Abstract
The hepatitis B virus (HBV) PreS mutations C1653T, T1753V, and A1762T/G1764A were reported as a strong risk factor of hepatocellular carcinoma (HCC) in a meta-analysis. HBV core promoter overlaps partially with HBx coding sequence, so the nucleotide 1762 and 1764 mutations induce HBV X protein (HBx) 130 and 131 substitutions. We sought to elucidate the impact of HBx mutations on HCC development. Chronically HBV-infected patients were enrolled in this study: 42 chronic hepatitis B (CHB) patients, 23 liver cirrhosis (LC) patients, and 31 HCC patients. Direct sequencing showed HBx131, HBx130, HBx5, HBx94, and HBx38 amino acid mutations were common in HCC patients. Of various mutations, HBx130+HBx131 (double) mutations and HBx5+HBx130+HBx131 (triple) mutations were significantly high in HCC patients. Double and triple mutations increased the risk for HCC by 3.75-fold (95% confidence interval [CI] = 1.101 to 12.768, P = 0.033) and 5.34-fold (95% CI = 1.65 to 17.309, P = 0.005), respectively, when HCC patients were compared to CHB patients. Functionally, there were significantly higher levels of NF-κB activity in cells with the HBx5 mutant and with the double mutants than that of wild-type cells and the triple-mutant cells. The triple mutation did not increase NF-κB activity. Other regulatory pathways seem to exist for NF-κB activation. In conclusion, a specific HBx mutation may contribute to HCC development by activating NF-κB activity. The HBx5 mutation in genotype C2 HBV appears to be a risk factor for the development of HCC and may be used to predict the clinical outcomes of patients with chronic HBV infection.
INTRODUCTION
Hepatitis B virus (HBV) infection is a global health issue, with two billion people infected worldwide and 350 million suffering from chronic HBV infection (18). Chronic HBV infection is known as the most common underlying etiology of hepatocellular carcinoma (HCC) (2) and can further progress to liver cirrhosis (LC) and HCC. However, the pathogenesis of these HBV-related diseases has not been fully clarified. The disease progression may depend on complex and multistep mechanisms.
In an HBV management consensus report, the National Institutes of Health (NIH) suggested risk factors associated with the increased risk of HCC and cirrhosis in chronic HBV-infected patients (20). That report described the HCC risk based on demographic factors, environmental factors, and viral factors and classified them using the scoring system of evidence-based studies. The association between HBV mutations and HCC has not been fully investigated. Recently, the HBV PreS mutations C1653T, T1753V, and A1762T/G1764A were reported as strong risk factors for HCC in a meta-analysis (19). Mutations of HBV have been considered as an escape mechanism from the host immune system and may affect the oncogenic potential of chronic HBV diseases.
HBV X is the smallest of four kinds of HBV functional genes, but it expresses a 154-amino-acid multifunctional protein (HBx), with an N-terminal negative regulatory/antiapoptotic domain and a C-terminal transactivation/proapoptotic domain. The gene also seems to be associated with modulation of a wide range of cellular functions, leading to HCC (5). Moreover, of the four open reading frames, the X gene remains enigmatic. There are conflicting suggestions about the functional activity of HBx. HBx induced HCC in certain transgenic mice in vivo (15, 30) and in vitro (11, 25). Nevertheless, there are other transgenic lineages in which HBx does not lead to HCC development (3, 24). Many reports suggest that HBx is a multifunctional regulatory protein that directly or indirectly interacts with a variety of targets and mediates many cellular functions. Thus, the goals of the present study were to elucidate the characteristics of HBx mutations according to the clinical phase of HBV infection and to examine the functions of significant HBx mutations that appear to be related to HCC pathogenesis.
MATERIALS AND METHODS
Study subjects.
Patients with chronic HBV infection were enrolled in the present study. There were 42 chronic hepatitis B disease (CHB) patients, 23 LC patients, and 31 HCC patients. The HBV-related etiology was confirmed based on clinical and radiologic findings, laboratory tests, and pathological findings (i.e., serum alanine aminotransferase, HBsAg, HBeAg, HBV DNA, and liver biopsy). All patients' samples were divided into aliquots and kept frozen at −70°C until used.
The study protocol conformed to the 1975 Declaration of Helsinki and was approved by the Institutional Review Board of the Yonsei University Health System.
Sequencing of HBV DNA from the subjects.
HBV DNA was extracted from serum samples by using a DNA minikit (Qiagen, Hilden, Germany) or a QuickGene DNA Tissue Kit S (Fujifilm, Tokyo, Japan). Nested PCR was performed to amplify HBV X gene. The primers for HBV X gene amplification were as follows: sense (5′-CATGCGTGGAACCTTTGTG-3′; positions 1233 to 1251) and antisense (5′-CTTGCCTKAGTGCTGTATGG-3′; positions 2072 to 2053) for the first PCR amplification and sense (5′-TCCTCTGCCGATCCATACTG-3′; positions 1254 to 1263) and antisense (5′-CAGAAGCTCCAAATTCTTTATA-3′; positions 1937 to 1916) for the second PCR amplification.
PCR premix (Accupower Pfu PCR PreMix; Bioneer, Daejun, Korea) with a MyGenie 96 Gradient Thermal Block (Bioneer, Daejun, Korea) were used to produce an 839-bp amplicon. A 683-bp amplicon of the second nested PCR was identified in 1% agarose gel electrophoresis stained with ethidium bromide. PCR products were purified and then sequenced by an ABI 3730 automated sequencer (Applied Biosystems, Foster City, CA) with a BigDye Terminator v3.1 sequencing kit (Applied Biosystems). If primary PCR was not successful, the supplementary primer sets described by Kim et al. (17) were also used for direct sequencing.
All sequencing data were analyzed based on the consensus HBV sequence (GenBank accession no. GQ475316) of a chronic HBV carrier patient without antiviral therapy. The sequencing data were multiply aligned and analyzed by CLUSTAL W and Bioedit v7.0.5 software (Ibis Therapeutics, Carlsbad, CA).
HBV genotyping.
Deduced X gene base pairs were compared to several consensus sequences of HBV genotypes, and a phylogenetic tree was constructed by the neighbor-joining method. Phylogenetic and molecular evolutionary analyses were performed using MEGA version 4.0.2 (The Biodesign Institute, Tempe, AZ). The genetic distances were estimated by Kimura's two-parameter method. The reliability of the phylogenetic tree analysis was assessed by the interior branch test with 1,000 replicates.
Construction of HBx plasmid.
Three most frequent HBx mutations in HCC patients were HBx131, HBx130, and HBx5 mutations in that order. Therefore, four different types of plasmids were constructed: (i) wild-type HBx, (ii) single amino acid mutants of HBx (HBx5; G1386A and V5M), (iii) a double amino acid mutants of HBx (HBx130+HBx131; A1762T+G1764A and K130M+V131I), and (iv) triple amino acid mutants of HBx (HBx5+HBx130+HBx131; G1386A+A1762T+G1764A and V5M+K130M+V131I). All plasmids were produced based on a 1.2-mer HBV construct within the pGEM4Z vector (kindly provided by Kyun Hwan Kim) with the appropriate site-directed mutagenesis primers. PCR products were treated with the restriction enzyme DpnI or NcoI/HindIII and then transformed into XL10 Gold Ultra Competent cells (Stratagene, Cedar Creek, TX) according to the manufacturer's instructions. All mutant clones were verified by sequencing both strands of DNA within the regions of interest.
Cell cultures and transfection into Huh7 cells.
Nucleic acids were prepared by Escherichia coli transformation using Qiagen plasmid midikits, and Huh7 cells were cultured in Dulbecco modified Eagle medium (Gibco) at 37°C in 5% CO2. Huh7 cells were transiently transfected with pGEM4Z, HBx wild-type, HBx5 mutant, HBx double-mutant, and HBx triple-mutant constructs, using Lipofectamine LTX and Plus reagents (Invitrogen, Carlsbad, CA). All experiments were performed in triplicate.
Detection of HBV replication by using HBV DNA, HBsAg, and HBeAg expression.
Real-time PCR was used to determine HBV DNA replication activity in each group of transfected cells. DNA was extracted from transfected cells transfected with each construct, and then the purity and quantity of DNA were measured by using Nanodrop (Thermo Fisher Scientific, Inc., Wilmington, DE). Real-time PCR amplifications were carried out using the ABI 7000 system (Applied Biosystems). The 100-ng/μl template was processed with SYBR green I and the primers HBs-F (5′-CCTCTTCATCCTGCTGCT-3′) and HBs-R (5′-AACTGAAAGCCAAACAGTG-3′). Real-time PCR amplification was carried out by using an ABI 7000 system (Applied Biosystems) using 2× SYBR green mix (Applied Biosystems). The thermal conditions for SYBR real-time PCR were 95°C for 2 min, followed by 40 cycles of 15 s at 95°C and 1 min at 60°C. The values were normalized to GAPDH (glyceraldehyde-3-phosphate dehydrogenase) expression in each sample.
The total protein concentration in harvested cell lysates and in the supernatants of transfected Huh7 cells were determined by bicinchoninic acid kit (Pierce, Rockford, IL). The corrected HBsAg and HBeAg levels were determined by using an Architect i2000 analyzer (Abbott Diagnostics, Abbott Park, IL).
NF-κB luciferase reporter gene assay.
To determine the NF-κB activity, cotransfection with NF-κB luciferase vector and HBx constructs was performed using Lipofectamine LTX and Plus reagents (Invitrogen). Cells were harvested 2 days after transfection, and then the luciferase activity was detected by using the dual luciferase reporter assay system (Promega, San Luis Obispo, CA) on a Victor X4 multilabel plate reader (Perkin-Elmer Life Sciences, Shelton, CT). All luciferase data were normalized with the corresponding Renilla luciferase activity. Each experiment was performed in triplicate.
Statistical analysis.
SPSS 12.0 (SPSS, Inc., Chicago, IL) was used for statistical analyses. Statistical analysis were performed by using the chi-square test or the Fisher's exact test for categorical variables and the Mann-Whitney U-test or one-way analysis of variance for continuous variables. P values of <0.05 were considered significant.
RESULTS
Basic characteristics of study subjects.
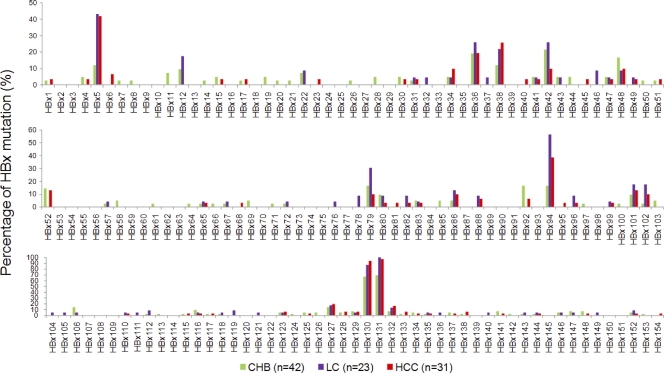
The basic characteristics and HBx mutations of enrolled patients are summarized in Tables 1 and 2, respectively. There were some significant differences according to the clinical phase of HBV infection. The average age of HCC patients was higher than that of CHB patients (P < 0.001) but was not different from that of LC patients (P = 0.275). The levels of HBV DNA in HCC patients were lower than those in CHB patients (P = 0.002) but were not different from those in LC patients (P = 0.872). HBeAg status was different among three clinical phases of HBV infection (P = 0.002). The most common HBx mutation in HCC patients was followed by HBx131, HBx130, HBx5, HBx94, and HBx38. The incidence of the HBx5+HBx130+HBx131 triple mutations in HCC patients increased with age (Table 3).
Table 1.
Characteristics of HBx mutations according to clinical diagnosisa
| Parameter | CHB | LC | HCC | Pb |
|---|---|---|---|---|
| No. of patients | 42 | 23 | 31 | |
| Mean age (yr) ± SD | 43.6 ± 10.7 | 51.7 ± 7.8 | 55.6 ± 7.8 | 0.007* |
| Sex ratio (males:females) | 2 (28:14) | 2.3 (16:7) | 5.2 (26:5) | 0.241† |
| HBV DNA (IU/ml)c ± SD | 162,785,477 ± 222,839,000 | 21,778,882 ± 77,209,447 | 841,788 ± 1,558,362 | <0.001* |
| HBeAg ratio (no. positive:no. negative) | 2.8 (31:11) | 1.1 (12:11) | 0.5 (10:21) | 0.002† |
| Mean no. of HBx mutations ± SD | 5.95 ± 5.04 | 7.26 ± 4.7 | 6 ± 2.37 | 0.455* |
| Range (no.) of HBx mutations | 1–25 | 3–27 | 1–11 |
CHB, chronic hepatitis B; LC, liver cirrhosis; HCC, hepatocellular carcinoma.
As determined by one-way analysis of variance (*) or chi-square test (†).
IU, international units.
Table 2.
Common HBx mutations (≥4 times incidence)
| Condition and mutation site | Total no. of mutations | Frequency (%) |
|---|---|---|
| CHB | ||
| HBx131 | 28 | 66.7 |
| HBx130 | 27 | 64.3 |
| HBx42 | 9 | 21.4 |
| HBx36 | 8 | 19.0 |
| HBx48 | 7 | 16.7 |
| HBx79 | 7 | 16.7 |
| HBx92 | 7 | 16.7 |
| HBx94 | 7 | 16.7 |
| HBx52 | 6 | 14.3 |
| HBx106 | 6 | 14.3 |
| HBx127 | 6 | 14.3 |
| HBx5 | 5 | 11.9 |
| HBx38 | 5 | 11.9 |
| HBx12 | 4 | 9.5 |
| HBx80 | 4 | 9.5 |
| HBx101 | 4 | 9.5 |
| HBx116 | 4 | 9.5 |
| LC | ||
| HBx131 | 23 | 100.0 |
| HBx130 | 20 | 87.0 |
| HBx94 | 13 | 56.5 |
| HBx5 | 10 | 43.5 |
| HBx79 | 7 | 30.4 |
| HBx42 | 6 | 26.1 |
| HBx36 | 6 | 26.1 |
| HBx38 | 5 | 21.7 |
| HBx127 | 4 | 17.4 |
| HBx12 | 4 | 17.4 |
| HBx101 | 4 | 17.4 |
| HBx102 | 4 | 17.4 |
| HCC | ||
| HBx131 | 28 | 90.3 |
| HBx130 | 27 | 87.1 |
| HBx5 | 13 | 41.9 |
| HBx94 | 12 | 38.7 |
| HBx38 | 8 | 25.8 |
| HBx36 | 6 | 19.4 |
| HBx127 | 5 | 16.1 |
| HBx101 | 4 | 12.9 |
| HBx52 | 4 | 12.9 |
Table 3.
Incidence of double and triple HBx mutations by clinical diagnosis
| Subject age range (yr) | No. (%) of HBx mutationsa |
||||||||
|---|---|---|---|---|---|---|---|---|---|
| CHB (n = 42) |
LC (n = 23) |
HCC (n = 31) |
|||||||
| Total | Double | Triple | Total | Double | Triple | Total | Double | Triple | |
| 20–30 | 5 | 3 (60) | 0 (0) | 0 | 0 | ||||
| 31–40 | 10 | 4 (40) | 1 (10) | 1 | 0 (0) | 1 (100) | 1 | 1 (100) | 0 (0) |
| 41–50 | 15 | 5 (33.3) | 4 (26.7) | 9 | 5 (55.6) | 3 (33.3) | 7 | 3 (42.9) | 3 (42.9) |
| 51–60 | 10 | 8 (80) | 0 (0) | 11 | 4 (36.4) | 5 (45.5) | 14 | 6 (42.9) | 5 (35.7) |
| 61–70 | 2 | 2 (100) | 0 (0) | 2 | 1 (50) | 1 (50) | 9 | 4 (44.4) | 5 (55.6) |
CHB, chronic hepatitis B; LC, liver cirrhosis; HCC, hepatocellular carcinoma; Double, HBx130+HBx131 mutations; Triple, HBx5+HBx130+HBx131 mutations.
HBx mutations according to the status of HBV infection.
Patients had at least 1 and a maximum of 27 amino acid mutations. The frequency of all HBx mutations was found in 6.28 ± 4.27 (means ± the standard deviation [SD]). The incidence of each HBx mutation related to the clinical diagnosis of HBV infections is presented in Fig. 1. A total of 603 HBx mutations were evaluated: 250 in CHB patients (n = 42), 167 in LC patients (n = 23), and 186 in HCC patients (n = 31). The mean frequencies for HBx mutations according to clinical diagnosis were 5.95 ± 5.04 in CHB patients, 7.26 ± 4.7 in LC patients, and 6 ± 2.37 in HCC patients. Amino acid deletions were noted in one CHB patient (deletion from HBx129 to HBx134) and two HCC patients (deletion from HBx127 to HBx131; deletion from HBx128 to HBx132). In HCC patients, the most common HBx mutations were HBx130+HBx131 double mutations (71%), followed by HBx5+HBx130+HBx131 triple mutations (41.9%) and HBx94+HBx130+HBx131 triple mutations (35.5%). The number of patients with single mutations (HBx130, HBx131, HBx5, and HBx94) was very low or zero. The quadruple mutations (HBx5+HBx94+HBx130+HBx131) were observed in 12.9% of HCC patients. Interestingly, HBx5+HBx130+HBx131 triple mutations were significantly more common in HCC patients than in CHB patients. HBx mutations showed significant differences by clinical phases.
Fig. 1.
Percentage of HBx mutations according to clinical diagnosis of HBV infection. Abbreviations: CHB, chronic hepatitis B; LC, liver cirrhosis; HCC, hepatocellular carcinoma.
Risk assessment of double mutations and triple mutations.
Odds ratios were calculated based on the number of patients with double or triple mutations (Table 4). Generally, the odds ratio of HCC patients with triple mutations was higher than that of HCC patients with double mutations. The odds ratio was not statistically significant when HCC patients were compared to CHB patients, including LC patients. However, double and triple mutations were significant risk factors for HCC incidence compared to CHB patients only. Considering LC as a pre-HCC disease status, double and triple mutations were also significantly associated with HCC incidence.
Table 4.
Odds ratios for HCC incidence of double and triple mutations
| Comparisona | No. of mutations for various conditionsb |
Odds ratio |
|||||
|---|---|---|---|---|---|---|---|
| HCC | No HCC | HCC+LC | No HCC+LC | Mean | 95% CIc | P | |
| HCC vs. CHB | |||||||
| Double mutations | 27 | 27 | |||||
| No double mutations | 4 | 15 | |||||
| Odds ratio | 3.75 | 1.101–12.768 | 0.033 | ||||
| Triple mutations | 13 | 5 | |||||
| No triple mutations | 18 | 37 | |||||
| Odds ratio | 5.344 | 1.65–17.309 | 0.005 | ||||
| HCC+LC vs. CHB | |||||||
| Double mutations | 47 | 22 | |||||
| No double mutations | 7 | 15 | |||||
| Odds ratio | 4.578 | 1.634–12.825 | 0.003 | ||||
| Triple mutations | 23 | 5 | |||||
| No triple mutations | 31 | 37 | |||||
| Odds ratio | 5.49 | 1.867–16.142 | 0.001 | ||||
| HCC vs. CHB+LC | |||||||
| Double mutations | 27 | 47 | |||||
| No double mutations | 4 | 18 | |||||
| Odds ratio | 2.585 | 0.793–8.432 | 0.126 | ||||
| Triple mutations | 13 | 15 | |||||
| No triple mutations | 18 | 50 | |||||
| Odds ratio | 2.407 | 0.962–6.026 | 0.091 | ||||
Double mutations, HBx130+HBx131; triple mutations, HBx5+HBx130+HBx131.
CHB, chronic hepatitis B; LC, liver cirrhosis; HCC, hepatocellular carcinoma.
CI, confidence interval.
HBV genotype analysis with phylogenetic tree constructs.
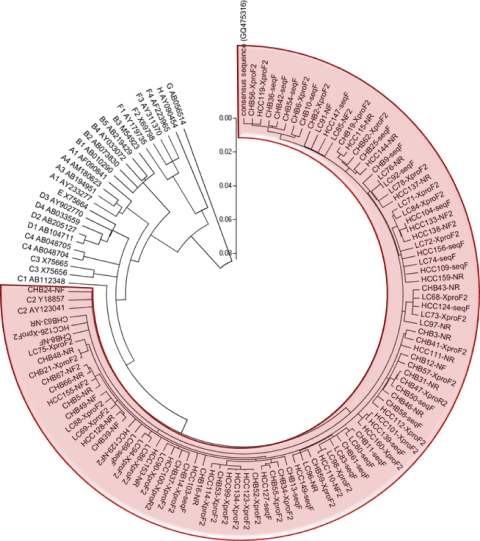
A neighbor-joining method was used to analyze HBV genotype. As expected, enrolled patients were determined as HBV genotype C2, which is known to be the most common genotype in Korea. A circular phylogenetic tree is presented in Fig. 2.
Fig. 2.
Phylogenetic tree of enrolled patients developed using the neighbor-joining method. Phylogenetic and molecular evolutionary analyses were performed using MEGA version 4.0.2 (The Biodesign Institute). The genetic distances were estimated by Kimura's two-parameter method. The reliability of the phylogenetic tree analysis was assessed by using the interior branch test with1,000 replicates. Abbreviations: CHB, chronic hepatitis B disease patient; LC, liver cirrhosis patient; HCC, hepatocellular carcinoma patient.
HBV DNA and viral protein assay of each construct.
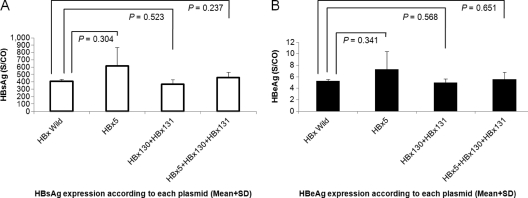
The total HBV DNA levels in cells transfected with HBx wild type and three mutant types were analyzed. There were no significant differences in HBV DNA levels between each HBx mutant and the HBx wild type (P > 0.05). HBV protein expression was analyzed by HBsAg and HBeAg assays in triplicate. A negative control yielded negative HBsAg and HBeAg results. There was no significant difference in the protein levels between the supernatants of the wild-type DNA transfected cells and the mutant DNA transfected cells (Fig. 3). Corrected levels of HBsAg and HBeAg were calculated according to the total protein concentration.
Fig. 3.
Protein expression of cells transiently transfected with wild-type and HBx mutant constructs. (A) HBsAg expression; (B) HBeAg expression.
NF-κB activity by luciferase reporter gene assay.
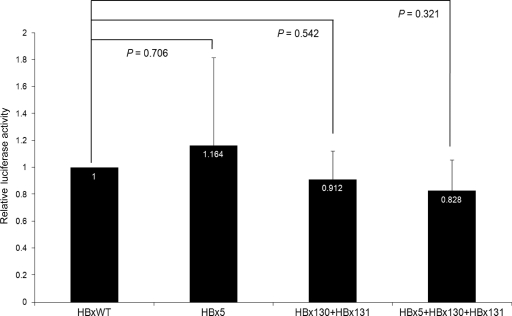
The luciferase activity was normalized by the activity of wild-type HBx-transfected cell lysates (relative luciferase activity). The results are expressed in Fig. 4 as means (bar) + the SD (line) of at least three experiments. There were significantly higher levels of NF-κB activity in cells with the HBx5 mutation and HBx130+HBx131 double mutations than in wild-type cells and those with the HBx5+HBx130+HBx131 triple mutations (Fig. 4). Cells with triple mutations did not show a significant difference in NF-κB activity compared to wild-type cells.
Fig. 4.
Relative luciferase assays showing the activity of NF-κB in wild-type and HBx mutant Huh7 cells.
DISCUSSION
HBx is known to be a multifunctional protein that not only activates transcriptional transactivation but also mediates cell growth via proliferation and apoptosis (5). Moreover, the HBx protein is potentially oncogenic via multistep carcinogenesis, modifies apoptosis, inhibits nucleotide excision, repairs damaged cellular DNA, and modulates transcriptional activation of cellular growth regulating genes (4).
To date, the clinical impact of HBx mutation has not been clearly defined. Specific regions of HBx mutations or deletions are reported to have an association with HBV pathogenesis in the sera (17, 28) or liver tissues (32) of CHB patients. However, the exact impact of specific HBx mutation on chronicity or carcinogenesis remains unclear.
We found that the incidence of HBx5 mutation was significantly higher in our HCC patients than in CHB patients. However, all patients with a HBx5 mutation had HBx130+HBx131 double mutations, too. The frequency and function of HBx5 mutation have not been determined in terms of the oncogenic potential. Further studies of the HBx5 mutation with or without HBx130+HBx131 double mutations are needed to understand the elucidate role of HBx5 mutation.
HBV DNA sequences differ between HBV genotypes or subgenotypes, so we believe that the HBx5 mutation might be characteristically related to genotype C2 in Korean HCC patients. Almost all Korean HBV genotypes are known to be HBV C2 (16) and, unfortunately, patients with HBV C2 genotype tend to rapidly progress to cirrhosis or HCC, and it can commonly cause complications in younger patients (12).
There have been several studies on the clinical correlations between HBx mutations and HCC. For example, Yeh et al. (29) reported that HBx codon 31 was detected more frequently in patients with HCC. Muroyama et al. (21) reported that an HBx codon 38 change in genotype C is an independent risk factor for HCC development. Shinkai et al. (26) reported that, regardless of HBeAg status, the T1653 mutation increases the risk of HCC in Japanese patients with HBV/C2. Choi et al. (6) reported that only the B1499 (C or G or T1499) mutation was significantly associated with HCC. However, there has been no established consensus on or mechanism of which HBx mutation or deletion directly affects the pathogenesis of HCC. This might be attributed to differences in HBV genotype, the level of HBV DNA, coinfection of other viruses, and effects of host factors.
Our study showed that double mutations of HBV at nucleotide 1762 (A→T) and 1764 (G→A) in the basal core promoter (BCP) region were the most common mutation types in HCC patients. Because double mutations in the BCP region also exist in the coding sequence of HBV X gene, these mutations convert K to M at position 130 and V to I at position 131 in the overlapping X-open reading frame gene product. To date, these double mutations have been suggested to arrest the transcription of the precore RNA but without a serious impact on the transcription of the pregenomic RNA (23). Cell transfection experiments on the T1762/A1764 double mutations showed conflicting results regarding the increase in viral replication (9). However, double mutations in the BCP region were suggested to be associated with HCC development (7, 10, 13), and this finding has been supported by a prospective cohort study (8).
CHB patients without cirrhosis but with HBx5+HBx130+HBx131 triple mutations showed a significant risk for HCC development. Also, CHB patients with HBx5+HBx130+HBx131 triple mutations showed a somewhat higher risk for HCC than those with HBx130+HBx131 double mutations.
The fifth amino acid of HBx is located in the negative regulatory region, which is known as a dispensable region for transactivation. HBx5 mutation cell line experiments showed no differences in viral replication activity whether they had double mutations or the HBx5 mutation. However, the relative transcriptional activity of NF-κB was significantly higher in patients with an HBx5 single mutation and HBx130+HBx131 double mutations than in patients with wild-type HBx and in those with HBx5+HBx130+HBx131 triple mutations. This might suggest that NF-κB was not significantly activated by triple mutations in HCC patients. Nevertheless, both the HBx5 mutation and the HBx130+HBx131 double mutation could activate the NF-κB signaling pathway. This phenomenon implies that HBx5 mutation does not directly influence HBV replication but might be associated with modulation of the cellular signaling pathway. In addition, NF-κB activity was characteristically affected by the existence of HBx5 mutation. In fact, HBx5 mutation was always found with double mutations in all clinical phases of HBV infection. This also implies that an independent outbreak of HBx5 mutation may burden HBV life cycle and may not be affordable for viral immune escape from the host defense system. However, HBx5+HBx130+HBx131 triple mutations did not increase NF-κB activity. We found through repeated experiments that the NF-κB activity in cells with triple mutations was almost the same as in cells with wild-type HBV. This result suggested that a specific HBx mutation might be related not only to the major NF-κB signaling pathway but also to other signaling pathways. Triple mutations seem to burden the life cycle of HBV and be associated with tumorigenesis. A triple mutation might also change the HBx protein structure, possibly affecting NF-κB activity. The present study focused on the NF-κB signaling pathway because it seems to have a central function in liver homeostasis, pathophysiology, and regulation of the inflammation-fibrosis-cancer axis (27). NF-κB also plays a role in the development of HCC (14). HBV and HBx have been known to activate NF-κB (22, 31), but it is unclear whether specific HBx mutation can affect NF-κB activity. So, we focused on determining whether a significant HBx mutation can affect NF-κB activity. Human NF-κB that regulates a number of oncogenic genes is associated with tumorigenesis in mammalian cells through the regulation of a number of oncogenic genes (1). Our findings suggest that a specific HBx mutation could modulate the NF-κB signaling pathway, which is possibly associated with tumorigenesis. The HBx5 mutation may enhance the oncogenic potential to progress from a chronic liver disease to HCC.
In conclusion, the HBx130+HBx131 double mutation was the most common HBx mutation occurring in HCC patients with chronic HBV infection, followed by the HBx5+HBx130+HBx131 triple mutation. The HBx5 mutation was more frequently observed in HCC patients than CHB patients. An HBV construct containing wild-type HBx and each of three types of HBx mutation was transiently transfected, and the effect of HBx mutation on the viral replication, HBV protein expression, and NF-κB activity was observed. As a result, the HBx5 single mutation, HBx130+HBx131 double mutations, and HBx5+HBx130+HBx131 triple mutations were not directly associated with either viral protein expression or viral replication activity. However, the HBx5 mutation and HBx130+HBx131 double mutations appear to modulate the transcriptional transactivating function of the NF-κB signaling pathway.
We showed here that the HBx5 single mutation and HBx130+HBx131 double mutation increased NF-κB activity and that the HBx5+HBx130+HBx131 triple mutation did not increase NF-κB activity. Thus, there seems to be another pathway that regulates NF-κB activation by HBx mutation. Also, older HCC patients were more likely to have the HBx130+HBx131 double mutations and HBx5+HBx130+HBx131 triple mutations than younger HCC patients. Therefore, patient age should be considered as a factor affecting the development of HCC. The HBx5 mutation in genotype C2 may be a risk factor for HCC development and appears to serve as a potential molecular marker for predicting the clinical outcomes of patients with chronic HBV infection.
ACKNOWLEDGMENTS
The 1.2-mer HBV construct within the pGEM4Z vector was kindly provided by Kyun Hwan Kim (Department of Pharmacology, School of Medicine, Konkuk University, Seoul, Korea).
This study was partially supported by grant A100117-1001-0000100 from the Korea Healthcare Technology R&D Project, Ministry of Health and Welfare, Seoul, Republic of Korea.
Footnotes
Published ahead of print on 13 April 2011.
REFERENCES
- 1. Aggarwal B. B. 2004. Nuclear factor-κB: the enemy within. Cancer Cell 6:203–208 [DOI] [PubMed] [Google Scholar]
- 2. Beasley R. P., Hwang L. Y., Lin C. C., Chien C. S. 1981. Hepatocellular carcinoma and hepatitis B virus: a prospective study of 22,707 men in Taiwan. Lancet ii:1129–1133 [DOI] [PubMed] [Google Scholar]
- 3. Billet O., et al. 1995. In vivo activity of the hepatitis B virus core promoter: tissue specificity and temporal regulation. J. Virol. 69:5912–5916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Birrer R. B., Birrer D., Klavins J. V. 2003. Hepatocellular carcinoma and hepatitis virus. Ann. Clin. Lab. Sci. 33:39–54 [PubMed] [Google Scholar]
- 5. Bouchard M. J., Schneider R. J. 2004. The enigmatic X gene of hepatitis B virus. J. Virol. 78:12725–12734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Choi C. S., et al. 2009. X gene mutations in hepatitis B patients with cirrhosis, with and without hepatocellular carcinoma. J. Med. Virol. 81:1721–1725 [DOI] [PubMed] [Google Scholar]
- 7. Chou Y. C., et al. 2008. Temporal relationship between hepatitis B virus enhancer II/basal core promoter sequence variation and risk of hepatocellular carcinoma. Gut 57:91–97 [DOI] [PubMed] [Google Scholar]
- 8. Fang Z. L., et al. 2008. HBV A1762T, G1764A mutations are a valuable biomarker for identifying a subset of male HBsAg carriers at extremely high risk of hepatocellular carcinoma: a prospective study. Am. J. Gastroenterol. 103:2254–2262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Fang Z. L., et al. 2009. The association of HBV core promoter double mutations (A1762T and G1764A) with viral load differs between HBeAg positive and anti-HBe positive individuals: a longitudinal analysis. J. Hepatol. 50:273–280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Fang Z. L., et al. 2002. Core promoter mutations [A(1762)T and G(1764)A] and viral genotype in chronic hepatitis B and hepatocellular carcinoma in Guangxi, China. J. Med. Virol. 68:33–40 [DOI] [PubMed] [Google Scholar]
- 11. Gottlob K., Pagano S., Levrero M., Graessmann A. 1998. Hepatitis B virus X protein transcription activation domains are neither required nor sufficient for cell transformation. Cancer Res. 58:3566–3570 [PubMed] [Google Scholar]
- 12. Kao J. H., Chen P. J., Lai M. Y., Chen D. S. 2000. Hepatitis B genotypes correlate with clinical outcomes in patients with chronic hepatitis B. Gastroenterology 118:554–559 [DOI] [PubMed] [Google Scholar]
- 13. Kao J. H., Chen P. J., Lai M. Y., Chen D. S. 2003. Basal core promoter mutations of hepatitis B virus increase the risk of hepatocellular carcinoma in hepatitis B carriers. Gastroenterology 124:327–334 [DOI] [PubMed] [Google Scholar]
- 14. Karin M. 2006. Nuclear factor-κB in cancer development and progression. Nature 441:431–436 [DOI] [PubMed] [Google Scholar]
- 15. Kim C. M., Koike K., Saito I., Miyamura T., Jay G. 1991. HBx gene of hepatitis B virus induces liver cancer in transgenic mice. Nature 351:317–320 [DOI] [PubMed] [Google Scholar]
- 16. Kim H., et al. 2007. Molecular epidemiology of hepatitis B virus (HBV) genotypes and serotypes in patients with chronic HBV infection in Korea. Intervirology 50:52–57 [DOI] [PubMed] [Google Scholar]
- 17. Kim H. J., et al. 2008. Hepatitis B virus X mutations occurring naturally associated with clinical severity of liver disease among Korean patients with chronic genotype C infection. J. Med. Virol. 80:1337–1343 [DOI] [PubMed] [Google Scholar]
- 18. Lavanchy D. 2004. Hepatitis B virus epidemiology, disease burden, treatment, and current and emerging prevention and control measures. J. Viral Hepat. 11:97–107 [DOI] [PubMed] [Google Scholar]
- 19. Liu S., et al. 2009. Associations between hepatitis B virus mutations and the risk of hepatocellular carcinoma: a meta-analysis. J. Natl. Cancer Inst. 101:1066–1082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. McMahon B. J. 2009. The natural history of chronic hepatitis B virus infection. Hepatology 49:S45–S55 [DOI] [PubMed] [Google Scholar]
- 21. Muroyama R., et al. 2006. Nucleotide change of codon 38 in the X gene of hepatitis B virus genotype C is associated with an increased risk of hepatocellular carcinoma. J. Hepatol. 45:805–812 [DOI] [PubMed] [Google Scholar]
- 22. Ohata K., et al. 2003. Interferon alpha inhibits the nuclear factor κB activation triggered by X gene product of hepatitis B virus in human hepatoma cells. FEBS Lett. 553:304–308 [DOI] [PubMed] [Google Scholar]
- 23. Okamoto H., et al. 1994. Hepatitis B virus with mutations in the core promoter for an e antigen-negative phenotype in carriers with antibody to e antigen. J. Virol. 68:8102–8110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Perfumo S., et al. 1992. Recognition efficiency of the hepatitis B virus polyadenylation signals is tissue specific in transgenic mice. J. Virol. 66:6819–6823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Schaefer S., et al. 1998. Properties of tumour suppressor p53 in murine hepatocyte lines transformed by hepatitis B virus X protein. J. Gen. Virol. 79(Pt. 4):767–777 [DOI] [PubMed] [Google Scholar]
- 26. Shinkai N., et al. 2007. Influence of hepatitis B virus X and core promoter mutations on hepatocellular carcinoma among patients infected with subgenotype C2. J. Clin. Microbiol. 45:3191–3197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Sun B., Karin M. 2008. NF-κB signaling, liver disease and hepatoprotective agents. Oncogene 27:6228–6244 [DOI] [PubMed] [Google Scholar]
- 28. Tanaka Y., et al. 2006. Specific mutations in enhancer II/core promoter of hepatitis B virus subgenotypes C1/C2 increase the risk of hepatocellular carcinoma. J. Hepatol. 45:646–653 [DOI] [PubMed] [Google Scholar]
- 29. Yeh C. T., Shen C. H., Tai D. I., Chu C. M., Liaw Y. F. 2000. Identification and characterization of a prevalent hepatitis B virus X protein mutant in Taiwanese patients with hepatocellular carcinoma. Oncogene 19:5213–5220 [DOI] [PubMed] [Google Scholar]
- 30. Yu D. Y., et al. 1999. Incidence of hepatocellular carcinoma in transgenic mice expressing the hepatitis B virus X-protein. J. Hepatol. 31:123–132 [DOI] [PubMed] [Google Scholar]
- 31. Yun C., et al. 2002. NF-κB activation by hepatitis B virus X (HBx) protein shifts the cellular fate toward survival. Cancer Lett. 184:97–104 [DOI] [PubMed] [Google Scholar]
- 32. Zhang H., et al. 2008. Identification of a natural mutant of HBV X protein truncated 27 amino acids at the COOH terminal and its effect on liver cell proliferation. Acta Pharmacol. Sin. 29:473–480 [DOI] [PubMed] [Google Scholar]