Abstract
In the peak of the 2009 Q fever outbreak in the Netherlands, we introduced a diagnostic algorithm for acute Q fever with an enzyme-linked immunosorbent assay for immunoglobulin M antibodies to Coxiella burnetii phase II antigens (MII screen) as an initial step. Subsequently, an immunofluorescence assay or PCR was performed depending on the MII screen outcome, date of onset of disease, and inpatient or outpatient setting. The impact of MII screen on the number of immunofluorescence assays performed and the contribution of PCR to diagnosis were retrospectively evaluated in 825 patients referred in a 17-day period. Acute Q fever was diagnosed in 256 patients. The introduction of MII screen reduced the number of immunofluorescence assays performed by more than 80%. In 103 patients, PCR analysis contributed to the diagnosis of acute Q fever. Q fever diagnostics were hampered by the fact that for a high number of patients the date of onset of disease was not provided and the requested follow-up serum samples were not received.
INTRODUCTION
Q fever is a ubiquitous zoonosis caused by the intracellular coccobacillus Coxiella burnetii. In 1937, it was first described in five slaughterhouse workers, two dairy farmers, and one sewage construction worker in Brisbane, Australia, by Edward Derrick (1). C. burnetii has an enormous range of hosts and can infect mammals (primarily sheep, cattle, and goats), birds, and arthropods, including ticks (2). The bacterium is shed in urine, feces, and milk and in especially high concentrations in placentas and birth fluids from infected animals. Inhalation of aerosols contaminated with the bacterium can infect humans (5). However, transmission through consuming contaminated milk and cheese, getting bitten by ticks, or having sex with an infected person has also been reported. Infection of humans results in either subclinical seroconversion or a flu-like syndrome with fever, headache, fatigue, malaise, pneumonia, or hepatitis known as acute Q fever. C. burnetii can be developed for use in biological warfare and is considered a potential bioterrorist threat (2).
Since 2007, the south of the Netherlands has been plagued by a large ongoing community outbreak of Q fever. From 1978 until 2006, between 1 and 32 Q fever cases were notified annually, with an average of 17 cases per year (5, 7). In 2007, 182 cases were notified, followed by 1,000 cases in 2008 and 2,361 cases in 2009. Seven patients were recorded to have died in 2008 and 2009 as a result of Q fever. The geographic spread of notified Q fever cases points to multiple sources and has been linked with high-intensity dairy goat farming in densely populated areas. Many patients, however, never had direct contact with animals (2). Increased awareness has led to a decrease in the interval between onset of disease and date of diagnosis from a median of 77 days in 2007 to 29 days in 2008 and 17 days in 2009 (6).
The Jeroen Bosch Hospital (JBH) in 's-Hertogenbosch is in the center of Noord-Brabant, the province hit hardest by the Q fever explosion. In 2009, the JBH diagnosed more than 1,300 Q fever cases and over 18,000 requests for Q fever diagnostics were received, with a maximum of 182 requests on a single day. To cope with the increasing demand for Q fever diagnostics, we implemented a diagnostic algorithm on 1 May 2009. The aim of this diagnostic algorithm was to provide accurate, fast, cost-effective, and standardized diagnostics for acute Q fever in an outbreak setting. Before May 2009, the immunofluorescence assay (IFA) for immunoglobulin M (IgM) and immunoglobulin G (IgG) antibodies to C. burnetii phase I and phase II antigens had been the cornerstone of Q fever diagnostics in the JBH. IFA, however, has disadvantages, as it is time-consuming, nonautomated, expensive, and subject to interobserver variability. In addition, the antibody response to C. burnetii takes time to develop, making serology less suitable for acute Q fever diagnostics during the first 2 weeks of the disease (6). Alternative diagnostic approaches incorporated in the algorithm included an enzyme-linked immunosorbent assay (ELISA) for IgM antibodies to phase II antigens as a screening assay and a highly specific real-time PCR, targeting IS1111A, for detection of C. burnetii DNA in serum during the very early stage of the disease. We have previously shown that this PCR detects C. burnetii DNA in virtually all sera of seronegative patients with acute Q fever but rapidly becomes negative when the antibody response develops (6). In this study, we retrospectively examined the impact of the ELISA on the number of IFA tests performed and the contribution of PCR to diagnosis with first serum samples.
MATERIALS AND METHODS
Inclusion criteria.
We retrospectively evaluated the outcome of the diagnostic algorithm for acute Q fever in patients referred in the peak of the 2009 epidemic from 15 to 31 May with a date of onset of disease ≤3 months earlier or without information on the date of onset of disease. Excluded from analysis were patients with follow-up serum samples referred from 15 to 31 May 2009 for whom initial acute Q fever diagnostics were requested from 1 January to 14 May 2009. However, patients referred for acute Q fever diagnostics from 15 to 31 May 2009 for whom an earlier negative outcome had been obtained in the 2007 or 2008 epidemic before 1 January 2009 were not excluded.
Diagnostic algorithm.
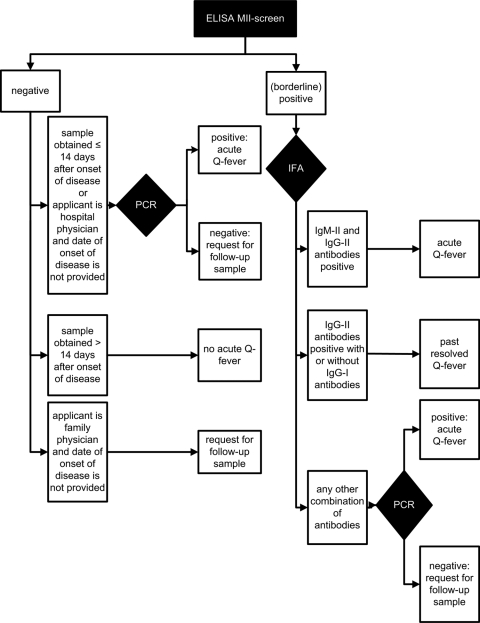
Figure 1 shows the flowchart of the diagnostic algorithm for acute Q fever. All serum samples were initially screened with a qualitative ELISA for IgM antibodies to phase II antigens (MII screen) according to the manufacturer's instructions (Institut Virion\Serion GmbH, Würzburg, Germany) on a DSX automated ELISA processing system (Dynex Technologies, Chantilly, VA).
Fig. 1.
Diagnostic algorithm for acute Q fever in an outbreak setting introduced 1 May 2009 in the Jeroen Bosch Hospital, 's-Hertogenbosch, Netherlands. Abbreviations: ELISA MII-screen, enzyme-linked immunosorbent assay for immunoglobulin M antibodies to Coxiella burnetii phase II antigens; IFA, immunofluorescence assay; IgM-II antibodies, immunoglobulin M antibodies to C. burnetii phase II antigens; IgG-II antibodies, immunoglobulin G antibodies to C. burnetii phase II antigens; IgG-I antibodies, immunoglobulin G antibodies to C. burnetii phase I antigens.
IFA for IgM and IgG antibodies to phase I and phase II antigens (Focus Diagnostics, Inc., Cypress, CA) was subsequently performed on all MII screen-positive and borderline-positive serum samples. IFA was performed according to the manufacturer's instructions with the adjustment that titers of 1:32 or greater were considered positive (instead of titers of 1:16 and greater). When IgM and IgG antibodies to phase II antigens were detected, the diagnosis of acute Q fever was made regardless of the presence or absence of IgM and IgG antibodies to phase I antigens. In cases in which only IgG antibodies to phase II antigens with or without IgG antibodies to phase I antigens were detected, the diagnosis of past resolved Q fever was made. When any other combination of antibodies was found, IFA was considered inconclusive and a PCR was performed on the serum sample as described previously (6). In the case of a positive PCR, the diagnosis of acute Q fever was made. If the PCR was negative, a follow-up serum sample after 14 days was requested.
When the MII screen was negative, PCR was performed on first serum samples obtained ≤14 days after the onset of disease and on serum samples referred by hospital physicians without information on the date of onset of disease. In the case of a positive PCR, the diagnosis of acute Q fever was made. If the PCR was negative, a follow-up serum sample after 14 days was requested. In cases in which serum samples were referred by family physicians without information on the date of onset of disease, PCR was not performed and a follow-up serum sample after 14 days was requested. Patients with serum samples obtained >14 days after onset of disease and a negative MII screen were considered not to have acute Q fever.
Table 1 lists the six possible outcomes of acute Q fever diagnostics in the diagnostic algorithm: (i) acute Q fever, (ii) past resolved Q fever, (iii) no acute Q fever, (iv) cross-reactivity (false-positive MII screen not confirmed by additional tests in first or follow-up serum samples), (v) inconclusive outcome (noncompliance with the request to refer a follow-up serum sample), and (vi) inconsistency with the diagnostic algorithm (nonadherence to the flowchart of the diagnostic algorithm).
Table 1.
Outcomes of the diagnostic algorithm for acute Q fever in an outbreak setting and definitions
| Outcome in the diagnostic algorithm | Definition |
|---|---|
| Acute Q fever | Presence of IgM and IgG antibodies to phase II antigens with or without IgM and IgG antibodies to phase I antigens or PCR positivity |
| Past resolved Q fever | Presence of IgG antibodies to phase II antigens with or without IgG antibodies to phase I antigens in the absence of IgM antibodies |
| No acute Q fever | Absence of a serologic response in serum samples obtained >14 days after onset of disease |
| Cross-reactivity | Presence of false-positive IgM antibodies to phase II antigens detected by ELISA but not confirmed by additional tests in first or follow-up serum samples |
| Inconclusive outcome | Noncompliance with the request to refer a follow-up serum sample |
| Inconsistency with the diagnostic algorithm | Nonadherence to the flowchart of the diagnostic algorithm |
Additional PCR analysis.
The outcome of Q fever diagnostics could be hampered by noncompliance with the request to refer follow-up serum samples leading to an inconclusive outcome. This may lead to underdiagnosis of acute Q fever, especially in the group of patients referred by family physicians without information on the date of onset of disease and a negative MII screen, in which case the diagnostic algorithm does not allow for PCR analysis. In the context of this evaluation, serum samples from this particular group of patients were retrospectively submitted to additional PCR analysis.
Statistical analysis.
The Mann-Whitney U test was used to compare the numbers of days from the onset of disease to acquisition of the first serum sample in patients with acute Q fever and either a positive or borderline-positive MII screen or a negative MII screen. Significance was designated at P values of <0.05. The test was performed with SPSS 18 software for Windows (SPSS Inc., Chicago, IL).
RESULTS
Study population.
Using the laboratory information system, 825 patients (421 [51.0%] males and 404 [49.0%] females) were identified who had been referred for acute Q fever diagnostics in the 2009 epidemic from 15 to 31 May and met the inclusion criteria. Hospital physicians referred 143 (17.3%) patients, while the other 682 (82.7%) patients had been referred by family physicians. The date of onset of disease was provided for 32/143 (22.4%) referrals from hospital physicians and 396/682 (58.1%) referrals from family physicians.
Outcome of diagnostic algorithm.
The outcome of acute Q fever diagnostics is presented in Table 2. The diagnosis of acute Q fever was made in 256/825 (31.0%) patients (161/256 [62.9%] males and 95/256 [37.1%] females), including 11 cases where this diagnosis was inconsistent with the diagnostic algorithm.
Table 2.
Outcome of acute Q fever diagnostics according to a diagnostic algorithm in 825 patients referred from 15 to 31 May 2009
| Status | No. of samples with definitive outcome |
Relative outcome overall (%) | ||
|---|---|---|---|---|
| In first serum sample | In follow-up serum sample | Overall | ||
| Acute Q fever | 190 | 55 | 245 | 29.7 |
| Past resolved Q fever | 2 | 1 | 3 | 0.4 |
| No acute Q fever | 116 | 129 | 245 | 29.7 |
| Cross-reactivity | 0 | 4 | 4 | 0.5 |
| Inconclusive outcome | 299 | 2 | 301 | 36.5 |
| Inconsistency with the diagnostic algorithm | ||||
| Acute Q fever | 7 | 4 | 11 | |
| No acute Q fever | 5 | 4 | 9 | |
| Cross-reactivity | 0 | 2 | 2 | |
| Inconclusive outcome | 5 | 0 | 5 | |
| Total with inconsistent result | 17 | 10 | 27 | 3.3 |
A positive or borderline-positive MII screen in the first serum sample was obtained in 143/825 (17.3%) and 13/825 (1.6%) of patients, respectively. Subsequent IFA on these serum samples confirmed the diagnosis of acute Q fever in 94 patients. In the other 62 patients, IFA was either indicative of past resolved Q fever (n = 2) or inconclusive (n = 60). Subsequent PCR on the 60 serum samples with inconclusive IFA confirmed the diagnosis of acute Q fever in another 11 patients. Thus, in 49 patients with a positive MII screen in the first serum sample no conclusive outcome could be made. Therefore, a follow-up serum sample was requested, which was received from 31/49 patients. Serologic analysis of these follow-up serum samples yielded the diagnosis of acute Q fever in 25 patients and indicated cross-reactivity in 6 patients. Q fever diagnostics were considered inconclusive in the 18 patients for whom the requested follow-up serum sample was not received.
A negative MII screen in the first serum sample was obtained in 669/825 (81.1%) patients. As a result, the number of IFA tests performed on first serum samples was reduced for more than 80% of patients. Subsequently, PCR was performed on 230 MII screen-negative first serum samples obtained ≤14 days after the onset of disease and 94 MII screen-negative first serum samples referred by hospital physicians without information on the date of onset of disease. In these groups, PCR yielded the diagnosis of acute Q fever in 75/230 (32.7%) and 17/94 (18.1%) patients, respectively. In 454 patients with a negative MII screen in the first serum sample, a follow-up serum sample was requested because no conclusive outcome could be made. A follow-up serum sample was received from 168/454 (37.0%) patients. Serologic analysis of these follow-up serum samples yielded the diagnoses of acute Q fever in 32 patients, no acute Q fever in 133 patients, past resolved Q fever in 1 patient, and an inconclusive outcome in 2 patients. In 11 of the 32 patients with acute Q fever diagnosed on the follow-up serum sample, the diagnosis was missed in the first serum sample despite the fact that PCR had been performed in addition to MII screen. Q fever diagnostics were considered inconclusive in the 286 patients in whom a follow-up serum sample was not received. In 123/669 (18.4%) patients with a negative MII screen, the first serum sample was obtained >14 days after onset of disease and these patients were considered not to have acute Q fever. However, unrequested follow-up serum samples from 26 patients in this group were received. Serologic analysis of these follow-up serum samples confirmed the diagnosis of no acute Q fever in 24 patients but yielded the diagnosis of acute Q fever in 2 patients. In these two patients, the first serum samples had been received 22 and 23 days after the onset of disease, with follow-up serum samples received at 42 and 145 days, respectively.
The date of onset of disease was available for 65/130 (50.0%) patients with a positive or borderline positive MII screen in the first serum sample and an outcome of acute Q fever. The number of days from onset of disease to acquisition of the first serum sample in this group was 17 ± 15 days (mean ± standard deviation [SD]). In the group of patients with a negative MII screen and an outcome of acute Q fever, the date of onset of disease was available for 84/126 (66.7%) patients. The number of days from onset of disease to acquisition of the first serum sample in this group was significantly lower (P < 0.01), being 5 ± 4 days.
In all, the diagnosis of acute Q fever was made solely on the first serum sample in 197/256 (77.0%) patients in whom the diagnosis was eventually made. In 103 patients, a positive PCR analysis on the first serum sample contributed to this diagnosis.
Inconsistency with the diagnostic algorithm was observed for 27/825 (3.3%) patients. Mainly, nonadherence to the flowchart resulted from misinterpretation of the diagnostic algorithm or miscalculation of the number of days following onset of disease. Other causes for nonadherence to the flowchart were personal requests by treating physicians for additional diagnostics, for instance because of special situations like pregnancy.
Outcome of additional PCR analysis.
In all, we identified 147 patients with an MII screen-negative first serum sample who had been referred by a family physician without information on date of onset of disease and for whom the requested follow-up serum sample was not received. For these patients, the diagnostic algorithm did not allow for PCR analysis on first serum samples. In the context of the evaluation of the diagnostic algorithm, PCR analysis was performed retrospectively on 145 of these 147 serum samples (two samples had insufficient volume for PCR analysis). A positive PCR result was obtained in 11 serum samples. Thus, the total number of patients in this cohort in whom the diagnosis of acute Q fever was made either prospectively or retrospectively was 267/825 (32.4%) patients.
DISCUSSION
Laboratory diagnosis of acute Q fever is important because of the atypical clinical presentation of the disease, its requirement of specific antibiotic treatment, and the needs for follow-up of the development of chronic disease, outbreak surveillance, and response in case of bioterrorist attack (2, 6). The microbiological diagnostic facilities of the JBH, which lies in the center of the epidemic area, had been confronted with an explosive increase in diagnostic requests for acute Q fever since the start of the epidemic in 2007. In 2009, the diagnostic demands on our laboratory were even further increased as a result of the foreseen influenza A (H1N1) pandemic. These factors urged us to implement a diagnostic algorithm that provided accurate, fast, cost-effective, and standardized diagnostics for acute Q fever in an outbreak setting.
Before May 2009, IFA had been the cornerstone of Q fever diagnostics in the JBH. An important drawback to serological diagnosis of acute Q fever, however, is the lag phase in antibody response following infection. Previously, we reported that real-time PCR on serum samples is a useful approach to diagnose acute Q fever before development of the antibody response (6). Specific drawbacks to IFA include the facts that it is time-consuming, nonautomated, expensive, and subject to interobserver variability. Introduction of the MII screen on an automated ELISA processing system was expected to significantly reduce the number of IFA tests performed and overcome the specific drawbacks of IFA. A diagnostic algorithm implementing both PCR and MII screen was implemented on 1 May 2009.
Evaluation of the outcome of the diagnostic algorithm in patients referred in a 17-day period in the peak of the 2009 epidemic revealed that introduction of the MII screen resulted in a reduction of IFA tests performed on first serum samples by more than 80%. In addition, PCR of the first serum samples contributed to the diagnosis of acute Q fever in 92 patients with a negative MII screen and in 11 patients with a positive MII screen. The diagnosis of acute Q fever was made solely on the first serum sample in 77.0% of patients in whom this diagnosis was eventually made. These results do not take into account the 11 patients referred by family physicians without information on the date of onset of disease and a negative MII screen for whom retrospective PCR analysis was positive. One could argue for the performance of PCR as the sole test on all first serum samples. However, C. burnetii DNA rapidly becomes undetectable in serum as the serological response develops (6). Extrapolation of reported rates of PCR positivity for specific serologic profiles to the group of 94 patients with positive MII screens and subsequent positive IFAs on the first serum sample indicates that PCR—if performed—would presumably have been positive in 8 patients and negative in 86 patients. Thus, the performance of PCR as the sole test would have presumably resulted in a missed diagnosis of acute Q fever on the first serum sample in 86 patients where our diagnostic algorithm did allow for this diagnosis.
Eleven patients were identified in whom the diagnosis of acute Q fever was initially missed on the first serum sample despite the fact that both MII screen and PCR had been performed. Our diagnostic algorithm did not allow for PCR and IFA (the latter being considered the gold standard) to be performed on all samples, making it not possible to calculate sensitivities and specificities based on this study. However, in a separate direct comparison to IFA, we observed a sensitivity of 85.7% for the MII screen (J.C.E. Meekelenkamp, unpublished data). In addition, PCR has a reported sensitivity of 98.0% in a retrospective analysis on selected seronegative serum samples (6). Thus, both tests have some limitations in sensitivity that might account for missed diagnosis of acute Q fever on first serum samples. Furthermore, it cannot be excluded that there is a window period following initial PCR positivity in which C. burnetii DNA has disappeared but IgM antibodies to phase II antigens have not yet appeared.
Cross-reactivity (false positivity) was documented in six patients with a positive MII screen but could not be excluded in 18 patients with a positive MII screen but no conclusive outcome of Q fever diagnostics because a requested follow-up serum sample was not received. In a separate direct comparison to IFA, we observed a specificity of 97.6% for the MII screen (Meekelenkamp, unpublished). Cross-reactivity in C. burnetii antibody tests has also been documented by others (4). For that reason, serological evidence of acute Q fever was defined in our laboratory as the presence of both IgM and IgG antibodies to phase II antigens as detected by IFA.
ELISA and IFA have been reported to detect seroconversion 10 to 15 days postinfection (4). Therefore, the diagnostic algorithm considered patients with serum samples obtained >14 days after onset of disease and a negative MII screen not to have acute Q fever. Consequently, follow-up serum samples were not requested from these patients. In 26 patients, however, an unrequested follow-up serum sample was received, and serologic analysis revealed acute Q fever in 2 of those patients. Although it cannot be excluded that in both patients seroconversion resulted from (asymptomatic) infection acquired after referral of the first serum sample, it also cannot be excluded that seroconversion occurred more than 3 weeks postinfection. These and similar observations not part of this evaluation have subsequently led to a change in the diagnostic algorithm: follow-up serum samples are now requested from all patients with a negative MII screen in a first serum sample received <28 days after onset of disease.
Hampering the overall outcome of Q fever diagnostics, but unrelated to the intrinsic usefulness of the diagnostic algorithm, were the facts that the date of onset of disease was not provided in more than half of the referrals and the requested follow-up serum samples were not received from a high number of patients. Hospital physicians did not provide the date of onset of disease in more than three of four referrals. It was anticipated that they would be less accurate in providing this date, which was one of the reasons that the diagnostic algorithm allowed for PCR to be performed on all MII screen-negative first serum samples referred by hospital physicians without information on the date of onset of disease. Moreover, we reasoned that hospitalized patients with acute Q fever would have a more severe disease course than nonhospitalized patients, justifying an additional PCR test on the first serum sample even if the date of onset of disease was not provided. Allowing PCR on the 94 MII screen-negative first serum samples referred by hospital physicians without information on the date of onset of disease resulted in the diagnosis of acute Q fever in 18.1% of these patients. In cases of MII screen-negative patients referred by family physicians without information on the onset of disease, it was anticipated that the disease course would be less severe, justifying a request for a follow-up serum sample after 14 days. This approach was recently justified by a retrospective analysis that demonstrated that in 2009 95% of family physicians in the epidemic area would immediately start antibiotic treatment in case of suspected acute Q fever without awaiting microbiological test results (3).
Requested follow-up serum samples were not received from 306 patients, leading to an inconclusive outcome. This implies that cases of acute Q fever might have been missed. Indeed, a retrospective PCR analysis on first serum samples revealed 11 cases of undiagnosed acute Q fever in patients for whom requested follow-up serum samples were not referred. Therefore, additional efforts will be made to inform both hospital and family physicians of the importance of accurately providing the date of onset of disease and referring requested follow-up serum samples.
Here, we show that introduction of a diagnostic algorithm for acute Q fever in an outbreak setting resulted in increased diagnostic yield from first serum samples, with a considerable reduction in IFA tests performed. Both hospital and family physicians need to be made more aware of the importance of providing the date of onset of disease and referring requested follow-up serum samples.
ACKNOWLEDGMENTS
We thank the laboratory technicians of the serology unit of the Department of Medical Microbiology and Infection Control and the Molecular Diagnostics Department of the Jeroen Bosch Hospital for their tremendous efforts in continuously providing high-quality microbiological services during the world's largest Q fever outbreak recorded to date.
Footnotes
Published ahead of print on 20 April 2011.
REFERENCES
- 1. Cooke R. A. 2008. Q fever. Was Edward Derrick's contribution undervalued? Med. J. Aust. 189:660–662 [DOI] [PubMed] [Google Scholar]
- 2. Enserink M. 2010. Infectious diseases. Questions abound in Q-fever explosion in the Netherlands. Science 327:266–267 [DOI] [PubMed] [Google Scholar]
- 3. Lassche S., Schrauwen M. M. W. P., Rietveld A., Wijkmans C. J. 2010. General practitioners aware of Q-fever. Infect. Bull. 21:A34–A38 [Google Scholar]
- 4. Maurin M., Raoult D. 1999. Q fever. Clin. Microbiol. Rev. 12:518–553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Schimmer B., et al. 2009. Sustained intensive transmission of Q fever in the south of the Netherlands, 2009. Euro Surveill. 14:(19):pii=19210 http://www.eurosurveillance.org/ViewArticle.aspx?ArticleId=19210 [DOI] [PubMed] [Google Scholar]
- 6. Schneeberger P. M., et al. 2010. Real-time PCR with serum samples is indispensable for early diagnosis of acute Q fever. Clin. Vaccine Immunol. 17:286–290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. van Gageldonk-Lafeber A. B., Koopmans M. P. G., Bosman A., Heijnen M. L. A. 2003. Surveillance of Q fever in the Netherlands. Infect. Bull. 14:173–177 [Google Scholar]