Abstract
A laboratory testing algorithm was evaluated to confirm West Nile virus (WNV) infection in human serum following the introduction of the virus in Puerto Rico in 2007. This testing algorithm used two standard diagnostic assays, the IgM antibody capture enzyme-linked immunosorbent assay (MAC ELISA) and real-time reverse transcriptase PCR (RT-PCR), along with two nonconventional assays, the nonstructural protein 1 (NS1) ELISA and a 90%-plaque-reduction neutralization test (PRNT90) with IgG depletion for dengue virus (DENV) and WNV. A total of 2,321 serum samples from suspected WNV human cases were submitted for testing. Approximately one-third (867, 37%) were cross-reactive for DENV and WNV by MAC ELISA and had negative RT-PCR results for both viruses. Of a subset of 43 samples tested, 31 (72%) of these cases were identified as positive for DENV in the PRNT90 with IgG depletion and 8 (19%) were positive in the DENV NS1 antigen ELISA. These two assays combined differentiated 36 (84%) of the samples that could not be diagnosed using the standard diagnostic testing methods.
INTRODUCTION
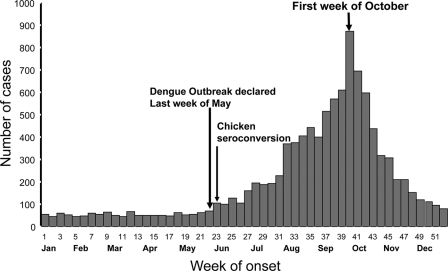
The introduction of West Nile virus (WNV) into the northeastern United States in 1999 and its subsequent rapid spread throughout the United States raised concerns about the potential for the introduction and spread of the virus in the Caribbean (4, 6, 7, 16). Since 1999, evidence of WNV transmission has been reported throughout the Caribbean, where diagnosis has been complicated by the cocirculation of other flaviviruses, including the dengue virus (DENV) (12). The continued spread of WNV through North America, Latin America, and the Caribbean has highlighted the need for disease-specific diagnostic tests for flaviviruses. Until recently, DENV has been the only circulating flavivirus in Puerto Rico; therefore, the surveillance system testing algorithm was not designed to detect other arboviruses. The first serological evidence of WNV in Puerto Rico was reported in wild birds in 2003. The first WNV isolate was obtained in mosquitoes in June 2007 in the municipalities of Ceiba and Naguabo along the northeastern coast of the island and coincided with the largest outbreaks of dengue since 1998 (Fig. 1) (1). The epidemic curve indicated that the dengue outbreak began May 2007, 1 week prior to the serological detection of WNV in sentinel chickens.
Fig. 1.
Epidemiology curve of the dengue outbreak during the introduction of WNV in Puerto Rico. The second arrow depicts the seroconversion of the sentinel chickens in the Ceiba region of Puerto Rico. The first arrow indicates the date the dengue outbreak was declared. The samples from this study correspond to July through December of 2007.
The Centers for Disease Control and Prevention (CDC) and the Puerto Rico Health Department have jointly managed an island-wide WNV surveillance system for humans since 2003. The data presented in this study are an evaluation of the samples obtained from the WNV surveillance from July through December 2007 following the detection of WNV in sentinel chickens (1). The purpose of this study was to evaluate a new testing algorithm to differentiate between WNV and DENV cases in IgM-cross-reactive samples. A new testing algorithm was developed to evaluate suspected WNV-positive serum samples using a 90%-plaque-reduction neutralization test (PRNT90) with IgG depletion. Further differentiation was achieved using the dengue nonstructural protein 1 (NS1) antigen enzyme-linked immunosorbent assay (ELISA). These results will likely prove useful in developing a better testing algorithm for DENV- and WNV-cross-reactive samples using IgM, PRNT90 with IgG depletion, and the NS1 antigen ELISA.
MATERIALS AND METHODS
Criteria for sample submission.
In 2003, a human encephalitis surveillance program which focused on suspected neuroinvasive WNV cases was established in Puerto Rico. Lectures and presentations on WNV and the importance of surveillance were provided to promote participation from health care providers. Health care providers were requested to submit serum and cerebrospinal fluid from patients with encephalitis-like syndrome, motor disorders associated with acute fever, and aseptic meningitis. Following the first detection of WNV in 2007, health providers were encouraged to submit samples from all patients suspected of having WNV fever and WNV neuroinvasive disease to the CDC Dengue Branch for WNV diagnostic testing. These samples were laboratory tested for both WNV and DENV using IgM antibody capture ELISA (MAC ELISA) and real-time reverse transcriptase PCR (RT-PCR) techniques upon submission. Samples that were negative by RT-PCR for both DENV and WNV and with cross-reactivity to both WNV and DENV in the MAC ELISA were selected for this study. These samples were then evaluated using the NS1 antigen ELISA and PRNT90 with IgG depletion to further evaluate the infecting virus.
Real-time RT-PCR.
A Singleplex RT-PCR was used for the detection of dengue virus serotypes 1 to 4 (DENV1 to -4) as previously described (11). Additionally, the samples were tested with a WNV real-time RT-PCR assay as previously described (15).
MAC ELISA.
Serum samples (n = 2,231) were submitted to the CDC's Dengue Branch in 2007 as a part of the island-wide surveillance system for WNV. The samples were initially tested using the MAC ELISA with WNV- and DENV (DENV1 to -4)-recombinant envelope and prM antigen, as previously described (17). The ratio of positive to negative results for DENV1 to -4 and WNV antigens was determined (P/N value). Samples with P/N values of ≥2 were considered positive for that antigen. For each sample positive by the MAC ELISA, we deemed the infecting virus that which had a P/N value 2-fold greater than that for the other antigen. This interpretation of the results was previously described by Martin et al. (17). Samples with equal reactivities for DENV and WNV antigens were interpreted as cross-reactive (CR). IgM results that had either equal reactivities to WNV and DENV or a >2-fold difference in their reactivities to the DENV and WNV antigens received further testing.
PRNT90.
A 90%-plaque-reduction (IgG depletion plaque reduction) neutralization test (PRNT90) was performed on samples that either expressed equal reactivities to DENV and WNV antigens in the MAC ELISA or did not express at least a 2-fold difference in IgM P/N values between the two antigens. Prior to neutralization, the serum was depleted of IgG using IgG depletion columns (PanBio, Queensland, Australia) in order to neutralize only with the IgM antibody fraction of the serum. Serum samples were diluted 1:16 and placed through PanBio IgG columns. The serum was then tested using the DENV IgG ELISA to confirm complete removal of IgG from the sample (18). The PRNT90 was performed following IgG depletion. Briefly, Vero cells (African green monkey kidney cells) were plated onto 6-well plates to confluence. Serum samples were heat inactivated at 56°C for 30 min and incubated with the virus reference strains (DEN1, Hawaii; DEN2, NGC; DEN3, H87; DEN4, H241 amplified in C6/36 cells; and WNV, ChimeriVax [donated by Acambis] amplified in Vero cells) for 2 h at room temperature. Control-virus-only wells were titrated to produce an average of 50 plaques per well. The starting dilution of the sample was 1:16 in phosphate-buffered saline with 30% fetal bovine serum. Following incubation, the sample virus mixture was inoculated onto the Vero cell monolayer and incubated for 1 h at room temperature. A 1% agarose medium mixture was added onto the plate, which was incubated at 37°C with 10% CO2 for 5 days. Following incubation, the plates were treated with neutral red solution overnight and the plaques were counted. Samples were tested in duplicate, and an average plaque count was determined for each dilution and sample. The endpoint titer was reported as the reciprocal of the titer in which there was a 90% reduction in the number of plaques compared to the number in the virus control for each sample. Endpoint titers were determined for each virus tested following five 2-fold serial dilutions of the serum sample starting with 1:16 and ending with 1:512 (19, 20).
Dengue virus NS1 antigen ELISA.
All IgM-cross-reactive samples were tested for dengue NS1 antigen with the Bio-Rad Platelia dengue NS1 antigen ELISA (Bio-Rad Laboratories, Marnes-La-Coquette, France), according to the manufacturer's instructions. This test has not been approved for use by the Food and Drug Administration (FDA) and can be used only for research purposes. Briefly, 100 μl of diluted horseradish peroxidase-labeled anti-NS1 monoclonal antibody was combined with 50 μl of serum, and positive and negative controls were diluted 1:2. Each sample was inoculated in duplicate on a 96-well plate precoated with an anti-NS1 capture antibody and incubated for 90 min at 37°C. Following incubation, the plate was washed 6 times, and 160 μl 3,3′,5,5′ tetramethylbenzidine (TMB) substrate was added for 30 min at room temperature. The reaction was stopped with 100 μl of stop solution, and the plate was read at 450/620 nm. Results were analyzed by comparing the average optical density of the sample to the cutoff control optical density. Samples were classified as negative, equivocal, and positive according to ratios of <0.5 unit, 0.5 to <1 unit, and >1 unit, respectively. All equivocal samples were repeated. The results for all positive samples are represented in Tables 1 and 2.
Table 1.
DENV and WNV IgM-positive samplesa
| Sample | DPO | DENV P/N | WNV P/N | PRNT90 titer of antigen to: |
PRNT90 interpretation | Presence of DENV NS1 | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| DENV1 | DENV2 | DENV3 | DENV4 | WNV | SLEV | ||||||
| 1A | 1 | 3.9 | 7.2 | 32 | <16 | 64 | <16 | 32 | 16 | CR | Neg |
| 1B | 1 | 3.2 | 2 | 64 | >512 | >512 | 32 | <32 | 32 | DENV | Neg |
| 1C | 1 | 5 | 5.5 | 512 | 512 | 512 | 64 | 32 | 32 | DENV | Pos |
| 1D | 2 | 3.2 | 3.3 | 64 | >512 | 128 | 16 | 64 | 32 | DENV2 | Neg |
| 1E | 4 | 1.9 | 2.1 | 64 | 32 | 32 | 64 | 16 | 16 | CR | Pos |
| 1F | 4 | 2.4 | 2.2 | 32 | 128 | 64 | 32 | 32 | 32 | CR | Pos |
| 1G | 4 | 3.6 | 2.2 | 128 | 128 | 64 | 16 | 16 | 32 | DENV | Neg |
| 1H | 4 | 2.1 | 3.4 | 32 | 256 | 128 | 32 | <32 | 64 | DENV | Neg |
| 1I | 4 | 2.3 | 4.1 | 16 | 32 | 64 | 32 | <16 | 16 | CR | Neg |
| 1J | 4 | 2.7 | 4.4 | 512 | >512 | >512 | 64 | 16 | 16 | DENV | Neg |
| 1K | 4 | 5.6 | 6.7 | 64 | 16 | 256 | 32 | 16 | 16 | DENV3 | Neg |
| 1L | 5 | 3.5 | 3.5 | >512 | >512 | 64 | 32 | 16 | 16 | DENV | Neg |
| 1M | 5 | 2.8 | 2.9 | 128 | 64 | 256 | 32 | <32 | 32 | DENV | Neg |
| 1N | 5 | 2.2 | 4.3 | 32 | 64 | 64 | 32 | 16 | 16 | CR | Pos |
| 1O | 5 | 3 | 4 | 64 | 64 | 128 | 32 | 16 | 16 | DENV | Neg |
| 1P | 5 | 5.4 | 3.9 | 128 | 256 | 256 | 32 | 32 | 32 | DENV | Neg |
| 1Q | 5 | 5.2 | 8.8 | 256 | 128 | 128 | 16 | <16 | 16 | DENV | Neg |
| 1R | 6 | 3.6 | 4.9 | 256 | 128 | 64 | 16 | 16 | 16 | DENV | Neg |
| 1S | 7 | 5.5 | 4 | >512 | 256 | 256 | 32 | 16 | 16 | DENV1 | Neg |
| 1T | 7 | 16.3 | 21 | >512 | >512 | 16 | 64 | 32 | 16 | DENV | Neg |
| 1U | 8 | 5 | 2.8 | 32 | 16 | 32 | 16 | 16 | 16 | CR | Neg |
| 1V | 8 | 6.5 | 5.3 | 256 | 128 | 64 | 64 | 16 | 16 | DENV | Neg |
| 1W | 10 | 5.1 | 4.3 | >512 | >512 | >512 | 64 | 64 | 16 | DENV | Neg |
The samples are listed according to day after onset of illness (DPO) and were tested with the MAC ELISA, PRNT90 IgG depletion, and NS1 ELISA. These samples had equal reactivities or less than 2-fold difference in reactivities to DENV and WNV antigen in the MAC ELISA. CR, cross-reactive; SLEV, St. Louis encephalitis virus; Neg, negative; Pos, positive. Boldface indicates the infecting virus.
Table 2.
DENV- and WNV-tested samples that had a >2-fold increase in their P/N value for WNV antigen than for DENV antigen or for which the MAC ELISA result for DENV antigen was negativea
| Sample | DPO | DENV P/N | WNV P/N | PRNT90 titer of antigen to: |
PRNT90 interpretation | Presence of DENV NS1 | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| DENV1 | DENV2 | DENV3 | DENV4 | WNV | SLEV | ||||||
| 2A | 1 | 0.9 | 2.5 | 64 | 128 | 256 | 64 | 32 | 64 | DENV | Neg |
| 2B | 1 | 0.2 | 3.3 | 16 | 16 | 16 | 16 | 64 | 16 | WNV | Neg |
| 2C | 2 | 2.8 | 10.5 | 256 | 128 | 64 | 32 | 128 | <16 | CR | Pos |
| 2D | 2 | 1.5 | 4.8 | 64 | 32 | 32 | 128 | <16 | <16 | DENV4 | Neg |
| 2E | 3 | 1.3 | 2.9 | 32 | 512 | 64 | 16 | 64 | 32 | DENV2 | Neg |
| 2F | 4 | 1.8 | 5.2 | 32 | 16 | <16 | 16 | 16 | 16 | CR | Pos |
| 2G | 4 | 1.6 | 3.1 | 128 | 32 | >512 | 16 | 32 | 16 | DENV3 | Neg |
| 2H | 5 | 2.4 | 14.6 | 64 | 64 | 64 | 16 | 16 | 16 | CR | Neg |
| 2I | 5 | 1.1 | 2.4 | 64 | 64 | 128 | >512 | 16 | 16 | DENV4 | Neg |
| 2J | 5 | 5.9 | 13.6 | 128 | >512 | 128 | 32 | 16 | 16 | DENV2 | Neg |
| 2K | 6 | 1.3 | 2.1 | 128 | 256 | >512 | 32 | 32 | <32 | DENV3 | Pos |
| 2L | 6 | 1.9 | 6.8 | >512 | >512 | >512 | 64 | 32 | 16 | DENV | Neg |
| 2M | 6 | 2.8 | 7.7 | >512 | >512 | 256 | 32 | 16 | 16 | DENV | Neg |
| 2N | 7 | 3.2 | 10.3 | 512 | 256 | 512 | 128 | 32 | 64 | DENV | Pos |
| 2O | 7 | 1.5 | 5.6 | >512 | 256 | >512 | 64 | 16 | 16 | DENV | Neg |
| 2P | 8 | 2.4 | 14.5 | 128 | 256 | >512 | 128 | 64 | 64 | DENV3 | Neg |
| 2Q | 8 | 3.4 | 10.4 | 32 | 32 | 64 | 32 | 16 | 16 | CR | Neg |
| 2R | 9 | 4.9 | 15.2 | >512 | >512 | 256 | 64 | 32 | 16 | DENV | Neg |
| 2S | 9 | 0.5 | 3.3 | 64 | 64 | 128 | 32 | 32 | 16 | CR | Neg |
| 2T | 10 | 2.1 | 5.8 | 32 | 64 | 128 | 32 | 32 | 16 | CR | Neg |
The samples are listed by day after onset of illness (DPO) and were tested with the MAC ELISA, PRNT90 IgG depletion, and NS1 ELISA. These samples represent those cases with greater reactivity to WNV antigen than to DENV antigen (or were DENV negative) in the MAC ELISA. SLEV, St. Louis encephalitis virus; CR, cross-reactive; Neg, negative; Pos, positive. Boldface indicates the infecting virus.
RESULTS
Of 2,321 serum samples that were tested using MAC ELISA, 867 (37%) were positive for WNV. Of the 867 samples that were positive by WNV MAC ELISA, 485 (56%) were cross-reactive to both the WNV and DENV antigens. Forty-four percent of the samples (373) were confirmed to be positive for DENV based on either (i) reactivity only to DENV antigen or (ii) no reactivity to WNV or NS1 but a DENV identification by RT-PCR and regular PRNT90. Of the remaining cross-reactive samples, a subset of 43 samples was tested using the PRNT90 with IgG depletion in order to neutralize with only IgM. This subset was determined based on the serum volume required for all the testing for the modified algorithm.
Reactivity to both the WNV and DENV antigens in the MAC ELISA.
Samples from suspected WNV cases that were reactive to both the WNV and DENV antigens in the MAC ELISA are presented in Tables 1 and 2. From the subset of 43 samples that underwent further testing, 23 specimens had equal reactivities to the WNV and DENV antigens or did not display a ≥2-fold difference between reactivities to the two sets of antigens (Tables 1 and 3); 20 specimens were at least twice as reactive to WNV antigen as to DENV antigen (Tables 2 and 3). Table 2 also shows specimens that were positive for WNV and negative for DENV in the MAC ELISA.
Table 3.
| IgM positivity | No. of cases | % (no.) of samples positive by: |
||
|---|---|---|---|---|
| PRNT90 with IgG depletion | DENV NS1 assay | PRNT90 with IgG depletion and NS1 assay | ||
| DENV = WNV | 23 | 74 (17) | 17 (4) | 4 (1) |
| WNV > DENV | 20 | 70 (14) | 20 (4) | 10 (2) |
| Total | 43 | 72 (31) | 19 (8) | 7 (3) |
This table presents the total numbers of cases that were either not interpretable by MAC ELISA alone or that were potentially false positive for WNV in this study. These cases were resolved by the PRNT90 with IgG depletion and/or the DENV NS1 ELISA.
Confirmation of the MAC ELISA by the PRNT90 with IgG depletion.
Of the 43 specimens tested by the PRNT90 with IgG depletion, 31 (72%) were identified as DENV positive and 10 provided a serotype-specific result (Table 1 to 3). Of the 20 samples in Table 2 that would have been categorized as WNV infections based on the results of the MAC ELISA, 14 (70%) were classified as DENV infections using the PRNT90 with IgG depletion (Table 3). Sample 2B was confirmed as WNV positive based on the PRNT90 with IgG depletion result (Table 2). Because serum was depleted of IgG before the PRNT90 was performed, the results in Tables 1 and 2 refer only to neutralization by IgM. As the example in Table 4 shows, all four dengue serotypes failed to reach an endpoint titer without IgG depletion. The results from PRNT90 with IgG depletion indicated that DENV3 was the most prevalent serotype in 2007 based on IgM reactivity (21).
Table 4.
Example of a single specimen that was cross-reactive in the MAC ELISA for DENV and WNV and subsequently tested by the PRNT90a
| Serum | PRNT90 titer of antigen to: |
||||
|---|---|---|---|---|---|
| DEN1 | DEN2 | DEN3 | DEN4 | WNV | |
| Nondepleted | >640 | >640 | >640 | >640 | 80 |
| Depleted | 64 | 256 | >512 | 16 | <16 |
The results presented for DENV1 to -4 and WNV demonstrate the difference in PRNT titers in IgG-depleted serum and nondepleted serum from the specimen. Using IgG depletion identified DENV as the infecting virus for this sample (in bold). The starting dilutions for the sample were 1:16 for IgG-depleted serum and 1:40 for nondepleted serum. (The sample endpoint was not determined due to insufficient sample volume.).
Differential diagnosis by NS1 antigen ELISA.
Using the NS1 antigen ELISA, we confirmed dengue infection in 4 (17%; samples 1C, 1E, 1F, and 1N) of the 23 samples in Table 1 (cross-reactive samples were identified by MAC ELISA) and 4 (20%; samples 2C, 2F, 2K, and 2N) of the 20 samples in Table 2 (WNV-infected samples identified by MAC ELISA). In this assay, the PRNT90 with IgG depletion corroborated the results for sample 1C in Table 1 and samples 2K and 2N in Table 2. Hence, the NS1 antigen ELISA was able to further differentiate samples 1E, 1F, 1N, 2C, and 2F, which were considered uninterpretable due to cross-reactivity in both the MAC ELISA and PRNT90 with IgG depletion (Tables 1 to 3).
Testing algorithm of WNV in regions where dengue is endemic.
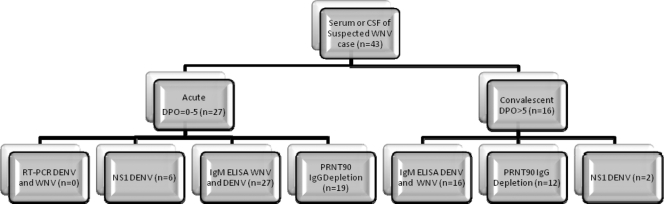
Based on our findings, we have developed a new testing algorithm that was capable of differentiating 36 of 43 cross-reactive samples. We recommend that this new algorithm be used in regions where dengue is endemic when serological results indicate that the serum sample is cross-reactive to both DENV and WNV antigens in the MAC ELISA. This new algorithm employs NS1 antigen ELISA and PRNT90 with IgG depletion to further differentiate the current infection in cross-reactive DENV and WNV samples (Fig. 2). With this algorithm, the results from laboratory testing can be interpreted using Table 5.
Fig. 2.
Algorithm of testing during the transmission season for WNV and DENV, separated by acute- and convalescent-phase serum or cerebral spinal fluid (CSF) samples. The n value for each box represents the number of positive samples within that test group from the 43 subset samples used in the study and described in Tables 1 to 2.
Table 5.
| Result(s) for acute-phase sample |
Result(s) for convalescent-phase sample |
Interpretation | ||||||
|---|---|---|---|---|---|---|---|---|
| IgM WNV positivity | IgM DENV positivity | NS1 positivity | RT-PCR DENV/WNV | IgM WNV positivity | IgM DENV positivity | NS1 positivity | PRNT90 with IgG depletion | |
| − | − | − | − | − | − | − | ND | Negative |
| +/− | +/− | − | + for WNV | +/− | +/− | − | ND | Confirmed WNV infection |
| +/− | +/− | +/− | + for DENV | +/− | +/− | +/− | ND | Confirmed DENV infection |
| + | + | + | − | + | + | +/− | + for DENV | Confirmed DENV infection |
| + | +2× >WNV | − | − | + | +2× >WNV | − | + for DENV | Presumed DENV infection |
| +2× >DENV | + | − | − | +2× >DENV | + | − | + for WNV | Presumed WNV infection |
With this table, a decision for the interpretation of the laboratory tests can be made for patients suspected of having WNV or DENV infection during cocirculation of both flaviviruses. Acute phase, 0 to 5 days after the onset of symptoms; convalescent phase, 6 to 14 days after the onset of symptoms; ND, not done; WNV, West Nile virus; DENV, dengue viruses; +/−, positive or negative; +2× >WNV, the sample's reactivity to the DENV antigen was >2-fold different from its reactivity to the WNV antigen; +2× >DENV, the sample's reactivity to the WNV antigen was >2-fold different from its reactivity to the DENV antigen.
DISCUSSION
The identification of West Nile fever cases in a country where dengue is endemic presents us with many challenges. These challenges are due to the extensive cross-reactivity of flavivirus antigen in serological assays. Without the identification of the infecting virus either through nucleic acid or virus isolation, serology results often cannot with confidence provide a correct diagnosis when more than one flavivirus is circulating in the population. In order to resolve this limitation, the laboratory testing algorithm was modified in this study to include PRNT90 with IgG depletion and an NS1 antigen test. The PRNT90 with IgG depletion allowed for the differentiation of 17 (74%) samples that were initially designated uninterpretable due to cross-reactivity in the MAC ELISA, as well as of 14 (70%) WNV samples that were false positive for WNV in the MAC ELISA (Tables 1 and 2). The dengue NS1 antigen ELISA further confirmed dengue infection in 8 (19%) of the total 43 samples.
NS1 antigen detection has been shown to be useful as a tool for the diagnosis of an acute dengue infection. Previous studies have demonstrated that the NS1 antigen ELISA is specific for DENV rather than for other flaviviruses, such as WNV, Japanese encephalitis virus (JEV), and yellow fever virus (2, 3, 8, 13). The advantage of NS1 antigen detection is that it appears as early as 1 day after the onset of symptoms (DPO) and up to 18 DPO and may bridge the gap in which viral nucleic acid and IgM antibody detection is less likely to be positive (DPO = 4 to 5) (24). Moreover, a previous study evaluating commercially available NS1 detection systems demonstrated the excellent sensitivity (83.2%) and specificity (100%) of the Bio-Rad Platelia dengue NS1 antigen test (2). Because the NS1 test has high specificity for DENV, it can be utilized for differential diagnosis in cases in which the antigen for the IgM ELISA cross-reacts and the infecting virus cannot be identified by conventional RT-PCR.
Cross-reactivity between DENV and WNV was not observed in previous years of WNV surveillance in Puerto Rico (2003 to 2006). This may have been because the earlier surveillance system was a neuroinvasive WNV reporting system, whereas the WNV fever surveillance system, which began in July of 2007, tested cases that were suspected of having WNV regardless of encephalitic symptoms. In 2007, the American Red Cross (ARC) screened blood donations in Puerto Rico for WNV and confirmed WNV transmission in humans (5). The goal of this study was to identify WNV cases using serological techniques that can differentiate between DENV and WNV despite the dengue immune background in the Puerto Rican population.
The background IgG reactivity of past DENV infections often cannot be distinguished from the current or most recent infection because of the high avidity of this antibody. A serosurvey conducted in 1982 in the municipality of Florida, Puerto Rico, indicated that 68 to 80% of individuals had past exposure to dengue; however, recent unpublished CDC data indicate that these levels are now much higher, demonstrating a high prevalence of dengue in Puerto Rico (22). In the case of multiple arboviral infections over time, the sequence of infecting viral species can be an important determinant in the resulting serological responses detected by immunological assays. Secondary flavivirus infections where DENV infection is followed by an infection with a nondengue flavivirus (e.g., WNV) are hypothesized to result in less common epitopes based on the neutralization test; however, the epitopes are not well understood in the secondary flavivirus infections. The anamnestic response to the second flavivirus infection is believed to be less potent than that to sequential DENV infections. Alternatively, previous studies in Asia demonstrated that exposure to Japanese encephalitis virus (a flavivirus in the same serocomplex group as WNV) followed by a natural DENV infection caused a positive serological response to both viruses, although DENV neutralization titers were measurably higher than those for JEV (10). However, sequential flavivirus infection in which the individual has been exposed to DENV on one or more occasions followed by a nondengue flavivirus results in a different immune response. In an animal study by Edelman et al. (9), animals subjected to sequential infections with multiple DENV and JEV infections responded with high neutralizing titers to both JEV and DENV. Fifty animals displayed “original antigenic sin” in which the highest neutralizing titer was to DENV despite the most recent infection being caused by JEV (9). In that study, the incidence of cases in which original antigenic sin occurred varied and was dependent on the first acquired flavivirus infection (14).
The PRNT is the gold standard for differential diagnosis of flavivirus infections in humans. However, due to background DENV IgG reactivity in a population in which the disease is endemic, this test is not always a useful tool for differential diagnosis of secondary flavivirus infections. IgM antibodies are more specific and less cross-reactive than IgG (9, 23). For example, for an individual who acquired a laboratory infection of DENV4 after previous vaccination with JEV, when the IgM fraction of the individual's serum was neutralized, DENV4-specific results were obtained, while the IgG was broadly reactive (3). In our study, the PRNT90 with IgG depletion used IgM antibodies to neutralize, and this resulted in the differentiation of 74% of the samples that were cross-reactive in the MAC ELISA for both WNV and DENV. The combination of PRNT90 with IgG depletion and the NS1 antigen test differentiated 36 (84%) of the samples in this study.
Cocirculation of flaviviruses such as DENV and WNV presents difficult challenges in the interpretation of results from standard serological assays, often complicating the diagnosis of the current infection. Frequently a series of confirmatory diagnostic tests is necessary, and our proposed testing algorithm was effective in determining the infecting virus in samples with a cross-reactive result for WNV and DENV in the MAC ELISA. Our findings show that the PRNT90 with IgG depletion, although technically difficult and time-consuming, is useful for differentiation of WNV and DENV, especially when used in combination with the NS1 antigen ELISA. Further evaluation of this algorithm is necessary for more complete information on the antibody response during the course of infection through the convalescent phase following a WNV infection in an individual that has past single or multiple exposures to DENV.
ACKNOWLEDGMENTS
We acknowledge D. Fermín Argüello for his assistance in developing the Enhanced Human WNV Surveillance System and for critical review of the manuscript and Heidi Acosta for technical assistance in the PRNT assay.
Footnotes
Published ahead of print on 20 April 2011.
REFERENCES
- 1. Barrera R., et al. 2008. First isolation of West Nile virus in the Caribbean. Am. J. Trop. Med. Hyg. 78:666–668 [PubMed] [Google Scholar]
- 2. Bessoff K., Delorey M., Sun W., Hunsperger E. 2008. Comparison of two commercially available dengue virus (DENV) NS1 capture enzyme-linked immunosorbent assays using a single clinical sample for diagnosis of acute DENV infection. Clin. Vaccine Immunol. 15:1513–1518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Blacksell S. D., et al. 2008. Evaluation of the Panbio dengue virus nonstructural 1 antigen detection and immunoglobulin M antibody enzyme-linked immunosorbent assays for the diagnosis of acute dengue infections in Laos. Diagn. Microbiol. Infect. Dis. 60:43–49 [DOI] [PubMed] [Google Scholar]
- 4. Blitvich B. J., et al. 2003. Serologic evidence of West Nile virus infection in horses, Coahuila State, Mexico. Emerg. Infect. Dis. 9:853–856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. CDC 2008. Detection of West Nile virus in blood donations—Puerto Rico, 2007. MMWR Morb. Mortal. Wkly. Rep. 57:577–580 [PubMed] [Google Scholar]
- 6. Dupuis A. P., II, Marra P. P., Kramer L. D. 2003. Serologic evidence of West Nile virus transmission, Jamaica, West Indies. Emerg. Infect. Dis. 9:860–863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Dupuis A. P., II, et al. 2005. Serologic evidence for West Nile virus transmission in Puerto Rico and Cuba. Am. J. Trop. Med. Hyg. 73:474–476 [PubMed] [Google Scholar]
- 8. Dussart P., et al. 2006. Evaluation of an enzyme immunoassay for detection of dengue virus NS1 antigen in human serum. Clin. Vaccine Immunol. 13:1185–1189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Edelman R., Nisalak A., Pariyanonda A., Udomsakdi S., Johnsen D. O. 1973. Immunoglobulin response and viremia in dengue-vaccinated gibbons repeatedly challenged with Japanese encephalitis virus. Am. J. Epidemiol. 97:208–218 [DOI] [PubMed] [Google Scholar]
- 10. Fukunaga T., Okuno Y., Tadano M., Fukai K. 1983. A retrospective serological study of Japanese who contracted dengue fever in Thailand. Biken J. 26:67–74 [PubMed] [Google Scholar]
- 11. Johnson B. W., Russell B. J., Lanciotti R. S. 2005. Serotype-specific detection of dengue viruses in a fourplex real-time reverse transcriptase PCR assay. J. Clin. Microbiol. 43:4977–4983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Komar N., Clark G. G. 2006. West Nile virus activity in Latin America and the Caribbean. Rev. Panam. Salud Publica 19:112–117 [DOI] [PubMed] [Google Scholar]
- 13. Kumarasamy V., et al. 2007. Evaluation of a commercial dengue NS1 antigen-capture ELISA for laboratory diagnosis of acute dengue virus infection. J. Virol. Methods 140:75–79 [DOI] [PubMed] [Google Scholar]
- 14. Kuno G., Gubler D. J., Oliver A. 1993. Use of ‘original antigenic sin’ theory to determine the serotypes of previous dengue infections. Trans. R. Soc. Trop. Med. Hyg. 87:103–105 [DOI] [PubMed] [Google Scholar]
- 15. Lanciotti R. S., et al. 2000. Rapid detection of West Nile virus from human clinical specimens, field-collected mosquitoes, and avian samples by a TaqMan reverse transcriptase PCR assay. J. Clin. Microbiol. 38:4066–4071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Lorono-Pino M. A., et al. 2003. Serologic evidence of West Nile virus infection in horses, Yucatan State, Mexico. Emerg. Infect. Dis. 9:857–859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Martin D. A., et al. 2000. Standardization of immunoglobulin M capture enzyme-linked immunosorbent assays for routine diagnosis of arboviral infections. J. Clin. Microbiol. 38:1823–1826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Miagostovich M. P., et al. 1999. Evaluation of an IgG enzyme-linked immunosorbent assay for dengue diagnosis. J. Clin. Virol. 14:183–189 [DOI] [PubMed] [Google Scholar]
- 19. Roehrig J. T., Hombach J., Barrett A. D. 2008. Guidelines for plaque-reduction neutralization testing of human antibodies to Dengue viruses. Viral Immunol. 21:123–132 [DOI] [PubMed] [Google Scholar]
- 20. Russell P. K., Nisalak A., Sukhavachana P., Vivona S. 1967. A plaque reduction test for dengue virus neutralizing antibodies. J. Immunol. 99:285–290 [PubMed] [Google Scholar]
- 21. Tomashek K. M., et al. 2009. Description of a large island-wide outbreak of dengue in Puerto Rico, 2007. Am. J. Trop. Med. Hyg. 81:467–474 [PubMed] [Google Scholar]
- 22. Waterman S. H., et al. 1985. Dengue transmission in two Puerto Rican communities in 1982. Am. J. Trop. Med. Hyg. 34:625–632 [DOI] [PubMed] [Google Scholar]
- 23. Westaway E. G. 1968. Greater specificity of 19S than 7S antibodies on haemagglutination-inhibition tests with closely related group B arboviruses. Nature 219:78–79 [DOI] [PubMed] [Google Scholar]
- 24. Xu H., et al. 2006. Serotype 1-specific monoclonal antibody-based antigen capture immunoassay for detection of circulating nonstructural protein NS1: implications for early diagnosis and serotyping of dengue virus infections. J. Clin. Microbiol. 44:2872–2878 [DOI] [PMC free article] [PubMed] [Google Scholar]