data. N.L.L. designed the research, analyzed data, and wrote the paper.
Footnotes
Published ahead of print on 16 February 2011.
REFERENCES
- 1. Baglioni C., Nilsen T. W. 1983. Mechanisms of antiviral action of interferon. Interferon 5:23–42 [PubMed] [Google Scholar]
- 2. Barouch D. H., et al. 2004. Immunogenicity of recombinant adenovirus serotype 35 vaccine in the presence of pre-existing anti-Ad5 immunity. J. Immunol. 172:6290–6297 [DOI] [PubMed] [Google Scholar]
- 3. Billy E., Brondani V., Zhang H., Muller U., Filipowicz W. 2001. Specific interference with gene expression induced by long, double-stranded RNA in mouse embryonal teratocarcinoma cell lines. Proc. Natl. Acad. Sci. U. S. A. 98:14428–14433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Caplen N. J. 2004. Gene therapy progress and prospects. Downregulating gene expression: the impact of RNA interference. Gene Ther. 11:1241–1248 [DOI] [PubMed] [Google Scholar]
- 5. Chattergoon M. A., et al. 2000. Targeted antigen delivery to antigen-presenting cells including dendritic cells by engineered Fas-mediated apoptosis. Nat. Biotechnol. 18:974–979 [DOI] [PubMed] [Google Scholar]
- 6. Condon C., Watkins S. C., Celluzzi C. M., Thompson K., Falo L. D., Jr 1996. DNA-based immunization by in vivo transfection of dendritic cells. Nat. Med. 2:1122–1128 [DOI] [PubMed] [Google Scholar]
- 7. Elbashir S. M., Martinez J., Patkaniowska A., Lendeckel W., Tuschl T. 2001. Functional anatomy of siRNAs for mediating efficient RNAi in Drosophila melanogaster embryo lysate. EMBO J. 20:6877–6888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Geiben-Lynn R., et al. 2008. CD4+ T lymphocytes mediate in vivo clearance of plasmid DNA vaccine antigen expression and potentiate CD8+ T-cell immune responses. Blood 112:4585–4590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Goebels N., Michaelis D., Wekerle H., Hohlfeld R. 1992. Human myoblasts as antigen-presenting cells. J. Immunol. 149:661–667 [PubMed] [Google Scholar]
- 10. Greenland J. R., Geiben R., Ghosh S., Pastor W. A., Letvin N. L. 2007. Plasmid DNA vaccine-elicited cellular immune responses limit in vivo vaccine antigen expression through Fas-mediated apoptosis. J. Immunol. 178:5652–5658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hovav A. H., et al. 2007. Duration of antigen expression in vivo following DNA immunization modifies the magnitude, contraction, and secondary responses of CD8+ T lymphocytes. J. Immunol. 179:6725–6733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kim T. W., et al. 2003. Enhancing DNA vaccine potency by combining a strategy to prolong dendritic cell life with intracellular targeting strategies. J. Immunol. 171:2970–2976 [DOI] [PubMed] [Google Scholar]
- 13. Kim T. W., et al. 2004. Enhancement of suicidal DNA vaccine potency by delaying suicidal DNA-induced cell death. Gene Ther. 11:336–342 [DOI] [PubMed] [Google Scholar]
- 14. Kim T. W., et al. 2003. Enhancing DNA vaccine potency by coadministration of DNA encoding antiapoptotic proteins. J. Clin. Invest. 112:109–117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kim T. W., et al. 2005. Modification of professional antigen-presenting cells with small interfering RNA in vivo to enhance cancer vaccine potency. Cancer Res. 65:309–316 [PubMed] [Google Scholar]
- 16. Leitner W. W., Restifo N. P. 2003. DNA vaccines and apoptosis: to kill or not to kill? J. Clin. Invest. 112:22–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Leitner W. W., Ying H., Driver D. A., Dubensky T. W., Restifo N. P. 2000. Enhancement of tumor-specific immune response with plasmid DNA replicon vectors. Cancer Res. 60:51–55 [PMC free article] [PubMed] [Google Scholar]
- 18. Leung R. K., Whittaker P. A. 2005. RNA interference: from gene silencing to gene-specific therapeutics. Pharmacol. Ther. 107:222–239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Liu M. A. 2003. DNA vaccines: a review. J. Intern. Med. 253:402–410 [DOI] [PubMed] [Google Scholar]
- 20. Liu M. A., Wahren B., Karlsson Hedestam G. B. 2006. DNA vaccines: recent developments and future possibilities. Hum. Gene Ther. 17:1051–1061 [DOI] [PubMed] [Google Scholar]
- 21. Mantegazza R., et al. 1991. Modulation of MHC class II antigen expression in human myoblasts after treatment with IFN-gamma. Neurology 41:1128–1132 [DOI] [PubMed] [Google Scholar]
- 22. Nakagawa T., et al. 2000. Caspase-12 mediates endoplasmic-reticulum-specific apoptosis and cytotoxicity by amyloid-beta. Nature 403:98–103 [DOI] [PubMed] [Google Scholar]
- 22a. National Researach Council 1996. Guide for the care and use of laboratory animals. National Academy Press, Washington, DC [Google Scholar]
- 23. Nkolola J. P., et al. 2010. Breadth of neutralizing antibodies elicited by stable, homogeneous clade A and clade C HIV-1 gp140 envelope trimers in guinea pigs. J. Virol. 84:3270–3279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Orlans F. B. 1997. Ethical decision making about animal experiments. Ethics Behav. 7:163–171 [DOI] [PubMed] [Google Scholar]
- 25. Paddison P. J., Caudy A. A., Hannon G. J. 2002. Stable suppression of gene expression by RNAi in mammalian cells. Proc. Natl. Acad. Sci. U. S. A. 99:1443–1448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Pietersz G. A., Tang C. K., Apostolopoulos V. 2006. Structure and design of polycationic carriers for gene delivery. Mini Rev. Med. Chem. 6:1285–1298 [DOI] [PubMed] [Google Scholar]
- 27. Restifo N. P. 2000. Building better vaccines: how apoptotic cell death can induce inflammation and activate innate and adaptive immunity. Curr. Opin. Immunol. 12:597–603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Roberts D. M., et al. 2006. Hexon-chimaeric adenovirus serotype 5 vectors circumvent pre-existing anti-vector immunity. Nature 441:239–243 [DOI] [PubMed] [Google Scholar]
- 29. Ruiz-Vela A., Opferman J. T., Cheng E. H., Korsmeyer S. J. 2005. Proapoptotic BAX and BAK control multiple initiator caspases. EMBO Rep. 6:379–385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Saleh M., et al. 2006. Enhanced bacterial clearance and sepsis resistance in caspase-12-deficient mice. Nature 440:1064–1068 [DOI] [PubMed] [Google Scholar]
- 31. Sasaki S., Amara R. R., Oran A. E., Smith J. M., Robinson H. L. 2001. Apoptosis-mediated enhancement of DNA-raised immune responses by mutant caspases. Nat. Biotechnol. 19:543–547 [DOI] [PubMed] [Google Scholar]
- 32. Shankar P., Manjunath N., Lieberman J. 2005. The prospect of silencing disease using RNA interference. JAMA 293:1367–1373 [DOI] [PubMed] [Google Scholar]
- 33. Shata M. T., Stevceva L., Agwale S., Lewis G. K., Hone D. M. 2000. Recent advances with recombinant bacterial vaccine vectors. Mol. Med. Today 6:66–71 [DOI] [PubMed] [Google Scholar]
- 34. Sledz C. A., Holko M., de Veer M. J., Silverman R. H., Williams B. R. 2003. Activation of the interferon system by short-interfering RNAs. Nat. Cell Biol. 5:834–839 [DOI] [PubMed] [Google Scholar]
- 35. Stan A. C., Casares S., Brumeanu T. D., Klinman D. M., Bona C. A. 2001. CpG motifs of DNA vaccines induce the expression of chemokines and MHC class II molecules on myocytes. Eur. J. Immunol. 31:301–310 [DOI] [PubMed] [Google Scholar]
- 36. Terabe M., et al. 2000. NKT cell-mediated repression of tumor immunosurveillance by IL-13 and the IL-4R-STAT6 pathway. Nat. Immunol. 1:515–520 [DOI] [PubMed] [Google Scholar]
- 37. Whitmire J. K., Tan J. T., Whitton J. L. 2005. Interferon-gamma acts directly on CD8+ T cells to increase their abundance during virus infection. J. Exp. Med. 201:1053–1059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Williams B. R. 1997. Role of the double-stranded RNA-activated protein kinase (PKR) in cell regulation. Biochem. Soc. Trans. 25:509–513 [DOI] [PubMed] [Google Scholar]
- 39. Wootz H., Hansson I., Korhonen L., Napankangas U., Lindholm D. 2004. Caspase-12 cleavage and increased oxidative stress during motoneuron degeneration in transgenic mouse model of ALS. Biochem. Biophys. Res. Commun. 322:281–286 [DOI] [PubMed] [Google Scholar]
- 40. Xu L., Anchordoquy T. 2011. Drug delivery trends in clinical trials and translational medicine: challenges and opportunities in the delivery of nucleic acid-based therapeutics. J. Pharm. Sci. 100:38–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Zong W. X., et al. 2003. Bax and Bak can localize to the endoplasmic reticulum to initiate apoptosis. J. Cell Biol. 162:59–69 [DOI] [PMC free article] [PubMed] [Google Scholar]