Abstract
Dengue is a mosquito-borne infection caused by four distinct serotypes of dengue virus, each appearing cyclically in the tropics and subtropics along the equator. Although vaccines are currently under development, none are available to the general population. One of the main impediments to the successful advancement of these vaccines is the lack of well-defined immune correlates of protection. Here, we describe a protein microarray approach for measuring antibody responses to the complete viral proteome comprised of the structural (capsid, membrane, and envelope) and nonstructural (NS1, NS2A, NS2B, NS3, NS4A, NS4B, and NS5) components of all four dengue virus serotypes (1 to 4). We examined rhesus macaques vaccinated with tetravalent vaccines consisting of live-attenuated virus (LAV) or purified inactivated virus (PIV), followed by boosting with LAV and challenging with wild-type dengue virus. We detected temporal increases in antibodies against envelope proteins in response to either vaccine, while only the PIV/LAV vaccination strategy resulted in anticapsid antibodies. In contrast to results from vaccination, naïve macaques challenged with wild-type viruses of each serotype demonstrated a balanced response to nonstructural and structural components, including responses against the membrane protein. Our results demonstrate discriminating details concerning the nature of antibody responses to dengue virus at the proteomic level and suggest the usefulness of this information for vaccine development.
INTRODUCTION
Dengue virus (DENV) is considered by the World Health Organization to be the greatest threat to world health among mosquito-borne viral diseases (41) and is classified by the Centers for Disease Control as a category A biothreat. Isolates of DENV are segregated into serotypes 1 to 4 (DENV-1 to DENV-4). The zoonotic transfer of dengue from sylvatic nonhuman primate hosts to sustained human transmission is estimated to have occurred between 125 and 320 years ago (38). Yellow fever, Japanese encephalitis, and West Nile viruses are other flaviviruses that are phylogenetically related to DENV and also are recently emerged human pathogens. With the exception of asymptomatic cases, the most common clinical presentation of infection by DENV is dengue fever (DF), which is characterized by acute febrile episodes and headaches, usually followed by generalized joint and muscular pain, leucopenia, and petechiae. A maculopapular rash is common in younger patients. The viremic phase of DF usually lasts for about 7 days. Although DENV serotypes 1 to 4 are all currently circulating in the world, coinfections with more than one serotype are not common. Usually resolving after a few days without major consequences, DF induces homotypic and long-lasting immunity along with temporary antibody cross-reactivity against other DENV serotypes (1, 35). In contrast to DF, dengue hemorrhagic fever (DHF) is an infrequent but far more serious consequence of infection. The association between DHF and dengue virus was first described in Manila in 1953 (15). DHF is strictly defined by the WHO as a condition of severe symptoms, including high fever, hepatomegaly, generalized hemorrhage, and circulatory failure. The major manifestation of DHF is plasma leakage, which may progress to dengue-shock syndrome (DSS), hypotension, and death (40). Although the precise etiology of DHF is not clear, peak viremia levels are 100 to 1,000 times higher than in DF (39), and proinflammatory cytokines are found at pathological levels in the blood (27). The geographical spread of both the mosquito vectors and DENV has led to a global resurgence of epidemic DF and DHF in the past 25 years, with areas of hyperendemicity developing in many urban centers of the tropics. Further, dengue cases have increased 30-fold during the last 50 years to 50 million new infections every year, including approximately 500,000 cases of DHF. DENV is now endemic in over 100 countries along the equatorial line, with 2.5 billion people at risk of contracting dengue (21).
The development of vaccines to prevent infection is part of the overall strategy for controlling dengue. In one approach for preparing vaccine candidates, live-attenuated virus (LAV) was produced by serial passages of wild-type (wt) isolates of DENV in cultured cells, resulting in viruses that are sufficiently immunogenic but reduced in reactogenicity (10, 37). Alternatively, purified inactivated viral (PIV) vaccines were produced by chemical (e.g., formalin) treatment to inactivate infectivity while preserving viral antigenicity and structural integrity. Each vaccine approach may offer certain advantages, such as stimulating neutralizing antibody responses or reduced reactogenicity, but it is not clear if all positive attributes are contained by any one of the current vaccine candidates. Further, a tetravalent vaccine formulation (containing serotypes 1 to 4) is required for coverage of infections caused by most isolates in circulation. This is especially important for mitigating the risk for antibody-dependent enhancement (ADE) of dengue infection, which is proposed to increase disease severity and may lead to DHF/DSS (16, 25) in DF patients who have previously acquired immunity to other dengue serotypes and in infants carrying subneutralizing maternal antibodies (20).
As the overall effectiveness of the current generation of dengue vaccine candidates is evaluated, dissection of the host antibody response may provide vital information concerning the relationship between humoral immunity to DENV and antigens displayed by the different vaccines. The primary method used to assess serological responses to dengue vaccines is based on plaque-reduction neutralization tests (PRNTs) of antibody titers. While DENV neutralizing antibody is an important component of protection, agreement between PRNT assays and protection is sometimes weak, rendering PRNT inadequate for evaluating sera from recipients of the tetravalent DENV vaccine or from dengue cases owing to antibody cross-reactivity among serotypes (29). Furthermore, as results from PRNTs may not detect subtle differences in immunogenicity between vaccines, new approaches for measuring dengue immunity are clearly necessary. We previously explored the use of protein microarrays to capture and analyze antibodies resulting from specific vaccination or disease (19). Microarrays are especially advantageous as a high-throughput method for studying antibody and protein interactions (23, 28, 33, 43, 44) with complex proteomes. For example, antibody recognition of specific proteins on a microarray of the vaccinia virus proteome (approximately 274 proteins) from a previous report (32) correlated with PRNTs measured in clinical responses to smallpox vaccination. Although comprised of only 10 mature proteins, the DENV proteome must be considered in context with protein variations present within multiple strains. During DENV replication, a single polypeptide translated from the 11-kb RNA (positive-sense) genome is cleaved by viral and cellular proteases into seven nonstructural (NS) proteins and three structural proteins: the capsid protein (C), the precursor membrane glycoprotein (prM), and the envelope glycoprotein (E). The propeptide is further cleaved from the prM protein by cellular furin protease to form the M protein of the mature virion. The surface of the mature DENV is comprised of 180 copies (each) of the E and M proteins and is encapsulated by a host-derived lipid membrane (26). Though each of these proteins synthesized during infection are potential targets of host defense, current knowledge of the immune response to the DENV proteome is limited. We now report the development of a protein microarray comprised of the complete proteomes from strains representing all four DENV serotypes. The DENV proteome microarray was used to obtain a detailed analysis of the antibody response to vaccination and DENV challenge of rhesus macaques. In the previously reported rhesus study (36), immunogenicity was independently assessed by measuring neutralizing antibody titers by PRNT, and efficacy was assessed by measuring protection against viral replication after challenge with wt DENV. Our microarray analyses of the rhesus sera indicate substantial differences among antibody specificities in response to each vaccine and wt virus challenge, distinguishable by subsets of DENV proteins.
MATERIALS AND METHODS
Vaccinations and DENV challenges.
The dengue vaccines used were described in detail by Simmons et al. (36). Briefly, the LAV vaccines were comprised of tetravalent formulations of DENV-1 West Pac 74 45AZ5, DENV-2 S16803, DENV-3 CH53489, and DENV-4 341750 strains, all attenuated by passage in PDK cells. The PIV vaccine was based on a tetravalent formulation of nonattenuated viruses of the same strains used in the LAV (except that DENV-4 341750 was replaced by TVP360), grown in Vero cells, purified, and inactivated with formalin. Study groups, each consisting of four rhesus macaques, were vaccinated (day 0) with one dose of either the PIV or the LAV vaccine. A control group (n = 4) received phosphate-buffered saline (PBS) solution. On day 60, both the LAV and the PIV groups received an LAV booster dose. All groups received a wt DENV-3 challenge dose on day 240 (Fig. 1). Serum samples were obtained from peripheral blood collected on days 0, 60, 90, 210, and 261 of the study and stored (−20°C) before use. To evaluate immune responses after infection, groups of four naïve, unvaccinated macaques each received 5 log10 PFU of DENV-1 (West Pac 74), DENV-2 (S16803), DENV-3 (CH53489), or DENV-4 (341750). Serum samples were collected 15 days after the infection and stored until analyzed.
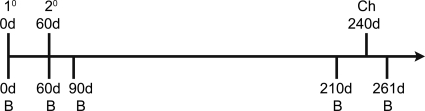
Fig. 1.
Vaccination protocol for primary (1°, at day 0 [0d]) and secondary vaccinations (2°, at day 60), serum collection dates (B, at days 0, 60, 90, 210 and 261), and challenge with wt DENV-3 (Ch, at day 240).
DENV proteome microarrays.
Genomic RNA was isolated from the Southeast Asian/Pacific prototype strains DENV-1 HAW (Hawaii), DENV-2 New Guinea C, DENV-3 H87, and DENV-4 H241 and used as templates for PCR amplification. Genomic clones were obtained for all 10 open-reading frames (ORFs) per serotype, consisting of the structural proteins premembrane (prM), C, and E and the nonstructural proteins NS1, NS2A, NS2B, NS3, NS4A, NS4B, and NS5. Gateway recombination cloning (pDEST20; Invitrogen, Carlsbad, CA) was used to facilitate high-throughput production of proteins from all ORF clones, and the proteome microarrays were assembled as previously described. In brief, all DNA clones were sequence verified throughout the insert length. Baculovirus-based expression was used to produce the recombinant viral proteins as glutathione S-transferase (GST)-tagged fusions. The DENV proteins were affinity purified from insect cell lysates in 96-well plates using glutathione-agarose, sized and measured by capillary electrophoresis by using an AMS90 SE system (Caliper Life Sciences, Mountain View, CA), and spotted with controls on glass slides coated with ultrathin nitrocellulose (GenTel BioSciences, Madison, WI). Protein spot intensities of representative slides were measured by using an anti-GST antibody and compared to a dilution series of known quantities of protein printed on each slide. Further quality control of printed slides was performed as previously described. The NS5 proteins from DENV-1 and DENV-4 genomes were not included in the final microarray due to poor stability in solution.
Antibody assay.
Microarray slides were preincubated (1 h, 22°C) with a blocking buffer (50 mM HEPES, 200 mM NaCl, 0.08% Triton X-100, and 25% glycerol at pH 7.5 to 8). Serum samples were diluted 1:50 in probe buffer (1× PBS, 1% Tween 20, 1% bovine serum albumin [BSA]). Diluted sera were overlaid (400 μl) on the slides, covered with glass coverslips, and incubated in a slide incubator (2 h, 26°C, 80% relative humidity). After incubation, the slides were washed four times with wash buffer (1× PBS, 0.2% Tween 20, 1% BSA). Antibody binding was detected by incubation (1 h) with a 1:2,000 dilution of fluorescent, goat anti-human IgG (Invitrogen, Carlsbad, CA). The slides were washed four times after incubation with the secondary antibody and dried before analysis. Microarray slides were imaged using a GenePix laser confocal 4000B scanner (Molecular Devices, CA), and image analysis was performed using GenePix Pro 6.0 software (Molecular Devices). Data acquired from GenePix software were analyzed using ProtoArray Prospector version 3.1 (Invitrogen, CA) in immune response profiling mode. The results represent fluorescence intensities of protein spots in pixel counts as a measure of interaction between serum DENV-specific IgG and proteins on the microarray.
Statistical analyses.
For specific comparisons between experimental groups, data were analyzed by paired-sample t tests with step-down Bonferroni correction for multiple comparisons and plotted as the geometric mean ± standard error of the mean. Significance was assigned to each group by comparison to day 0 results.
RESULTS
Microarray of the DENV proteome.
The DENV isolates selected for construction of the microarray represented strains isolated in the Caribbean region (DENV-1 HAW, DENV-2 New Guinea C, DENV-3 H87, DENV-4 H241). The protein microarray used for our study was comprised of the proteomes of all four dengue serotypes, consisting of the seven NS proteins and three structural proteins. Protein controls included human IgG, mouse anti-human IgG, and unrelated viral and nonviral proteins for calibration of signals. All DNA clones were fully sequenced to confirm identity before being used for protein expression, and the correct size and stability were verified for all proteins included in the microarray. Fig. S1 in the supplemental material shows representative GST-tagged dengue virus proteins that have been separated by SDS-PAGE and blotted using an anti-GST antibody (panel A), as well as a laser scanner image of a complete microarray and grids showing representative dengue virus and control proteins (panel B).
Antibody response to dengue vaccination.
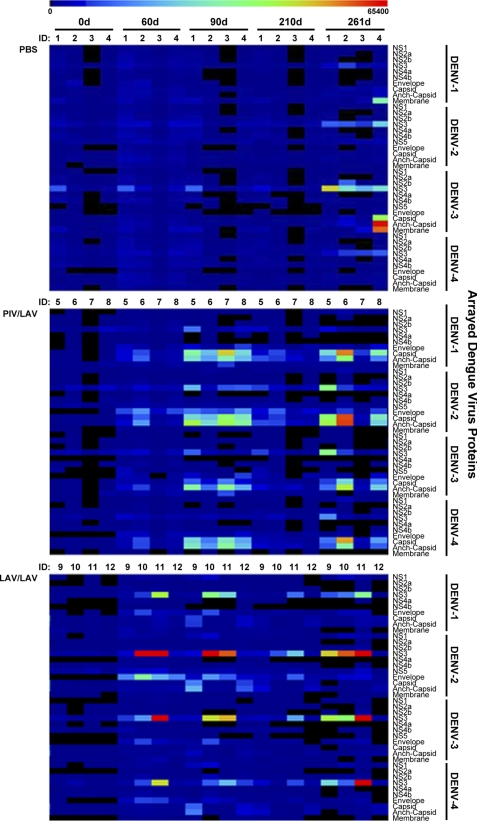
We first examined the antibody response in rhesus macaques to a primary vaccination with PIV vaccine followed by boosting with LAV vaccine (PIV/LAV). This response was compared to that of animals that received two doses of LAV vaccine (LAV/LAV). Figure 1 shows the schedule for vaccinations and the serum collection time points used in our study (36). One group of macaques was vaccinated (day 0) with the PIV vaccine and boosted with LAV on day 60. A second group was vaccinated with LAV (day 0) and boosted with LAV on day 60. A control group received only PBS without vaccine. All groups were challenged on day 240 with wt DENV-3. Preliminary analysis of all sera collected by using microarrays containing whole, fixed viruses of each serotype demonstrated that there were no significant levels of antiviral IgM in sera from any of the vaccinated animals (data not shown), whereas large amounts of IgG were observed for the PIV/LAV and LAV/LAV groups. This prescreening of the serum samples was necessary to ascertain that only IgG binding data were collected when probing the serum samples with the DENV proteome microarray. The heat map in Fig. 2 depicts a summary of IgG responses to arrayed DENV (serotypes 1 through 4) proteins by vaccinated and control subjects across sampling dates. The humoral immune responses to each vaccination followed a rise-and-fall pattern corresponding to the vaccination schedule (day 0, primary vaccination; day 60, secondary vaccination). In both vaccination groups, overall increases in protein-specific IgG levels were evident by day 60, with these levels peaking by day 90. However, a substantial part of this response returned to basal levels by day 210. Challenging at day 240 with the wt DENV-3 resulted in recall responses in the vaccinated groups (day 261). Neither vaccination strategy generated detectable IgG responses to the DENV proteins NS2A, NS2B, NS4A, NS4B, and NS5 or membrane across any of the examined sampling times. No detectable antibody recognition occurred in the PBS controls before virus challenge, while challenge with wt DENV-3 elicited IgG responses against select DENV proteins (day 261).
Fig. 2.
Summary of IgG recognition of DENV proteome. The heat map depicts global IgG binding to proteomes of DENV-1 to -4 for each vaccinated or control macaque. The analysis includes sera from the PBS control (upper panel), PIV/LAV-vaccinated (center panel), and LAV/LAV-vaccinated (lower panel) macaques, collected on days 0, 60, 90, 210, and 261. Color key at the top of the heat map represents signal intensity (relative fluorescent units). Anch, anchor.
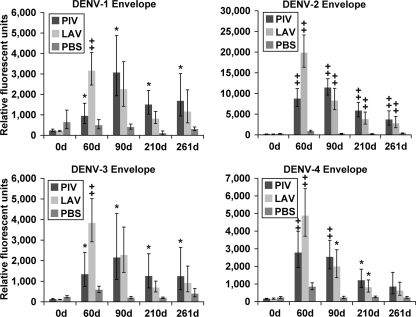
Envelope protein.
Both vaccination strategies resulted in significant IgG responses against the E protein of all DENV serotypes by day 60 and 90 compared to levels at day 0 (Fig. 3, prevaccination). However, anti-E IgG levels for all serotypes diminished in both vaccination groups by day 210, though in some cases these continued to be significantly higher than at day 0. Challenging with wt DENV-3 virus at day 240 did not lead to a detectable increase in IgG levels against E of any serotype, including DENV-3, compared to the levels at day 210. In the PBS group, the virus challenge at day 240 induced a slightly higher (but not significant) anti-DENV-3 E IgG level by day 261 (compared to levels at day 0).
Fig. 3.
IgG binding to DENV envelope proteins. Sera from PIV/LAV-, LAV/LAV-, or PBS-vaccinated macaques (four in each group) were collected at days 0, 60, 90, 210, and 261 and analyzed for binding to the DENV proteome microarray. The figure shows IgG binding to DENV-1 E (upper left), binding to DENV-2 E (upper right), binding to DENV-3 E (lower left), and binding to DENV-4 E (lower right). The y axis represents the geometric mean of the average pixel value for all four subjects in each group. Error bars represent the standard error of the mean in each group. Significance was assigned to each group compared to day 0 (0d) as follows: *, P ≤ 0.05; , P ≤ 0.01.
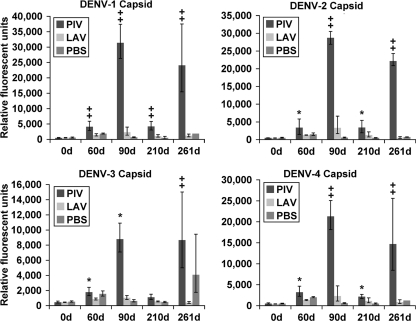
Capsid protein.
In contrast to results observed with the E protein, the PIV/LAV vaccination induced a substantially greater anti-C IgG response than that induced by LAV/LAV vaccination (Fig. 4). The response for all serotypes was highest in the PIV/LAV group at day 90 compared to that at day 0 or to results for the PBS group, but there were also significant responses after receiving only PIV (day 60). The LAV/LAV vaccination induced only slightly elevated, but not significant, anti-C IgG levels by day 60 and 90 compared to those at day 0 for all serotypes. However, these levels were substantially below those found in the PIV/LAV group. Anti-C IgG levels in the PIV/LAV group decreased substantially by day 210 but remained significantly higher for C of all serotypes except DENV-3 C. The wt DENV-3 challenge resulted in significant increases of anti-C IgG levels for all serotypes in the PIV/LAV group but not in the LAV/LAV group. The same challenge in the PBS group induced an increase of IgG levels by day 261 against the C proteins of DENV-3. Results identical to the C protein data were obtained for responses to anchor-C (data not shown), which contains an additional 12 amino acid residues in the carboxyl terminus; these are not present in mature virions but are required for translocation into the endoplasmic reticulum of the infected host cell during viral replication.
Fig. 4.
IgG binding to DENV capsid proteins. Sera from PIV/LAV-, LAV/LAV- or PBS-vaccinated macaques (four in each group) were collected at days 0, 60, 90, 210, and 261 and analyzed for binding to DENV proteome microarray. The figure shows IgG binding to DENV-1 C (upper left), binding to DENV-2 C (upper right), binding to DENV-3 C (lower left), and binding to DENV-4 C (lower right). The y axis represents the geometric mean of the average pixel value for all four subjects in each group. Error bars represent the standard error of the mean in each group. Significance was assigned to each group compared to day 0 (0d) as follows: *, P ≤ 0.05; , P ≤ 0.01.
Nonstructural proteins.
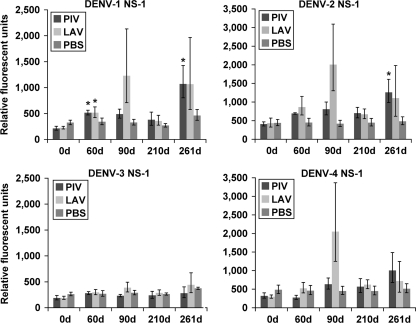
While the PIV/LAV vaccine group, and to a lesser degree the LAV/LAV vaccine group, exhibited sustained IgG responses against DENV structural proteins, the responses against NS1 and NS3 elicited by the two groups were inconsistent. The PIV/LAV vaccination produced slightly elevated levels of anti-DENV-1, -2, and -4 NS1 IgG by day 90, compared to levels at day 0 (Fig. 5). These anti-NS1 antibody levels remained low through day 210. Interestingly, the highest response obtained in the PIV/LAV group came after the challenge with wt DENV-3, resulting in elevated levels of heterotypic IgG against DENV-1, -2, and -4 NS1, with little or no response against NS1 of DENV-3. In contrast, the LAV/LAV group presented elevated levels of IgG against DENV-1, -2, and -4 NS1 by day 90, without detectable levels of anti-DENV-3 NS1 IgG. By day 210, anti-NS1 IgG levels decreased to levels similar to that of the PIV/LAV group. Challenge of the PIV/LAV group with wt DENV-3 resulted in increased levels of anti-NS1 IgG for all serotypes, excluding DENV-3. Viral challenge of the PBS control group resulted in only a slight elevation of anti-DENV-3 NS1 antibodies.
Fig. 5.
IgG binding to DENV NS1 proteins. Sera from PIV/LAV-, LAV/LAV-, or PBS-vaccinated macaques (four in each group) were collected at days 0, 60, 90, 210, and 261 and analyzed for binding to the DENV proteome microarray. The figure shows IgG binding to DENV-1 NS1 (upper left), binding to DENV-2 NS1 (upper right), binding to DENV-3 NS1 (lower left), and binding to DENV-4 NS1 (lower right). The y axis represents the geometric mean of the average pixel value for all four subjects in each group. Error bars represent the standard error of the mean in each group. Significance (*) was assigned to each group compared to results for day 0 (0d) (P ≤ 0.05).
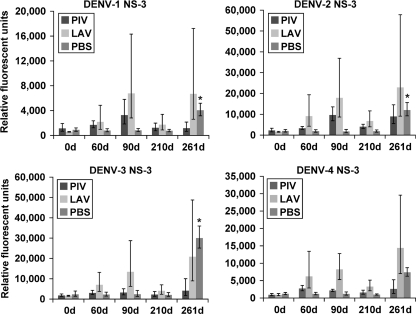
The LAV/LAV vaccination induced levels of anti-NS3 IgG higher than those induced by PIV/LAV (Fig. 6). In the LAV/LAV group, the levels of IgG against NS3 of all serotypes were higher by day 90 than at day 0, but by day 210, they had decreased significantly, remaining slightly higher than at day 0. The challenge with wt DENV-3 resulted in increased levels of IgG recognizing NS3 of all four serotypes. The PIV/LAV vaccination produced only slightly elevated serum IgG against NS3 of DENV-1 and -2 and no response against NS3 of DENV-4. The wt DENV-3 challenge did not result in robust elevation of IgG levels against NS3 of any serotype. As expected, the PBS group responded to the wt DENV-3 challenge with sharp increases in serum anti-DENV-3 NS3 IgG. Significant levels of anti-DENV-1 and -2 IgG, and to a lesser degree DENV-4, were also found in the PBS group after the challenge.
Fig. 6.
IgG binding to DENV NS3 proteins. Sera from PIV/LAV-, LAV/LAV-, or PBS-vaccinated macaques (four in each group) were collected at days 0, 60, 90, 210, and 261 and analyzed for binding to the DENV proteome microarray. The figure shows IgG binding to DENV-1 NS3 (upper left), binding to DENV-2 NS3 (upper right), binding to DENV-3 NS3 (lower left), and binding to DENV-4 NS3 (lower right). The y axis represents the geometric mean of the average pixel value for all four subjects in each group. Error bars represent the standard error of the mean in each group. Statistical significance (*) was assigned to each group compared to results for day 0 (0d) (P ≤ 0.05).
Antibody response to wt DENV infection.
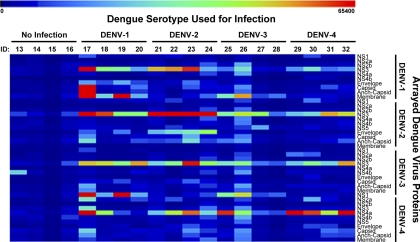
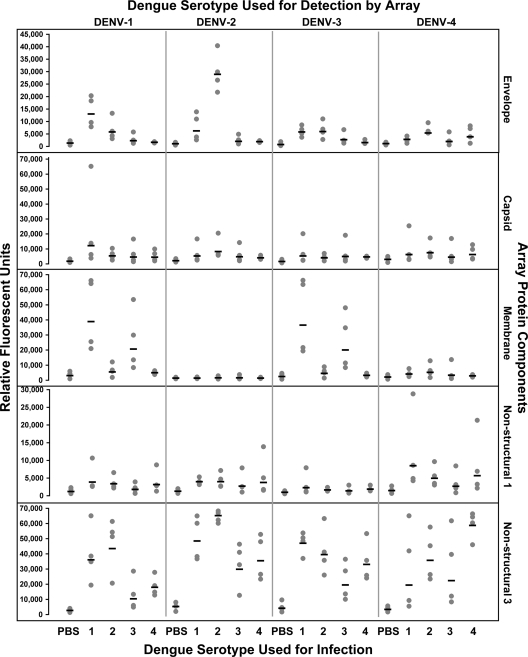
To compare the immune profiles observed after vaccination to those responses that might be observed after a natural infection, we analyzed sera collected from unvaccinated rhesus macaques infected with wt DENV representing each serotype. These dengue-naïve rhesus macaques were divided into four groups (n = 4 per group), and each group was infected with a different wt DENV serotype. Sera were collected 15 days after the infection, and antibody responses were compared to those for sera from dengue-naïve animals. Results describing the viral loads in the infected monkeys' sera are published elsewhere (36). The heat map in Fig. 7 depicts overall IgG responses to the arrayed DENV-1 to -4 proteins by all subjects, and a detailed analysis is shown in Fig. 8. There was a robust overall response in the infected groups to several viral structural and nonstructural antigens compared to results from the negative-control baseline sera. However, the response was cross-reactive, as serum IgG from the infected macaques not only recognized proteins from the same infecting strain but also bound to proteins from other strains. Unlike the PIV/LAV vaccinated group, but similar to the LAV/LAV group, dengue-naïve macaques infected with wt DENV developed significant responses against NS3 from all DENV serotypes compared to the controls (P ≤ 0.005), regardless of the infecting serotype. There was also an elevated response against NS1 of other serotypes, but this was limited to only a few macaques. The DENV-1 and DENV-2 infection groups developed elevated antibody responses against the homotypic E protein compared to results with the preinfected sera from the same group (P ≤ 0.0001). We detected levels of anti-E antibody in the DENV-3 and DENV-4 infection groups that were only marginally higher than those of the controls. The IgG response to C was similar to the response to E, with the DENV-1 and DENV-2 challenge groups developing significant levels (P ≤ 0.005), while fewer macaques in the DENV-3 and DENV-4 groups developed IgG responses to C. Interestingly, although we did not detect anti-M IgG in the vaccinated groups, antibody responses to this protein were observed for DENV-1- and DENV-3-infected macaques, with significant IgG responses against homotypic M protein (P ≤ 0.005) and weak cross-reactive antibodies. Finally, anti-M antibody responses were insignificantly low in the DENV-4 and DENV-2 infection groups.
Fig. 7.
Serum IgG binding to DENV proteome after infection. The heat map depicts global IgG binding to the complete DENV-1 to -4 proteome for each infected macaque. Samples running from left to right represent the uninfected and infected macaques (four in each group). Color key at the top of the heat map represents signal intensity. The y axis label specifies the arrayed viral protein. ID, sample identifier.
Fig. 8.
Scatter plot analyses of individual IgG responses to infection. The DENV serotype used for the infection is at the bottom of the graph, and the DENV serotype that was analyzed is at the top. Specific viral proteins analyzed are labeled at the right of the graph. The response (in relative fluorescent units) of each macaque against each DENV component is represented as a dot. The mean value in each group is represented by the horizontal bar.
DISCUSSION
A detailed understanding of the host response to individual components of the DENV proteome is important for the overall disease control strategy as a means of identifying immune responses to specific strains in circulation and assessing vaccine efficacy in relationship to these strains. We used a protein microarray encompassing representative proteomes of all four dengue serotypes to compare immune responses to two types of dengue vaccines with infection. Our results demonstrated that the macaque antibody response to a PIV prime, LAV boost vaccination differed from the response to an LAV prime, LAV boost vaccination in recognition of specific viral proteins. The LAV/LAV vaccination elicited serum IgG against E, and to a lesser extent against NS1 and NS3 proteins, while antibody responses induced by the PIV/LAV vaccine were limited to the E and C structural components, failing to activate a robust IgG response against nonstructural proteins. It is noteworthy that boosting the PIV recipients with a subsequent LAV dose did not change the immune profile originally generated by the PIV vaccine, as no significant increases in anti-NS1 and -NS3 IgG were observed by day 90 or thereafter. Further, dengue-naïve macaques infected with wt DENV developed an antibody response profile distinct from those of the vaccinated groups in that there were high levels of antibodies reacting with NS, structural antigens, and M. In a manner similar to that for LAV/LAV recipients, DENV-challenged macaques developed IgG responses against NS1, NS3, and E proteins, while some macaques infected with DENV resembled PIV/LAV recipients by directing immunity to the C protein. Unlike any of the vaccinated monkeys, DENV-1- and DENV-3-infected macaques developed IgG responses against M proteins. It is possible that differences in the virulence of the infecting virus, as evidenced by the mean number of viremic days postchallenge (36), contributed to the lower antibody responses to M proteins recorded for the macaques infected with DENV-2 and -4.
Both the LAV/LAV and PIV/LAV vaccinations in our study induced significant anti-E antibody against all serotypes by day 90 and to lesser degree by day 210. The E protein forms a sheath around the assembled virion, mediating cell attachment and fusion, and is thus an important target for neutralizing antibodies. Important epitopes are found in domain III of E; these have been shown to be targets of neutralization, presumably by blocking the site of cellular attachment (17, 24). However, immunity can drive selection of new infective isolates, such as structural variants of the E protein (4), targeted by antibodies that inhibit plaque formation and increase mouse survival in dengue challenge models (2, 8, 18) or increase protection in primate models (17). A second important structural component of DENV is C, previously reported to generate neutralizing anti-C monoclonal antibodies directed toward the N terminus (14, 30). Our results indicate that levels of anti-C antibody provide the most striking contrast between the different vaccination regimens and natural immunity. The PIV/LAV vaccination resulted in robust levels of anti-C IgG by day 90, which subsided by day 210. Infection with the wt DENV-3 restored the previous levels of anti-C IgG in the PIV/LAV group across all serotypes, while LAV/LAV vaccination resulted in substantially lower antibody responses to C, suggesting that primary vaccination with PIV was important for inducing antibodies to the C protein. Nonetheless, the strong anti-C antibody responses induced by the PIV priming may not necessarily translate into protective immunity as a result of the poor affinity maturation of antibodies noted with other formalin-inactivated vaccines (9).
We also detected important differences in humoral responses to NS proteins. LAV/LAV vaccination or DENV infection induced higher levels of antibodies against NS1 and NS3 across all serotypes than those induced by vaccination with PIV/LAV. It is interesting that increased anti-NS1 IgG responses were only evident in the PIV/LAV group after challenge with wt DENV-3. Detection of responses against NS1 and NS3 is relevant in this context, as both proteins appear to play important roles in dengue immunity. It was previously established that NS1 inoculation protects mice against viral challenge, though it is not clear if protection is exclusively antibody mediated (13, 31). Anti-dengue NS1 antibodies may serve as a mediator of phagocytic clearance (3, 31), a phenomenon corroborated by other closely related flaviviruses (5). More importantly, anti-NS1 antibodies are an important indicator of past infection that can be detected in convalescent human cases (7) or directly in the serum of acute and/or convalescent human cases of primary or secondary DF and DHF (6, 34, 42). Nonetheless, some anti-NS1 monoclonal antibodies can also be implicated in ADE (12). Furthermore, anti-NS3 antibodies found in the convalescence sera from primary and secondary cases (6) may also be important biomarkers. In contrast to NS1 antibodies, anti-NS3 IgG was reported to play no role in virus neutralization, but rather was shown to be a mediator of CD8+ cytotoxic T lymphocyte (CTL) activity (22). Finally, we did not detect any anti-M antibodies (data not shown) among either vaccinated group, while anti-M IgG was evident in the infected group. The role of the antibody response to M proteins in immunity is not clear, but it may provide passive protection from infection (11).
The protein microarray used in our study contains proteomes that are representative of dominant DENV serotypes, but our results do not address potential heterogeneity in circulating strains. The vast geographical and genomic diversity of the virus and relationships to disease and immunity remain a challenge, requiring a more comprehensive analysis of many additional strains. In addition, infectious DENV exists as a cluster of quasispecies within the host as a result of mutations generated during genome replication (38), complicating the analysis of host immunity. In spite of these shortcomings, the information provided by protein microarrays is more comprehensive than results obtained by plaque neutralization alone. A similar approach can be utilized to examine vaccine efficacy and aid in the establishment of immune correlates of vaccination or disease.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the Joint Science and Technology Office W81XWH-05-2-0077 (B.S., R.G.U.).
We thank Jorge Munoz and Wellington Sun for providing virus isolates for preparation of the microarrays and Sarah L. Norris for statistical analyses.
The views expressed in this paper are those of the authors and do not purport to reflect official policy of the U. S. Government.
A.P.T. and B.S. were employees of Life Technologies during this study. All other authors have no financial conflicts of interest.
Footnotes
Supplemental material for this article may be found at http://cvi.asm.org/.
Published ahead of print on 26 January 2011.
REFERENCES
- 1. Adams B., et al. 2006. Cross-protective immunity can account for the alternating epidemic pattern of dengue virus serotypes circulating in Bangkok. Proc. Natl. Acad. Sci. U. S. A. 103:14234–14239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Amexis G., Young N. S. 2006. Parvovirus B19 empty capsids as antigen carriers for presentation of antigenic determinants of dengue 2 virus. J. Infect. Dis. 194:790–794 [DOI] [PubMed] [Google Scholar]
- 3. Calvert A. E., Huang C. Y., Kinney R. M., Roehrig J. T. 2006. Non-structural proteins of dengue 2 virus offer limited protection to interferon-deficient mice after dengue 2 virus challenge. J. Gen. Virol. 87:339–346 [DOI] [PubMed] [Google Scholar]
- 4. Chao D. Y., et al. 2005. Strategically examining the full-genome of dengue virus type 3 in clinical isolates reveals its mutation spectra. Virol. J. 2:72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Chung K. M., Thompson B. S., Fremont D. H., Diamond M. S. 2007. Antibody recognition of cell surface-associated NS1 triggers Fc-gamma receptor-mediated phagocytosis and clearance of West Nile virus-infected cells. J. Virol. 81:9551–9555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Churdboonchart V., Bhamarapravati N., Peampramprecha S., Sirinavin S. 1991. Antibodies against dengue viral proteins in primary and secondary dengue hemorrhagic fever. Am. J. Trop. Med. Hyg. 44:481–493 [DOI] [PubMed] [Google Scholar]
- 7. Dejnirattisai W., et al. 2010. Cross-reacting antibodies enhance dengue virus infection in humans. Science 328:745–748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Delenda C., Frenkiel M. P., Deubel V. 1994. Protective efficacy in mice of a secreted form of recombinant dengue-2 virus envelope protein produced in baculovirus infected insect cells. Arch. Virol. 139:197–207 [DOI] [PubMed] [Google Scholar]
- 9. Delgado M. F., et al. 2009. Lack of antibody affinity maturation due to poor Toll-like receptor stimulation leads to enhanced respiratory syncytial virus disease. Nat. Med. 15:34–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Eckels K. H., et al. 2003. Modification of dengue virus strains by passage in primary dog kidney cells: preparation of candidate vaccines and immunization of monkeys. Am. J. Trop. Med. Hyg. 69:12–16 [DOI] [PubMed] [Google Scholar]
- 11. Falconar A. K. 1999. Identification of an epitope on the dengue virus membrane (M) protein defined by cross-protective monoclonal antibodies: design of an improved epitope sequence based on common determinants present in both envelope (E and M) proteins. Arch. Virol. 144:2313–2330 [DOI] [PubMed] [Google Scholar]
- 12. Falconar A. K. 2008. Monoclonal antibodies that bind to common epitopes on the dengue virus type 2 nonstructural-1 and envelope glycoproteins display weak neutralizing activity and differentiated responses to virulent strains: implications for pathogenesis and vaccines. Clin. Vaccine Immunol. 15:549–561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Falgout B., Bray M., Schlesinger J. J., Lai C. J. 1990. Immunization of mice with recombinant vaccinia virus expressing authentic dengue virus nonstructural protein NS1 protects against lethal dengue virus encephalitis. J. Virol. 64:4356–4363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Gagnon S. J., Zeng W., Kurane I., Ennis F. A. 1996. Identification of two epitopes on the dengue 4 virus capsid protein recognized by a serotype-specific and a panel of serotype-cross-reactive human CD4+ cytotoxic T-lymphocyte clones. J. Virol. 70:141–147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Gubler D. J. 1998. Dengue and dengue hemorrhagic fever. Clin. Microbiol. Rev. 11:480–496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Halstead S. B., Venkateshan C. N., Gentry M. K., Larsen L. K. 1984. Heterogeneity of infection enhancement of dengue 2 strains by monoclonal antibodies. J. Immunol. 132:1529–1532 [PubMed] [Google Scholar]
- 17. Hermida L., et al. 2006. A recombinant fusion protein containing the domain III of the dengue-2 envelope protein is immunogenic and protective in nonhuman primates. Vaccine 24:3165–3171 [DOI] [PubMed] [Google Scholar]
- 18. Kaufman B. M., Summers P. L., Dubois D. R., Eckels K. H. 1987. Monoclonal antibodies against dengue 2 virus E-glycoprotein protect mice against lethal dengue infection. Am. J. Trop. Med. Hyg. 36:427–434 [DOI] [PubMed] [Google Scholar]
- 19. Keasey S. L., et al. 2009. Extensive antibody cross-reactivity among infectious gram-negative bacteria revealed by proteome microarray analysis. Mol. Cell. Proteomics 8:924–935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kliks S. C., Nimmanitya S., Nisalak A., Burke D. S. 1988. Evidence that maternal dengue antibodies are important in the development of dengue hemorrhagic fever in infants. Am. J. Trop. Med. Hyg. 38:411–419 [DOI] [PubMed] [Google Scholar]
- 21. Kroeger A., Nathan M., Hombach J. 2004. Dengue. Nat. Rev. Microbiol. 2:360–361 [DOI] [PubMed] [Google Scholar]
- 22. Mathew A., et al. 1996. Dominant recognition by human CD8+ cytotoxic T lymphocytes of dengue virus nonstructural proteins NS3 and NS1.2a. J. Clin. Invest. 98:1684–1691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Michaud G. A., et al. 2003. Analyzing antibody specificity with whole proteome microarrays. Nat. Biotechnol. 21:1509–1512 [DOI] [PubMed] [Google Scholar]
- 24. Modis Y., Ogata S., Clements D., Harrison S. C. 2005. Variable surface epitopes in the crystal structure of dengue virus type 3 envelope glycoprotein. J. Virol. 79:1223–1231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Morens D. M., Halstead S. B. 1987. Disease severity-related antigenic differences in dengue 2 strains detected by dengue 4 monoclonal antibodies. J. Med. Virol. 22:169–174 [DOI] [PubMed] [Google Scholar]
- 26. Mukhopadhyay S., Kuhn R. J., Rossmann M. G. 2005. A structural perspective of the flavivirus life cycle. Nat. Rev. Microbiol. 3:13–22 [DOI] [PubMed] [Google Scholar]
- 27. Noisakran S., Perng G. C. 2008. Alternate hypothesis on the pathogenesis of dengue hemorrhagic fever (DHF)/dengue shock syndrome (DSS) in dengue virus infection. Exp. Biol. Med. (Maywood) 233:401–408 [DOI] [PubMed] [Google Scholar]
- 28. Predki P. F., Mattoon D., Bangham R., Schweitzer B., Michaud G. 2005. Protein microarrays: a new tool for profiling antibody cross-reactivity. Hum. Antibodies 14:7–15 [PubMed] [Google Scholar]
- 29. Putnak J. R., et al. 2008. Comparative evaluation of three assays for measurement of dengue virus neutralizing antibodies. Am. J. Trop. Med. Hyg. 79:115–122 [PubMed] [Google Scholar]
- 30. Puttikhunt C., et al. 2009. Production and characterization of anti-dengue capsid antibodies suggesting the N terminus region covering the first 20 amino acids of dengue virus capsid protein is predominantly immunogenic in mice. Arch. Virol. 154:1211–1221 [DOI] [PubMed] [Google Scholar]
- 31. Schlesinger J. J., Brandriss M. W., Walsh E. E. 1987. Protection of mice against dengue 2 virus encephalitis by immunization with the dengue 2 virus non-structural glycoprotein NS1. J. Gen. Virol. 68(Pt. 3):853–857 [DOI] [PubMed] [Google Scholar]
- 32. Schmid K., et al. 2008. Analysis of the human immune response to vaccinia by use of a novel protein microarray suggests that antibodies recognize less than 10% of the total viral proteome. Proteomics Clin. Appl. 2:1528–2538 [DOI] [PubMed] [Google Scholar]
- 33. Schweitzer B., Predki P., Snyder M. 2003. Microarrays to characterize protein interactions on a whole-proteome scale. Proteomics 3:2190–2199 [DOI] [PubMed] [Google Scholar]
- 34. Shu P. Y., et al. 2000. Dengue NS1-specific antibody responses: isotype distribution and serotyping in patients with Dengue fever and Dengue hemorrhagic fever. J. Med. Virol. 62:224–232 [DOI] [PubMed] [Google Scholar]
- 35. Sierra B., et al. 2002. Long-term memory cellular immune response to dengue virus after a natural primary infection. Int. J. Infect. Dis. 6:125–128 [DOI] [PubMed] [Google Scholar]
- 36. Simmons M., Burgess T., Lynch J., Putnak R. 2010. Protection against dengue virus by non-replicating and live attenuated vaccines used together in a prime boost vaccination strategy. Virology 396:280–288 [DOI] [PubMed] [Google Scholar]
- 37. Sun W., et al. 2003. Vaccination of human volunteers with monovalent and tetravalent live-attenuated dengue vaccine candidates. Am. J. Trop. Med. Hyg. 69(Suppl. 6):24–31 [DOI] [PubMed] [Google Scholar]
- 38. Twiddy S. S., Holmes E. C., Rambaut A. 2003. Inferring the rate and time-scale of dengue virus evolution. Mol. Biol. Evol. 20:122–129 [DOI] [PubMed] [Google Scholar]
- 39. Vaughn D. W., et al. 2000. Dengue viremia titer, antibody response pattern, and virus serotype correlate with disease severity. J. Infect. Dis. 181:2–9 [DOI] [PubMed] [Google Scholar]
- 40. World Health Organization 1997. Dengue haemorrhagic fever: diagnosis, treatment, prevention and control, 2nd ed. World Health Organization, Geneva, Switzerland: http://www.who.int/csr/resources/publications/dengue/Denguepublication/en/ [Google Scholar]
- 41. World Health Organization 2006. Scientific working group report on dengue. World Health Organization, Geneva, Switzerland: http://apps.who.int/tdr/svc/publications/tdr-research-publications/swg-report-dengue [Google Scholar]
- 42. Young P. R., Hilditch P. A., Bletchly C., Halloran W. 2000. An antigen capture enzyme-linked immunosorbent assay reveals high levels of the dengue virus protein NS1 in the sera of infected patients. J. Clin. Microbiol. 38:1053–1057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Zhu H., et al. 2001. Global analysis of protein activities using proteome chips. Science 293:2101–2105 [DOI] [PubMed] [Google Scholar]
- 44. Zhu H., et al. 2000. Analysis of yeast protein kinases using protein chips. Nat. Genet. 26:283–289 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.