Abstract
Summary: Atomic force microscopy (AFM) can visualize almost everything pertinent to structural virology and at resolutions that approach those for electron microscopy (EM). Membranes have been identified, RNA and DNA have been visualized, and large protein assemblies have been resolved into component substructures. Capsids of icosahedral viruses and the icosahedral capsids of enveloped viruses have been seen at high resolution, in some cases sufficiently high to deduce the arrangement of proteins in the capsomeres as well as the triangulation number (T). Viruses have been recorded budding from infected cells and suffering the consequences of a variety of stresses. Mutant viruses have been examined and phenotypes described. Unusual structural features have appeared, and the unexpectedly great amount of structural nonconformity within populations of particles has been documented. Samples may be imaged in air or in fluids (including culture medium or buffer), in situ on cell surfaces, or after histological procedures. AFM is nonintrusive and nondestructive, and it can be applied to soft biological samples, particularly when the tapping mode is employed. In principle, only a single cell or virion need be imaged to learn of its structure, though normally images of as many as is practical are collected. While lateral resolution, limited by the width of the cantilever tip, is a few nanometers, height resolution is exceptional, at approximately 0.5 nm. AFM produces three-dimensional, topological images that accurately depict the surface features of the virus or cell under study. The images resemble common light photographic images and require little interpretation. The structures of viruses observed by AFM are consistent with models derived by X-ray crystallography and cryo-EM.
INTRODUCTION
There are currently two principal techniques in common use for the direct visualization of virus structure, and these are X-ray diffraction analysis and electron microscopy (EM). X-ray diffraction from single crystals is extraordinary in the resolution of structural detail that it can reveal, approaching the atomic level. In many cases intimate details of the capsid architecture, as well as, in some cases, the protein-nucleic acid interactions, become clear with X-ray crystallography. It is unsurpassed in this regard, and it is unlikely ever to be challenged. It will remain the technique of choice, whenever it can be applied.
X-RAY DIFFRACTION AND ELECTRON MICROSCOPY
X-ray diffraction as applied to viruses does, however, have some limitations. First, it requires that the virus be crystallized. This is possible only for viruses having very regular shapes and uniform exterior surfaces, i.e., icosahedral viruses or, conceivably, some virus-like particles (VLP) produced in recombinant systems or reconstructed in vitro. Thus, most complex viruses, incorporating membranes or exhibiting structurally complicated external features such as tail assemblies or fibers, are excluded from X-ray analysis. In addition, viruses beyond a certain size limit, probably about 1,000 Å in diameter, exceed our current technologies for recording X-ray data from very large unit cells. Finally, interior components of viruses, especially encapsidated nucleic acid, generally do not share the external symmetry of the particles. Therefore, they are not usually seen in electron density maps. Those components are orientationally randomized in the crystallization process. X-ray diffraction is an “averaging method” that does not reveal the characteristics of individual particles. It assumes that all particles are exactly the same, or it makes them so.
In addition to allowing determination of the structures of intact, symmetrical viruses, X-ray diffraction can be useful in another way. It can also be used to determine, very precisely, the structures of individual proteins, and sometimes complexes of proteins, that make up viral substructures. These can then be fitted to lower-resolution models of the particles obtained by other means (electron microscopy, for example), and a “pseudo”-high-resolution structure of a virus can be created (45, 46). This has proven successful in a number of cases. X-ray diffraction analysis is a powerful approach, and can be made even more powerful when it is combined with information from other sources, as it has been with electron microscopy and potentially can be with atomic force microscopy (AFM).
Electron microscopy (EM) has been applied to the analysis of virus structure along two principal lines. Transmission EM of viruses spread on substrates, such as carbon-coated grids (13, 14), or of virus-infected cells after thin sectioning is one method. Reconstruction of particles obtained by acquisition of many low-dose images from cryo-electron microscopy (cryo-EM) is the other. While the former once predominated, in more recent times cryo-EM has certainly proven to be the more powerful for elucidating virus architecture (4) and has been shown to be capable of revealing detail at resolutions in the range of 3.5 to 4 Å. In some cases it has even permitted the tracing of a polypeptide chain.
Transmission EM is, however, problematic because it generally requires heavy metal staining or shadowing to increase contrast. It usually requires complete dehydration, and it often involves fixatives as well. Finally, it produces projections of entire particles onto the substrate plane. Elimination of one dimension obscures spatial relationships among substructures, results in loss of chirality, creates artifacts, and produces images that are frequently open to multiple interpretations. In the past decade, cryo-electron microscopy has been made increasingly powerful by automation in image collection and cataloging and by elaborate mathematical techniques for analyzing the low-dose images and reconstructing viruses from collections of particles observed in multiple orientations. The major advantage of cryo-EM over X-ray diffraction, of course, is that it does not require that the virus be crystallized.
Cryo-EM (1), however, also has limitations. Like X-ray diffraction, it is useful chiefly for analyzing regular viruses having icosahedral symmetry. It produces the “average” structure for the entire virus population and does not illuminate eccentricities of individuals. More advanced applications of cryo-EM (7) can deal with particles lacking any symmetry (ribosomes, for example), but even here the assumption is that all of the particles are identical. It is ineffective when the viruses are pleiomorphic and lack architectural uniformity. Cryo-EM is also likely to be technically limited in its application to very large viruses, even icosahedral viruses such as iridoviruses, because of the physical thickness of the particles and the difficulty of simply penetrating their bulk. Just as the power of X-ray diffraction has been amplified by its combination with cryo-EM, the converse is equally true. It is important to recognize, however, that even when combined, the pictures these techniques yield, for the same reasons as for X-ray crystallography alone, can still often be incomplete, particularly regarding the interior structural features of the viruses, and both can produce only static images of an averaged structure.
A third, direct imaging technology that promises to have a significant impact on structural biology and which is, in most ways, complementary to X-ray diffraction and electron microscopy is atomic force microscopy (AFM). An immediate advantage of AFM is that it is based on relatively simple physical principles, unlike X-ray crystallography, and AFM instruments are mechanically and electronically rather straightforward, unlike those used for electron microscopy. Unlike both of the other technologies, AFM is fairly inexpensive to institute and apply, even to biological specimens. The microscope itself is a small device, which is about the size of a common household coffee pot.
HOW DOES AFM WORK?
Atomic force microscopy was invented in the mid-1980s (5). The instrument is one of a family of scanning probe microscopes that had such a significant impact on imaging and surface science that the original incarnation, the scanning tunneling microscope, won Binnig a share of the 1996 Nobel Prize in Physics. For several years AFM was used almost exclusively to characterize the surfaces of nonbiological materials, and even now its chief applications are in the visualization of microcircuits on silicon chips and in nanotechnology. Its principle is remarkably simple, surprisingly so given its acuity, but is made useful only by sophisticated electronic image processing and particularly piezoelectric technology. Its application to biological materials and systems has been rather slow to develop (2, 6). This is curious because it can be used in physiological fluids as well as air, and its noninvasive and nonintrusive features make it an ideal tool for the study of sensitive, living systems. The chief difficulty with its use on biological materials is that they tend to be very soft. AFM is at its best on hard surfaces.
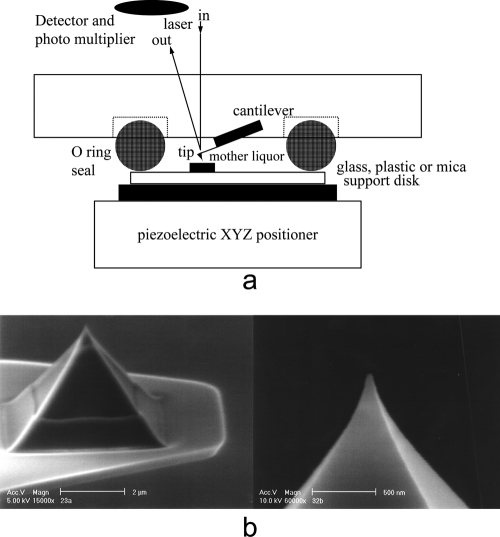
An AFM device and the principles by which it operates are shown schematically in Fig. 1a. AFM instruments can be operated in either contact mode or what is referred to as tapping mode. In contact mode, a probe made of silicon or silicon nitride is placed in near contact with the surface of interest, say the capsid of a virus, and then translated in a systematic raster mode over the surface (or substrate). The AFM probe is a sharp stylus, like those pictured in Fig. 1b, similar in function to a minute phonograph needle. The tip ideally has a single point, with a very small radius of curvature. The probe is mounted at the end of a short cantilever, typically 100 to 250 μm in length, which has a low spring constant to minimize the force between the tip and the sample.
Fig. 1.
(a) Schematic drawing illustrating the principles of an atomic force microscope. The vertical deflection that the cantilever tip experiences upon encountering some topological feature on a specimen is amplified through a reflected laser beam, which is tracked and reported by a split-diode photoelectric detector. Scanning takes place in a fluid-filled (or dry if preferred) cell of about 75 μl in volume. The sample is translated in a raster manner by piezoelectric positioners upon which the fluid cell is mounted. (b) Scanning electron micrographs of high-quality AFM tips etched from silicon.
Scanning is achieved by translating the sample beneath the probe, using a piezoelectric crystal-positioned x-y stage, along a continuous sequence of raster lines. As the probe tip passes over the surface, it does not actually touch the structural feature on the surface in a physical sense but interacts through “aggregate atomic forces,” which remain somewhat mysterious. Encounters with substructures cause the probe to be displaced vertically as the tip rides across. Exceedingly small displacements of the tip are amplified by deflection of a laser beam that is reflected from the upper surface of the cantilever, and these deflections are detected and tracked by a split photodiode. Photoelectric circuitry converts the deflections into height information. The resulting scan data, recorded as a digital topographical image, can then be presented in a number of visual formats.
In contact mode of operation, the data may be acquired as either “height” or “deflection” information, or the data from both modes may be obtained simultaneously. In “height” mode the sample surface is maintained at a constant distance from the probe tip by the piezoelectric positioner below, through a feedback mechanism. The cantilever deflection in this case is very small. In “deflection” mode the sample is stationary and actual cantilever deflection data are collected. Microfabricated cantilevers exert a force on the substrate surface of about 10−9 to 10−12 N/m, and, as might be anticipated, the resolution of the technique depends on the degree of force employed. Up to a point, the greater the force between probe and surface, the more sensitive the probe is to surface variations. On the other hand, the greater the force, the more the probe may perturb the surface.
Sample perturbation and other problems arising from unfavorable probe-surface interactions have been obviated to a great extent by the development of “tapping” mode instruments (12). With the tapping mode, the probe tip is not in continuous contact with the sample surface but rapidly oscillates up and down as it is scanned over the surface, essentially “tapping” its way and gently sensing the heights of obstacles it encounters. In tapping mode, the vertical position of the sample is continually adjusted by a feedback mechanism to maintain the amplitude of the freely oscillating probe constant. Image acquisition times range from 0.5 to 4 min, with shorter scan times usually associated with greater tip-specimen interaction.
The tapping mode minimizes contact between the probe tip and the sample surface and greatly reduces lateral forces. An even more sensitive means of scanning in tapping mode is called phase modulation scanning. Here, phase changes are introduced into the tip oscillations due not only to height differences but also to changes in the nature of the aggregate atomic interactions, caused in turn by variations in the physical or chemical properties of the sample surface. This approach has been shown to be useful for imaging very thin and delicate materials such as biological membranes (43).
The “tapping mode” approach has been a significant boon in biological investigations, as it has allowed the characterization of samples that would otherwise be too soft or too fragile to withstand contact mode examination. Operating with the tapping mode in a liquid environment presents some complications due to fluid dynamics, but these are not severe. A constraint that sometimes presents obstacles during analysis in a liquid medium is that the specimen under study must be fixed to or made to adhere firmly to the substrate surface of the fluid cell, which may be glass, cleaved mica, plastic, or any other hard material. To achieve this, it may be necessary to treat the substrate with various reagents, such as poly-l-lysine, to induce adhesion of samples. If this condition is not met, then the specimen can move due to interaction with the probe, and no useful information is gathered.
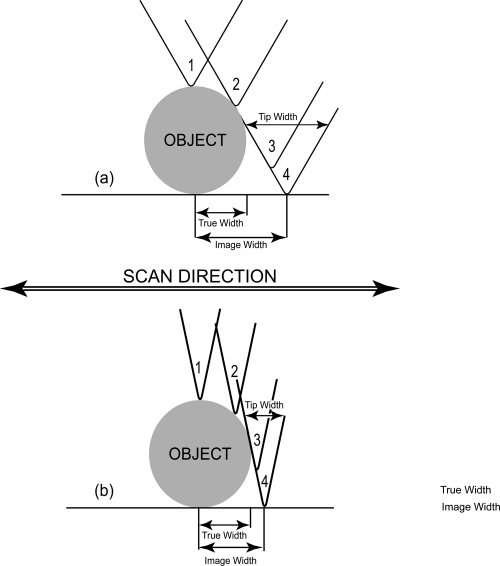
A particular feature of AFM must be borne in mind whenever images are interpreted. The one- or two-dimensional trace obtained for any object or surface substructure is the convolution of the tip shape with that of the feature being scanned. This is illustrated in Fig. 2. An image of an object scanned with a broad, dull tip is not the same as that acquired with a sharper tip. In particular, while the height of the object will be the same regardless of the tip shape (because the maximum vertical deflection of the cantilever tip would be the same), the lateral dimensions will not. The broader tip yields a broader object, and the sharper tip produces a more accurate size. Because in general, the tip shape being utilized at the time is not known, the image cannot be easily deconvoluted to provide the true dimensions. Hence, height information is almost always trustworthy, but lateral measurements are suspect. The reliability of lateral measurements can, however, be increased if some standard having defined spatial features is first scanned and its known spacings or cell dimensions compared with those in the image. Such standards may be etched grids on silicon or the surfaces of protein crystals (28).
Fig. 2.
Schematic illustration of the convolution of the shape of the AFM tip with the shape of the feature or particle being scanned. The side of the cantilever tip contacts the object and begins to produce a deflection of the cantilever before the tip apex actually reaches the object. Similarly, the opposite side of the tip is still in contact with the object even after the apex itself has passed. Thus, the total deflection implies a virtual lateral dimension for the object that is greater than its actual dimension. The difference between the virtual and actual dimensions is a function of the width of the cantilever tip. The sharper the tip, the more accurate the observed dimensions and the greater the resolution attainable.
The areas of scanning fields may range from 20 nm2 to 150 μm2, with vertical resolutions on biological samples of a fraction of a nanometer. Thus, this method provides precise topographical detail over a size range that eludes most other techniques. Lateral resolution varies depending on the prominence of features and the deformability of the specimen. For small isolated samples on mica, such as macromolecular assemblies and single virus particles, the resolution is most limited by the sharpness and structure of the tip. Commercially available tips have radii of curvature in the 5- to 20-nm range and provide resolution at fractions of those dimensions. For regular arrays of identical molecules and very small height differences, lateral resolution of a few nanometers can be achieved by imaging with small tip asperities. AFM application extends over the range of individual macromolecules, which are accessible by X-ray crystallography, to macromolecular assemblies amenable to electron microscopy, to living cells, which can just be seen using light microscopy.
On large soft samples, such as living animal cells (25), lateral resolution may be more limited by the motion and deformation of the cell surface in response to tip pressure than by tip structure or sharpness. Because visualization can be carried out in a fluid environment, specimens may suffer no dehydration as is generally the case with electron microscopy, and they require no staining. Indeed, specimens can be observed over long periods, as long as they stay relatively unchanged and immobilized during a single frame interval. For the most part, even living cells seem oblivious to the presence of the probe tip (25).
Specimens, however, are not always best visualized under strict physiological conditions, particularly when high resolution is desired. Because cantilever tip pressure, even in “tapping mode,” may produce deformation (for example, of a cell membrane), in some cases fixation is the better option. As with light microscopy histological procedures, this usually relies on glutaraldehyde, paraformaldehyde, or osmium tetroxide fixation, followed by dehydration and imaging in water-alcohol mixtures or in air. These methods have been developed by microscopists for more than a century to preserve the natural morphology of a sample but still allow high-resolution imaging. The methods remain applicable with AFM. While not as ideal as in situ observation, as the cells of course are no longer alive or viruses infective, fine details of their structures can be visualized that would otherwise be obscured by membrane flexion.
Atomic force microscopy (AFM) has both virtues and limitations, but as already noted, they tend to be complementary to those of X-ray diffraction and cryo-EM. The resolution of AFM, in the best of cases, is roughly that of current cryo-EM models (4), and like EM techniques, it does not require that the virus be crystallized. It is applied to individual particles and does not yield an average structure over an entire population. It does not require that the virus have a symmetrical or uniform architecture or even that all particles be the same in structure. Thus, it is equally applicable to small icosahedral viruses such as tomato bushy stunt virus, to helical viruses such as tobacco mosaic virus, and to completely irregular, complex viruses like vaccinia virus or the retroviruses. There is no size restriction. It has been used to analyze small plant viruses such as turnip yellow mosaic virus (TYMV) (26) and satellite tobacco mosaic virus (20), massive icosahedral viruses such as the algal virus Paramecium bursaria Chlorella virus 1 (PBCV-1) (21), and mimivirus (30, 48), the largest virus known.
We must note at the outset that there are other applications of AFM to the study of viruses in addition to their visualization, to which we have restricted the scope of this review. In particular, AFM has been utilized to measure mechanical and material properties of particles (11, 15, 41), and these of course often have structural implications. This field has recently been the subject of an excellent review (3), which is recommended as a complement to the one presented here.
VISUALIZATION OF VIRUSES BY AFM
Viruses were first visualized by AFM in their crystalline form, rather than as single isolated particles, in an investigation of the growth of orthorhombic crystals of satellite tobacco mosaic virus (35). Because they were immobilized on the surfaces of crystals, conditions were suitable for direct imaging of even these small, 17-nm-diameter virions. Larger icosahedral plant viruses in crystalline form were studied subsequently (24, 34, 37). The first AFM studies of noncrystalline viruses were of retroviruses on cell surfaces (19, 29), again principally because they were immobilized by their association with cell surfaces. Single particles of larger viruses, and helical viruses, were eventually visualized by AFM, and these included tobacco mosaic virus, cauliflower mosaic virus, Tipula iridescent virus (24), herpes simplex virus (44), vaccinia virus (17, 36), and the largest virus of all, mimivirus (30, 48). Although virus crystals were investigated using both the contact and tapping modes, noncrystalline specimens were imaged exclusively with the tapping mode, both in air and in buffer.
Because AFM images the surfaces of specimens, it might be thought that AFM would be of little use in visualizing the interior features of viruses or cells. This, however, is not the case. As has been shown in AFM investigations of a number of viruses, it is, in fact, an invaluable tool for deducing the interior architecture of virions, regardless of their external form or size. This is because it is possible to systematically strip away layers of structure by chemical, physical, and enzymatic means (16) and to accompany this process of dissection by AFM visualization. Using the same strategy as used by conventional anatomists, it has proven possible to disassemble viral specimens, see what is inside, and ascertain how the components are linked.
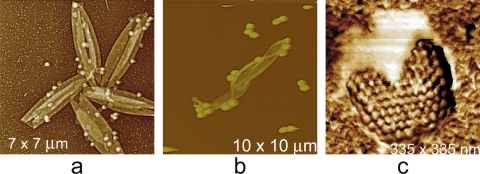
A valuable qualitative result that emerges almost immediately from AFM images is what the virus looks like, its overall architecture, and how similar particles are to one another. Are they uniformly the same in appearance, or are they present in a variety of forms? Thus, even a cursory investigation may quickly reveal certain general features that allow rapid classification. This is illustrated by the various structural classes of viruses shown in Fig. 3 and 4. The virions may be spherical, cylindrical, or filamentous. They may have symmetrically arranged capsomeres or other surface units, fibers, protruding vertices, prolate or icosahedral shapes, unusual morphologies, pleiomorphic character, etc. Tail assemblies may be observed directly, as on phages, for example. AFM is therefore a useful tool for simply deducing the kind of virus with which one is dealing, whether more than one kind of virus is present in a population, and the general level of contamination that may accompany the virus as a consequence (cellular material, degraded virions, and macromolecular impurities of all sorts).
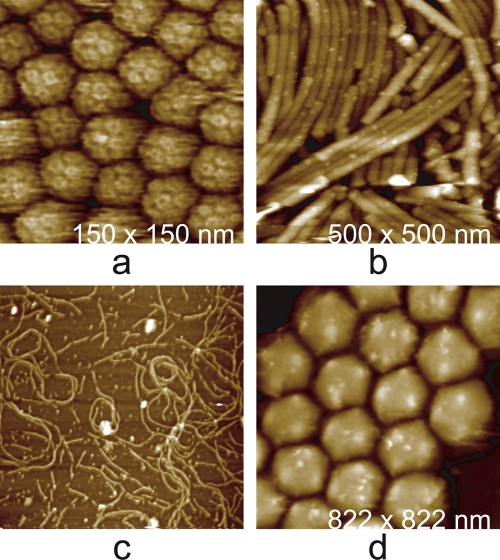
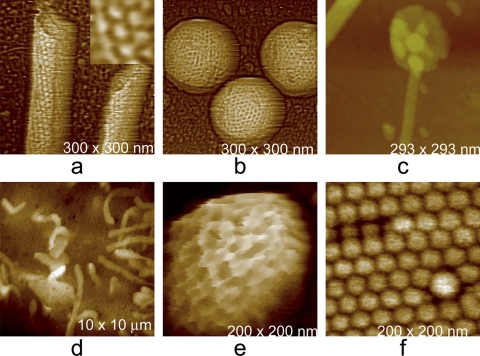
Fig. 3.
AFM images. (a) Condensed mass of brome mosaic virus (BMV), a T = 3 icosahedral virus that infects grasses such as barley. (b) Helical, rod-shaped tobacco mosaic virus (TMV), a ubiquitous pathogen throughout the plant world. (c) Tangles of marine filamentous bacteriophage and their broken fragments scattered on the AFM substrate. (d) Virions of Tipula iridescent virus, a very large icosahedral virus that infects insects. The virions of BMV have a diameter of 30 nm, TMV is about 20 nm in diameter and1,000 nm in length, and the adenovirus and iridovirus have diameters of about 100 nm and 200 nm, respectively.
Fig. 4.
AFM images. (a) A single virion of Moloney murine leukemia virus, an animal retrovirus having a diameter of about 150 nm. Clusters of envelope protein are clearly evident on its surface. (b) Surface of a virion of vaccinia virus, whose largest dimension is 250 to 300 nm. (c) A bacteriophage that infects cyanobacteria from marine environments. The icosahedral structure of the capsid and the distribution of its capsomeres are just becoming evident at this magnification. The components of the tail assembly are also clearly defined. (d) Mimivirus, the largest virus known in nature; its surface is covered by a forest of fine protein fibers with attached oligosaccharide.
QUANTITATIVE PROPERTIES OF VIRUSES
A fundamental parameter for virus particles is their diameter if they are spherical viruses or their diameter and length if they are helical. AFM can provide measures of these in both the hydrated and dried states, which also gives an estimate of the degree of shrinkage they undergo as a result of dehydration. Because of the finite tip size and tip-to-tip variation in radius of curvature, it is risky to measure linear dimensions directly by AFM. This was discussed above. It is, however, safe to measure the heights of objects above the substrate plane and the distances between the points of maximum elevation (e.g., capsomere to capsomere) on particles or center-to-center distances (e.g., particles in a crystal or in a cluster). This last approach can be applied to noncrystalline or paracrystalline arrays of viruses, including helical, rod-shaped, and spherical viruses.
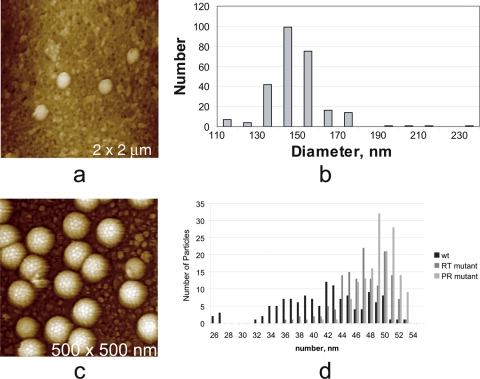
As has been emphasized already, for spherical and cylindrically symmetric particles, measurements of particle heights above the substrate plane yield accurate values for their diameters, and individual measurements are usually accompanied by rather modest error, generally on the order of 5% or less. By repeating measurements for a number of particles in the field and using different scan directions (which is facile for AFM), good statistics can be obtained and histograms of size distributions compiled. Precision of a few angstroms is possible. Histograms of particle sizes are often informative. If the distribution is a simple Gaussian one, then it can be presumed that particles of only one general morphology or icosahedra of only one triangulation number are present but that their diameters vary to some degree about the mean, perhaps due to the physiological state or degree of maturation. In Fig. 5a and b is an example where this approach was quite successful for Moloney murine leukemia virus (MuLV) (18, 23). If, on the other hand, a more complex distribution is observed, such as one having multiple peaks and shoulders, then particles of separate classes may be present. From such a histogram analysis (Fig. 5d), it was deduced that Ty3 retrotransposon particles seen in Fig. 5c existed as icosahedra having triangulation numbers 3, 4, and 7 (23, 31).
Fig. 5.
(a and b) Histogram (b) of diameters, determined solely by height measurements, taken from over 250 virions of Moloney murine leukemia virus that were still attached to the surfaces of their 3T3 host cells (a). The very small and very large particles at the extremities of the distribution are not due to errors in measurements, which are at most a few nanometers, but represent true deviations of particle sizes from the norm. (c and d) Histogram (d) of similarly measured diameters of the retrotransposon Ty3 from yeast (c). A careful analysis of the data revealed the presence of T = 7, 4, and 3 icosahedral particles making up the population. These had respective diameters of 155 nm, 140 nm, and 125 nm.
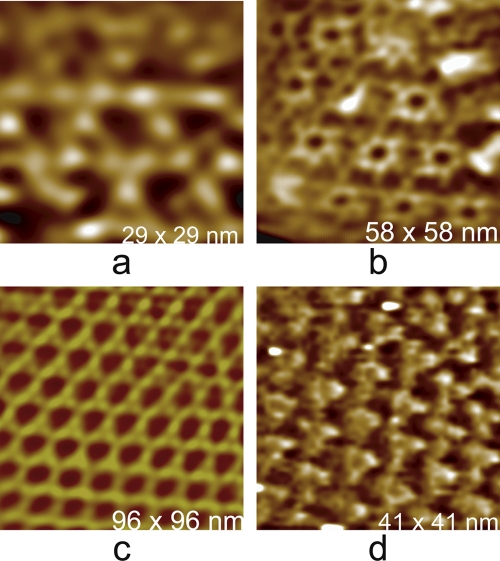
Even if crystals cannot be grown, if the virions can simply be closely packed so that there are few spaces between them, then center-to-center distances may be quite adequate. If the virions, be they spherical or helical, can be crystallized, then the high degree of periodicity makes crystals ideal specimens for measuring particle size. The image of a regular array may be Fourier transformed, filtered free of non-Bragg intensities, and transformed again to yield a clearer image of the specimen (39, 40). The initial transform, in addition, provides accurate values for interparticle distances and hence diameters. An example of this for an ideal case of a protein crystal, that of fungal lipase, is shown in Fig. 6.
Fig. 6.
(a) Raw AFM image of the surface of a crystal of fungal lipase, a protein of 30 kDa. The lattice spacings are clearly evident, but little more detail is apparent. (b) Fourier transform of the image of the crystal in panel a. It contains both Bragg reflections arising from the regular features of the image, which fall on a regular lattice, and randomly distributed intensity arising from noise in the image. (c) Image obtained when only the discrete Bragg reflections (intensities) are included in a reverse Fourier transform, i.e., the noise components are filtered out. (d) Enlargement of a small area from panel c in which individual protein molecules can be seen as bright globules arranged in the crystalline lattice.
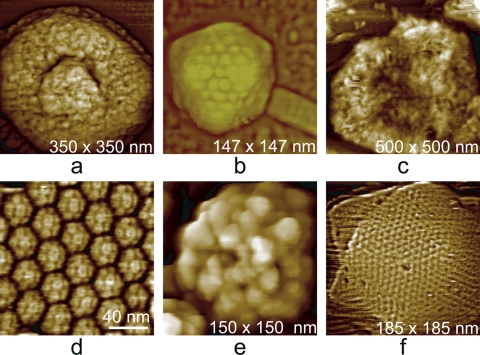
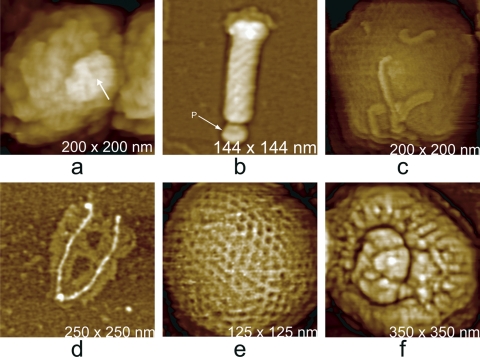
The surfaces of virus particles vary topographically as a function of their composition and architectures. Some examples illustrating this point are shown in Fig. 7 for six different kinds of viruses. Plant viruses, for example, generally exhibit protein capsids with few embellishments, and this is true of many animal viruses and bacteriophage capsids as well. These are generally based on icosahedral architectures, and clusters of coat protein subunits, or capsomeres, are symmetrically distributed (13; D. L. D. Caspar and A. Klug, presented at the Cold Spring Harbor Symposium on Quantitative Biology, 1962). More complex animal viruses, on the other hand, though they may contain an icosahedral capsid in their interior, often have either a lipid membrane over their surface, a covering of protein clusters (the Soc and Hoc proteins of T4 bacteriophage), or even a fur-like coating of fibers (mimivirus). These various surfaces are identifiable by AFM, and with the aid of some histological procedures, such as osmium tetroxide fixation or protease treatment, can be delineated with a considerable degree of precision.
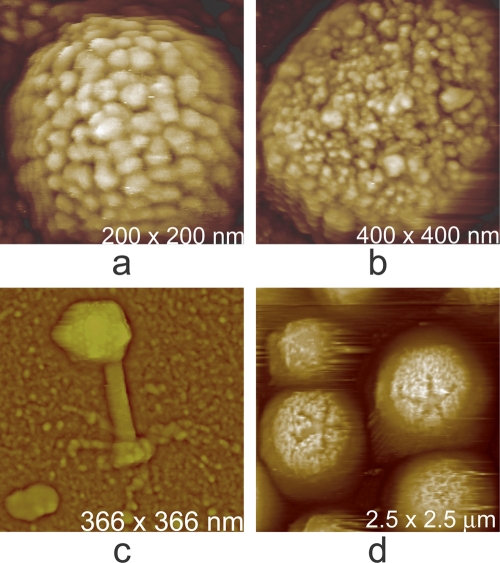
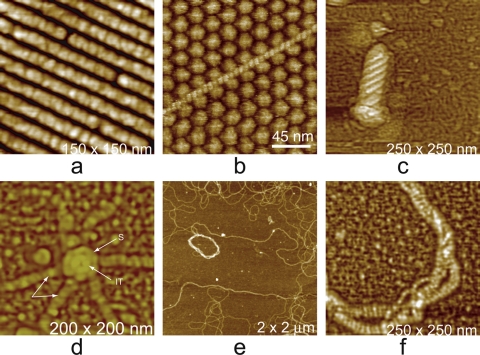
Fig. 7.
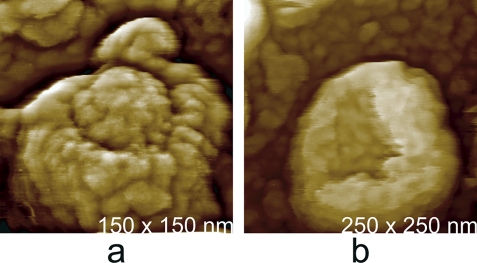
(a) A vaccinia virion which was dried and scanned in air. The body of the virus, in shrinking, has retracted from the external membrane, which remains on the mica substrate as a continuous palisade. (b) The icosahedral head and part of the tail assembly of a cyanophage. The distribution of capsomeres is clearly evident on the faces. (c) A virion of mimivirus thickly coated with clusters of glycoprotein fibers. (d) Surface of a crystal of brome mosaic virus. The protein capsomeres are icosahedrally distributed on the surfaces of the virions and are clearly evident. (e) A virion of human immunodeficiency virus with characteristic clusters of envelope protein (gp 120) distributed in an apparently arbitrary pattern over its surface. (f) Large triangular plates assembled to create the capsid of PBCV-1, a large (200-nm-diameter) icosahedral iridovirus that infects algae. The regular honeycomb arrangement of capsomeres on the surface can be seen even at a fairly low magnification.
ICOSAHEDRAL CAPSIDS
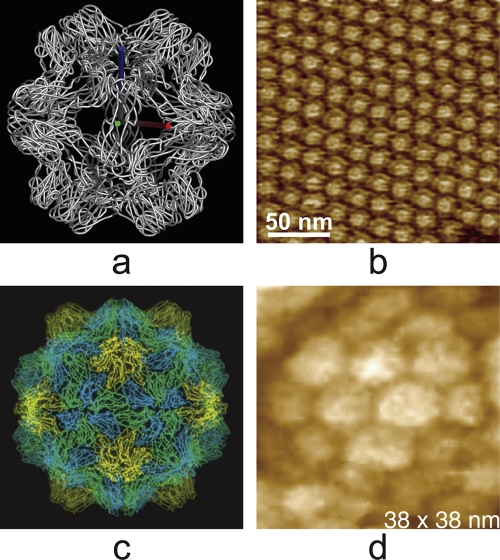
Icosahedral capsids, or bullet-shaped or elongated capsids based on that symmetry, can be characterized in terms of the structure of the fundamental capsid protein(s), along with the icosahedral triangulation number, T (Caspar and Klug, presented at the Cold Spring Harbor Symposium on Quantitative Biology, 1962). The triangulation number T can take on only integral values and only according to a defined scheme for subdividing the surface of a sphere (9). It essentially describes the polygonal distribution of pentameric and hexameric protein clusters on the surfaces of virus particles. This will vary from small numbers like T = 1 for satellite viruses or the T = 1 reassembly particles of brome mosaic virus (33) seen in Fig. 8a and b to T = 3 and higher for more conventional, small icosahedral viruses such as poliovirus or turnip yellow mosaic virus, which is presented in Fig. 8c, to very large numbers for complex viruses such as the iridoviruses (Fig. 3d), algal viruses (Fig. 7f) like PBCV-1 (T = 169), and mimivirus (Fig. 7c).
Fig. 8.
(a) Polypeptide backbone structure, determined by X-ray crystallography, of the T = 1 particle that forms when the T = 3 virion of brome mosaic virus is treated with high salt and neutral pH. It is seen looking along a 2-fold axis. (b) Surface of a crystal of the BMV T = 1 particles. (c) Polypeptide backbone structure of the T = 3 icosahedral turnip yellow mosaic virus, also determined by X-ray diffraction analysis. (d) A single virion of TYMV, imaged by AFM, which was incorporated into the surface of a crystal of the virus. The pentameric and hexameric capsomeres are evident in the AFM image.
In many cases the exterior shell of a virus may not be icosahedral, but it might possess an inner capsid that is. For example, though membrane covered and with a pleiomorphic external shape, herpes simplex virus possesses a nucleic acid-containing capsid of icosahedral form T = 16. Mimivirus (Fig. 7c) exhibits a complex outer surface coated with a forest of fibers, but it too contains an icosahedral core with a triangulation number of between T = 324 and 381 (48). The T number, then, contains a good part of the information needed to describe an icosahedral capsid.
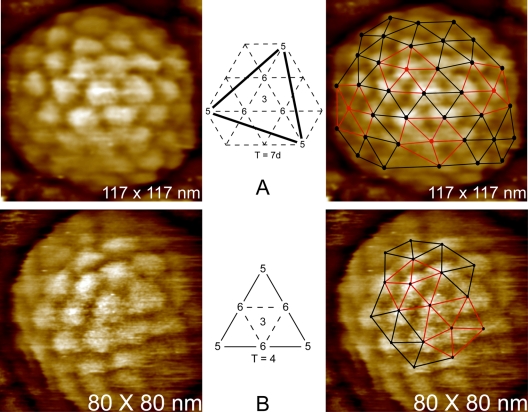
The triangulation numbers of icosahedral viruses can frequently be deduced from AFM images. Examples were seen in Fig. 7 and 8, but even more strikingly in the case of Ty3 retrotransposons, a protoretrovirus, as illustrated by Fig. 9. It was found for Ty3 (31) that virions existed in three different sizes and three different architectures corresponding to T numbers 7 (the largest fraction of the population), 4, and 3 (the smallest component). AFM images of the surface capsomere distribution could be triangulated visually, as in Fig. 9, by defining the arrangement of hexamers with respect to pentamers. A particularly good description of this approach is given by Rayment (44), where it is illustrated for surface lattices of T = 1, 3, 4, and 7, the four lattices with the lowest T number. The T number was thus defined. Of particular interest were the T = 7 particles, which could conceivably exist in either of two enantiomorphs, d and l. Because height information is preserved in AFM images, so is handedness. Thus, it was possible in the case of the T = 7 Ty3 virions to determine that their actual T number was 7d.
Fig. 9.
The icosahedral symmetry of the Ty3 retrotransposon, a protoretrovirus that infects yeast, was established by AFM. It was further shown that Ty3 existed in three different icosahedral forms corresponding to T numbers of 3, 4, and 7. (A) The left panel shows a Ty3 particle, and the right panel shows the same particle with a T = 7 icosahedral net overlay. Pentagonal vertices are in red. The center diagram is that of a triangular face of an icosahedron showing positions of pentagonal and hexagonal capsomeres for T = 7. (B) Another, smaller particle of Ty3 and the equivalent overlay and diagram showing it to have T = 4 icosahedral symmetry.
A somewhat different approach must be taken with very large icosahedral capsids, like those in Fig. 7c and f, which include mimivirus and PBCV-1. This is achieved by determining the two indices h and k, which define the triangulation number (T = h2 + hk + k2), by following a row of hexagonal capsomeres from one pentagonal vertex to the next icosahedral edge and simply counting the number of capsomeres along one edge h and the other k (the h and k coordinates of the intersection point on the icosahedral edge) needed to traverse (8; Caspar and Klug, presented at the Cold Spring Harbor Symposium on Quantitative Biology, 1962; Viperdb.scripps.edu). Precisely this method was used to determine the T numbers of the iridovirus PBCV-1 (49) and the capsid architecture of mimivirus (48).
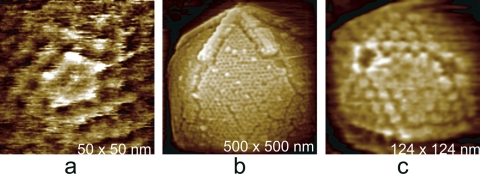
While the T number describes the overall distribution of capsomeres on the surface of an icosahedral capsid, a more complete description of a virus structure would require the distribution of protein units in the individual capsomeres, and ultimately the coordinates of the atoms comprising the virus coat proteins, to be defined. The last can be obtained only by X-ray crystallography, but the distribution of subunits within capsomeres can sometimes be determined or deduced by AFM analysis. Some examples are shown in Fig. 10.
Fig. 10.
High-magnification AFM images of the protein distributions comprising the surfaces of a number of viruses. (a) The honeycomb, open network of protein subunits on particles reassembled from the truncated Gag protein of Mason-Pfizer monkey virus. (b) Hexameric capsomeres are separated from one another by trimeric proteins on the surface of the icosahedral capsid of a cyanophage. (c) A honeycomb arrangement of trimeric major capsid protein making up the capsid of mimivirus. (d) Trimeric protein capsomeres composing the large faces of the icosahedral PBCV-1 algal virus.
In the case of Mason-Pfizer monkey virus (M-PMV) in Fig. 10a, for example, virus-like particles (VLP) of its truncated Gag protein were reassembled in vitro and imaged at high magnification by AFM. From the images, individual protein subunits were visible, and this allowed discrimination of two possible models for the capsomeres (28). A similar analysis was used in the case of the large algal virus PBCV-1 (21), which is shown in Fig. 10d. Knowing the diameters of capsomeres is often of considerable importance, even when individual subunits cannot be resolved. In mimivirus (Fig. 10c), for example, the capsomere diameter provided a crucial clue in delineating the capsid architecture and permitting subsequent detailed analysis and reconstruction by cryo-EM (30, 48). Although capsids of native HIV have yet to be visualized by AFM, images of helical tubes of capsid protein reassembled in vitro have been recorded (28), and one is shown in Fig. 11a. In these tubes a hexagonal arrangement of coat proteins could be clearly seen, and this provided support for a capsid model based on modified icosahedral architecture (10).
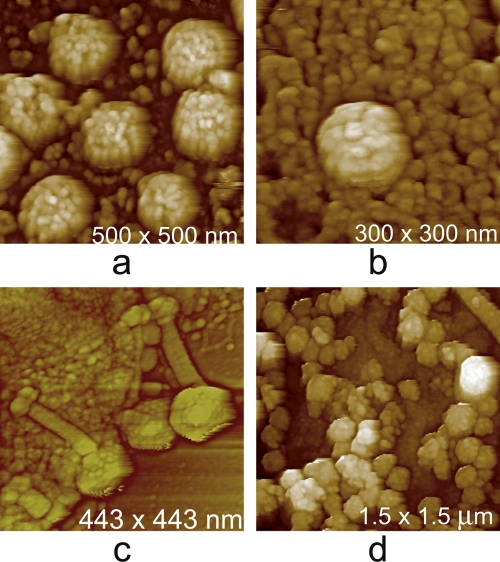
Fig. 11.
Two in vitro, reassembled, virus-like particles produced in bacteria as imaged by AFM. (a) Helical tubes, a product of in vitro self-assembly of the Gag protein of human immunodeficiency virus. (b) Particles reassembled in vitro from a truncated form of the Gag protein from Mason-Pfizer monkey virus. (c to f) Mutant and aberrant virus particles. (c) A mutant cyanophage that lacks any protein sheath about its tail assembly and fully exposes the injection tube normally found in its interior. (d) AFM image of 3T3 cells in culture that are infected with a mutant form (gPr80gag) of Moloney murine leukemia virus. The virus, upon budding, is unable to separate completely from the host cell membrane and forms long, comet-like extensions from the cell surface. (e) A mutant of Moloney murine leukemia virus that, genetically, lacks the capacity to make envelope protein. As a consequence, it appears as a “bald” virus that exposes its limiting lipid membrane to the exterior. The undulations and variations of the membrane surface are an effect produced by local movement of the membrane in response to AFM tip pressure. (f) A crystal of brome mosaic virus. Though most of the crystal is composed of conformist T = 3 particles with the standard diameter of 30 nm, the arrows point out the presence of two exceptionally small virions, probably T = 1 icosahedra lacking RNA, and one exceptionally large virion, probably T = 4 or 7, likely containing multiple copies of RNA.
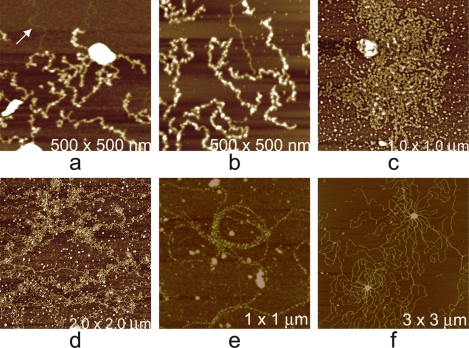
The helical tubes reassembled from HIV Gag protein should remind us that though we have emphasized spherical particles to this point, helical, rod-shaped, and fibrous structures having a periodic substructure are also ideal specimens for AFM analysis. These frequently appear in investigations of intact viruses and even in studies of spherical viruses when their interiors are explored. Figure 12 provides a few examples where fibrous and helical objects have been encountered in investigations of viruses and visualized by AFM. As with other virus particles and structures, accurate quantitative measures can be obtained by AFM. The lengths (if not indeterminately long), the diameter of the cylindrical cross section, and the periodicity of substructures are accessible. Qualitative features such as persistence length, flexibility, and tensile strength may also be evident from AFM images.
Fig. 12.
Various helical, fibrous, and rod-like structures observed on or associated with viruses. (a) Paracrystalline mass of tobacco mosaic virus whose helical repeat is just visible, though indistinct. (b) A helical fiber, possibly a bacterial cytoskeletal fiber, lying atop the surface of a crystal of satellite tobacco mosaic virus. (c) Helical tail assembly from bacteriophage T4 that is responsible for injection of the phage DNA into its host. (d) A view along the axis of a bottom segment of a cyanophage tail assembly that is standing upright on the AFM substrate. The tail fibers are splayed out, showing their segmented character. (e and f) Low- and high-magnification images of unique fibers having a pronounced 7-nm repeat along their lengths that were observed when mimivirus capsids were disrupted.
VIRUSES IN CRUDE SAMPLES AND ON CELLS
With AFM, it is not essential that highly purified virus particles be used as samples (24), although that might be ideal. MuLV is accompanied by a background of protein molecules in Fig. 13a, for example, but is nonetheless distinct in its features. Because individual particles can be investigated whenever a good specimen is spatially distinct from the surrounding rubble of proteins, cellular debris, and biological detritus, it may still yield excellent images. If a virus particle rises above the substrate more than any accompanying impurity, its upper surface may provide most of the information that is sought, and what is around it at the level of the substrate plane becomes irrelevant. A problem, however, is that biological debris often adheres to and fouls the AFM tip, and this can severely degrade the quality of images. Contamination of tips is one of the most frustrating and annoying issues with biological AFM.
Fig. 13.
(a) A cluster of Moloney murine leukemia virus budding from the surface of an infected 3T3 cell. (b) A single Moloney murine leukemia virus particle emerging from the protein matrix characterizing the external surface of a 3T3 cell. (c) Newly made cyanophage spilling from a lysed bacterium. (d) Cascade of human immunodeficiency virus budding from the surface of a heavily infected human lymphocyte in culture.
Viruses on the surfaces of host cells may be visualized as well as free particles and sometimes with superior results because the particles are better immobilized (23). MuLV emerging from an infected 3T3 cell is clearly delineated in Fig. 13a and b. They may be seen entering cells upon infection or budding from cells after replication and assembly. This often provides valuable insights into which cells in a population are producing virus, the distribution of virus particles on the surface of the cells (e.g., are there preferred sites for budding?), and some details of the budding process itself.
In Fig. 13d, for example, a human lymphocyte in culture is seen virtually coated with HIV budding from its plasma membrane. Investigation of a large population of cells, however, showed that this was an unusual occurrence, in that most cells exhibited only a dozen or fewer HIV particles on their surfaces. Thus, only a few cells seemed to account for the great majority of virus (23). Algal cells have both a cell membrane and a cell wall that must be breached in order for newly made virions to escape from the cell. PBCV-1 (Fig. 14a) is visualized by AFM to be entangled in fragments of cellular membranes as it tries to exit disintegrating cells. In Fig. 14b, mimivirus is seen attached to fragments of host amoeba plasma membrane, and in Fig. 14c, the membrane coating of a herpesvirus is seen shorn from the capsid.
Fig. 14.
(a) Five shards of plasma membrane from a disrupted, virus-infected Chlorella cell form a flowerlike aggregate. Attached to the membrane fragments is PBCV-1. (b) Giant mimivirus is seen still attached to fragments of the host amoeba plasma membrane. (c) The white sheet of the membrane coating a herpes simplex virus has “splashed” to one side of the capsid and lies on the substrate plane.
MUTANT VIRUSES
Mutant viruses, naturally occurring or produced in the laboratory, can be imaged as well as native virions and VLP created in vitro from capsid proteins. In some cases, the phenotype of the mutant can be revealed by observing infected host cells for unique or anomalous features. This was done, as shown in Fig. 11d, in a study of Moloney murine leukemia virus (MuLV)-infected 3T3 cells, where the mutant gPr80gag lacked glycosylated Gag protein (19, 32, 42). Prior evidence suggested that such mutants failed in some stage of viral budding. This was amply confirmed by AFM visualization of infected, virus-producing cells.
As seen in Fig. 11d, instead of normal, spherical virus emerging from the cell surface, bullet- and comet-shaped protrusions were found distributed over all of the plasma membrane of host cells. The comets were viruses that were trying to escape but were unable to terminate association with the host cell. From this it was concluded that the failure of glycosylation produced a defect in late stages of the budding process.
Other mutations in virus genomes may produce alterations in external features of virus particles that are readily observable by AFM. MuLV particles that failed to make envelope protein (gp120 protein), one of which is seen in Fig. 11e, were examined in another study (23). While normal particles, as were seen in Fig. 4a and 13a and b, are characterized by a coating of protein tufts, about 100 to 150 in number, mutant particles were “bald” virions lacking any such protein clusters. Instead, only an outer lipid membrane was visible. A study of Ty3 retrotransposon having a defective protease gene mutant and reverse transcriptase (RT) mutant Ty3 particles was illuminating in a different way (31). While wild-type virus appeared as T = 3, 4, and 7 icosahedral particles, the RT and Pr mutants appeared to be exclusively T = 7 particles. This suggested that T = 4 and T = 3 particle forms might represent mature or late-stage virus forms. All T = 7 particles, no matter what their genome, exhibited the same general appearance.
SPECIALIZED FEATURES ON VIRUSES
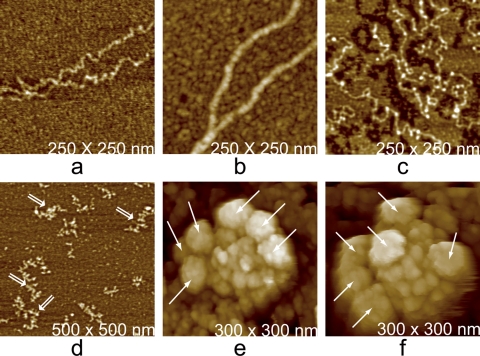
Some viruses exhibit specialized external structures, or deviations from their general architectures. A collection of these drawn from several different viruses and visualized by AFM are shown in Fig. 15. For example, the MuLV particles in Fig. 15a generally have, somewhere on their otherwise uniformly crenulated surfaces, a single small bump or brief protrusion. These are likely to be “budding scars” resulting from breaking away from the host cell (18). Some MuLV particles, perhaps defective, exhibited small sectors on their surfaces where protein was absent and a channel into the interior appeared (19). Other, more prominent features are the thick fibers on the surfaces of PBCV-1 (22) and the lateral bodies of vaccinia virus, seen in Fig. 15c and f, respectively (17, 36).
Fig. 15.
Distinctive accessory features observed on the surfaces of viruses. (a) A budding scar (arrow), a small protrusion seen on most virions of Moloney murine leukemia virus that is produced from infected 3T3 cells in culture. The bulges are thought to arise from pinching off the virus from the host cell membrane. (b) Helical tail assembly of a cyanophage. The tail fibers have been broken off, but at the head end of the sheath is a unique entity that has pulled from the head along with the tail. Denoted by an arrow and the letter P, this is the portal assembly responsible for drawing the phage DNA into the capsid. (c) A virion of PBCV-1 exhibiting several thick fibers, of unknown function, from its surface. (d) Typical example of the protein fibers, usually appearing like hairs from a basal tuft, that thickly coat the outer surface of mimivirus. The lighter colored material that shadows the two fibers is believed to be associated oligosaccharide. (e) A particle reassembled in vitro from truncated Gag protein of Mason-Pfizer monkey virus. The particles invariably display a number of dislocations and defects on their surfaces. The defects allow the particles rough sphericity without icosahedral or any other exact symmetry. (f) Air-dried vaccinia virus. Retraction of the membrane and overall shrinkage emphasize the lateral body at center. A unique feature of vaccinia virus is the presence of two such lateral bodies associated with each virion.
PBCV-1, a large algal virus, exhibited a unique pentagonal assembly of proteins (21) at every 5-fold vertex of its icosahedral capsid. This is illustrated in Fig. 16a. The assembly had a single protein in the center that could “push in” and “pull out” as demonstrated by the application of AFM tip pressure. Its exact function is speculative. Many bacteriophages have tail assemblies of one sort or another for packaging and injecting their DNA. Mimivirus is, in a sense, similar to these phages and has an assembly, seen in Fig. 16b, with a presumably similar function at a single unique, 5-fold vertex. This star-shaped structure is analogous to the tail assemblies of phages and is, as is evident in those figures, a distinctive feature of the virions. It is a complex structure, presumably composed of many proteins, and AFM reveals much of that complexity.
Fig. 16.
Some additional external features that characterize the pentameric vertices of some large icosahedral viruses. (a) A specialized cluster of five proteins, with another protein at the center, that occupy the 12 unique vertices on the surface of PBCV-1. All of the proteins in these clusters are clearly different in structure from the normal capsid protein. (b) Image of the “stargate,” five-vane apparatus found at only a single pentameric vertex of mimivirus. The apparatus provides a mechanism for the release of the encapsidated DNA. (c) AFM image of adenovirus. A noteworthy feature of adenovirus is that degradation of the virions invariably initiates and proceeds with loss of pentons at the 5-fold vertices, leaving particles perforated in an icosahedrally symmetric manner.
A point that deserves particular emphasis is that all of the particles within a population of virus are not absolutely identical, and often there are very significant differences in the detailed features of individual particles. The degree of structural diversity is evident in many of the AFM images presented here. This is a point often obscured by the results of X-ray crystallography or cryo-EM reconstructions. Those techniques rely completely on the assumption of structural conformity. They produce models that represent the average in time and space for the individuals that make up the population. AFM, on the other hand, reveals the eccentricities and unique features of the individuals, and these are instructive. They often define the extremes of what is structurally possible among a large population of viruses having, presumably, the same genome and the same environment for replication and assembly. What is seen with AFM is that anomalous and aberrant individuals are not only present but common.
DISSECTION OF COMPLEX VIRUSES
It might be thought that because AFM provides images of the surfaces of objects and does not peer into their interiors, as do X-ray diffraction and electron microscopy, AFM would be of little value in delineating the interior structure of viruses, i.e., the layers beneath the external surface. This is not true, however, as we can apply the same technique that has been used by anatomists for centuries: dissection. With the aid of chemical, enzymatic, and physical tools, we can systematically pare a complex entity, including a virus, down to its core, layer by layer. At each stage, AFM may then be used to visualize what remains and what has been removed as well.
This approach is particularly effective with large, complex viruses such as vaccinia virus (17, 36), as illustrated in Fig. 17, or mimivirus (30, 48), as illustrated in Fig. 18. For these complicated assemblies, ordered and disordered protein shells, lipid membranes, and the nucleic acid within can be revealed and analyzed. By deconstruction, the architecture of particles can be revealed, and at the same time, the kinds of biochemical interactions that maintain each level of structure may be delineated as well.
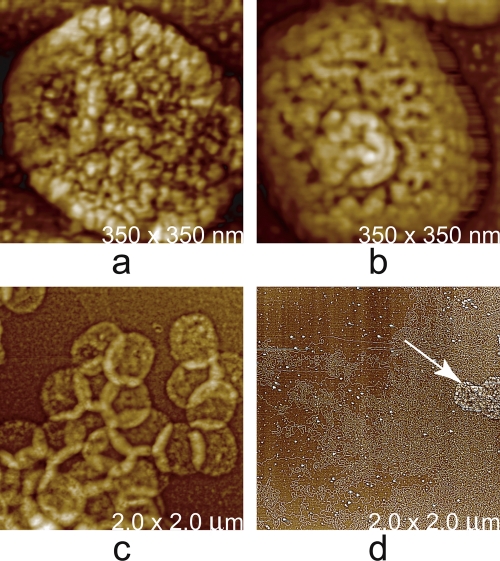
Fig. 17.
Four stages in the chemical and enzymatic dissection of vaccinia virus. (a) An intact vaccinia virus virion. (b) The core of the virus obtained by treatment with a nonionic detergent (NP-40) and dithiothreitol, a reducing agent. (c) The ghosts, or capsules, that remain after the cores have been treated with proteases, which also produces release of the viral nucleic acid. (d) Masses of DNA releases onto the AFM substrate by disrupted viral cores. The arrow indicates a disrupted vaccinia virus core adjacent to the DNA.
Fig. 18.
Stages in the enzymatic dissection of mimivirus, which infects protozoans and is the largest virus known. (a) A mass of intact virus still coated with glycoprotein fibers. The “starfish”-ornamented vertex is visible on many particles in spite of the fiber coating. (b) The fibrous coat has been removed by treatment with lysozyme and proteinase K, and the “starfish” apparatus has become more prominent. The geometric character of the underlying protein capsid is beginning to emerge. (c) The capsid has been further treated with proteases, and the icosahedral architecture of the protein capsid and the distribution of capsomeres have become evident. (d) The capsid has been breached, and the DNA contained within is seen spilling onto the mica substrate, which also contains many viral proteins released in the process.
Among the most useful agents for chemical dissection have been detergents, usually 0.5 to 2% of some nonionic detergent such as NP-40, and reducing agents such as dithiothreitol (DTT) or dithioerythritol (DTE). The former causes the protein structure to gradually unravel, and detergents strip away the lipid membrane. The latter reduce disulfide bonds and liberate polypeptides otherwise bonded to one another. Disulfide bond reduction appears to be particularly important in large, complex viruses where such covalent linkages cross-link coat proteins and stabilize capsids (36, 47).
In some cases, nonionic detergents are insufficient to disrupt structure, and more vigorous ionic detergents such as SDS must be used. There is difficulty with SDS, however. It tends to have an all-or-none effect, so that upon reaching a concentration sufficient to disrupt viruses, it completely degrades them uncontrollably.
The most effective enzymatic tools have been proteases that degrade polypeptides. These are particularly useful because they have a range of activities and a spectrum of specificities. As a consequence, a whole variety of proteases have been employed, and these include trypsin, bromelin, proteinase K, subtilisin, and mixtures of pancreatic proteases. Viruses are usually exposed to the proteases for anywhere from 15 min to several hours, or even overnight, at concentrations of from 0.5 mg/ml to as high as 5 mg/ml. The proteases must be washed from the virions with buffer or water before imaging, as they otherwise produce a dense, irregular background that make imaging problematic, and they foul the cantilever tip.
Physical forces have also been used to disrupt viruses, and often fortuitous perturbations, resulting simply from preparation and handling, have proven to be structurally illuminating. Heat, for example, was used to open TYMV (26) to release its encapsidated RNA, and direct physical pressure on mimivirus sandwiched between two layers of atomically smooth, cleaved mica was used as well (30). Cycles of freezing and thawing have been reported to disrupt viruses in some cases, but to this point, freezing has not proven useful in AFM studies. There are also instances where “hammering” of individual particles with the AFM tip has been utilized, taking advantage of the fact that AFM can serve as a tool as well as an imaging device.
In carrying out the dissection of a virus, or even in simply visualizing particles spread on a glass, plastic, or mica substrate, it is necessary to ensure that the virus particles adhere firmly to the substrate. Failure to do so allows the particles to move in response to close approach of the AFM tip and renders imaging impossible. Occasionally, altering the charge on the substrate is sufficient. Mica is negatively charged on its surface, but exposure to a nickel or magnesium salt such as MgCl2 coats it with divalent ions and leaves it positively charged. Some viruses or macromolecules, such as nucleic acids, if they are repelled by a negative surface, may then be firmly held by a positive surface or vice versa.
Charge is, however, frequently insufficient for virions where the contact area of the particle is relatively small. To fix most viruses to a substrate, as well as a wide variety of other biological entities and materials, an effective procedure is to coat the substrate with poly-l-lysine before depositing the virus. Presumably salt bridges between the ε amino groups of the lysines and the glutamic and aspartic acid carboxyl groups on the particles lock them in place. After such substrate-particle attachment, the substrate can be rinsed with water several times with acceptable loss of sample. The only serious disadvantage of coating with poly-l-lysine is that it produces a rather rough and irregular background. As a consequence, molecular objects, such as lipid membranes or nucleic acids, which rise only about a nanometer or two above the substrate plane, become difficult to identify and visualize. The method is excellent, however, for imaging cells and intact or partially degraded virions.
It is occasionally unnecessary to actually treat viruses with any chemical or biochemical agents to explore the interior, as the physical stress of preparation and especially structurally delicate virus purification may result in damaged or partially degraded particles. These may expose internal structural features otherwise not accessible to the cantilever tip. Retroviruses, in particular, are physically fragile. Some MuLVs, as shown in Fig. 19a, when subjected to the shear forces of even low-speed centrifugation lose portions of the shell surrounding the capsid. This permits direct visualization of the virus core still embedded within the layers of envelope and matrix protein (18).
Fig. 19.
Retroviruses are fragile and often are disrupted or damaged by the shear forces of preparative centrifugation alone. These damaged particles frequently expose, for AFM visualization, internal structure and reveal unique elements of the virus architecture. (a) A virion of Moloney murine leukemia virus where the outer shell of envelope protein, lipid membrane, and matrix protein have been partly torn away to reveal, like the kernel of a nut or the pit of a fruit, the nucleic acid-containing capsid that it enshrouds. From such images we can obtain quantitative measures of the thickness of the virus shells and the diameters and structural character of the capsid within. (b) A human immunodeficiency virus that has been shorn during centrifugation and has completely lost the protein capsid that it contained.
HIV is another example where even the mildest procedures produce some damaged virions. Although the cores of HIV have not yet been visualized by AFM, likely due to their fragility, the remainder of the virus without the cores has been (29). An example can be seen in Fig. 19b. Such partially disrobed particles, both MuLV and HIV, provide specimens that can be subjected to quantitative examination and thereby yield the thicknesses of internal layers of structure, and they give some clues as to their components as well.
Good examples of complete dissections of complex viruses using AFM are those of vaccinia virus, a poxvirus of about 300 nm diameter that is delimited by a lipid membrane (17, 36), and mimivirus (30, 48). Vaccinia virus contains a double-stranded DNA genome bounded by several protein shells. It also has associated with its inner core two unusual protein assemblies of still-unknown function, known as lateral bodies.
Vaccinia virus was sequentially degraded with 0.5% NP-40 nonionic detergent combined with 0.05 M DTT, followed by exposure to this same mixture but containing either trypsin or proteinase K or to the proteases alone. Four stages in this process are presented in Fig. 17. In the end, the innermost core was breached and the DNA was exposed. From images of the DNA emerging from the core, it was deduced that while some portion of the encapsidated DNA was heavily integrated with protein, the vast majority was largely naked, with only occasional associations with protein. The giant mimivirus was similarly dissected using essentially the same approach. A sampling of the results is presented in Fig. 18. Mimivirus, at 750 nm in diameter, was revealed to be a very complex particle with numerous layers of structure.
VISUALIZATION OF VIRAL RNA AND DNA
The nucleic acids of viruses, from a structural standpoint, are of considerable interest, in particular with regard to how they are condensed and packaged inside capsids and cores. Clearly, packaging is accomplished differently by specific families of viruses. It is unlikely, for example, that bacteriophages and poxviruses package their genomic double-stranded DNAs in the same way. The packing densities of the nucleic acid differ by more than 10-fold (17). Nor is it likely that large, single-stranded-RNA-containing viruses, such as retroviruses, package their genomes the same way as do T = 1 or T = 3 icosahedral plant viruses (38). Certainly, helical and filamentous viruses use entirely different mechanisms.
AFM investigations have been conducted on RNAs extracted by phenol from a series of small icosahedral viruses and from tobacco mosaic virus, the classical rod-shaped, helical virus (18). The spherical viruses included poliovirus, satellite tobacco mosaic virus, turnip yellow mosaic virus, and brome mosaic virus. Figure 20a shows the RNA from poliovirus. In this study, the gradual unraveling of the tertiary structure of the RNA, and ultimately the secondary structure as well, could be produced in stages simply by heating. A counterexample was provided by tobacco mosaic virus RNA (Fig. 20b), which appeared initially as a thread, a completely extended molecule lacking any secondary structure. With time, it began forming local secondary structural elements and eventually condensed into forms similar to those seen for the RNAs from the icosahedral viruses. The upshot of the study was that the single strands of RNA spontaneously condensed as linear arrangements of stem-loop substructures following synthesis, that the condensed RNA bound coat protein to it, and that the two cooperatively coalesced into the completed particle (20). In those studies, AFM proved itself an able technique for directly visualizing nucleic acid structure, demonstrating its fluidity and suggesting the mechanisms by which it is encapsidated.
Fig. 20.
(a) Loss of RNA from small icosahedral viruses, as illustrated here for poliovirus. The very bright object is a disrupted virion still in the process of discharging RNA. (b) Tobacco mosaic virus RNA illustrates the natural tendency of single-stranded viral RNA to form base-paired secondary structure and then tertiary structure and finally to condense and fold upon itself. (c to f) DNA visualized after extraction or release from a variety of viruses. (c) The DNA from ruptured vaccinia virus (a broken virion is the white object) is spread on the substrate. Evident here is that it is heavily complexed with proteins, presumably involved with transcription and replication. (d) Vaccinia virus DNA after several hours of exposure to proteases. The bound proteins have been stripped away. (e) Genomic DNA from a cyanobacterium shown for comparison. Like DNAs from vaccinia virus and other complex viruses, it too is heavily decorated with proteins over its entire length. (f) DNA from disrupted cyanophage heads. The DNA of bacteriophages is always seen to be naked and free of proteins.
DNA and RNA have quite different appearances in AFM images, and this can be seen by contrasting the images of RNA in Fig. 20a and b with the images of DNA from various viruses, as well as a bacterial cell, in Fig. 20c to f, which presents both kinds of nucleic acid. DNA looks like strands and coils of stiff rope lacking any higher levels of structure; RNA appears as complicated, linear sequences of self-involved secondary structure. Sometimes, however, the distinction is not entirely clear, and further evidence may be needed to show whether a filament, strand, or complex is DNA or RNA.
A method was devised for additional identification based on exposure of the nucleic acid to high concentrations of bovine RNase A (27). One example of results from that study is shown in Fig. 21. Intact RNA in Fig. 21c, as seen in Fig. 21d, was hydrolyzed to small pieces by RNase A and left only fragments on the substrate, presumably corresponding to protected stem-loops. DNA, seen in Fig. 21a, on the other hand, became coated with the protein, as shown in Fig. 21b, and the resulting strands exhibited thicknesses two to three times that of naked double-stranded DNA. Thus, it is possible with AFM to practice a kind of crude histology.
Fig. 21.
Consequences of treatment of viral nucleic acids with RNase A, an enzyme that binds to both RNA and DNA but degrades only RNA. (a) Two crossing double-stranded helices of DNA before treatment. (b) The same two strands after exposure to a high concentration of RNase A. The protein completely coats the DNA and increases the thickness of the strands from about 1.5 nm (dry) above the substrate to about 3.5 nm above the substrate. (c) Single-stranded RNA from poliovirus, showing its many secondary structural domains sequentially arranged along its length. (d) After treatment with RNase A, only short, disconnected secondary structural domains remain, apparently resistant to attack by virtue of conformational protection. (e and f) Images of two different Moloney murine leukemia viruses decorated with 15-nm-diameter gold particles coated with antibody whose antigen is the envelope protein of the virus. The arrows indicate the gold particles. The protein-tufted virions are seen beneath.
IMMUNE LABELING WITH AFM
A second example of histological AFM is the immunolabeling of viruses with antibodies specific for certain proteins. Although individual IgGs are not clearly identifiable by AFM when bound to a virion, IgGs conjugated with gold particles generally are. In a sense, these are used in the same way as they are used in transmission electron microscopy immunolabeling, except that instead of visualizing points of high electron density, one images with AFM objects having the size and shape of the immunogold particles.
Using gold-IgG conjugate particles against the envelope protein, as shown in Fig. 21e and f, it was possible to show that protein tufts on the surfaces of MuLV were indeed envelope protein (gp120) (19, 23). The major problem to this point with IgG-gold conjugates is that their physical size limits the resolution of the method. Conjugated gold particles can bind only as close as their diameters allow.
WHAT DOES AFM OFFER THE STRUCTURAL BIOLOGIST?
What can AFM visualize that is of value to structural virology? The answer is almost everything, and at resolutions that approach and in some cases surpass those with electron microscopy. At this time, lipid membranes have been identified, both RNA and DNA have been visualized, and large protein assemblies have been resolved into their component substructures. The capsids of icosahedral viruses and the icosahedral capsids of nonicosahedral viruses have been seen at high resolution, in some cases sufficiently high to deduce the arrangement of coat protein units in the capsomeres, as well as determine the triangulation number T.
In addition, viruses have been recorded budding from infected cells and suffering the consequences of a variety of stresses. Mutant viruses have been examined and phenotypes described. Unusual structural features have appeared, and very importantly, the unexpectedly great amount of structural nonconformity within populations of virus particles has been amply documented.
Virus samples may be imaged in air or in fluids having a range of viscosities, including culture medium or buffer, in situ on living cell surfaces, or after processing by histological procedures. AFM is nonintrusive and nondestructive, unless the intention is otherwise, and it can be applied to soft biological samples, particularly when the tapping mode is employed. In principle, only a single cell or virion need be imaged to learn of its structure, though normally images of as many as is practical are collected. While lateral resolution, limited by the finite size of the AFM cantilever tip, is a few nanometers, the height resolution is exceptional, approximately 0.5 nm. Finally, AFM produces three-dimensional, topographical images that accurately depict the surface features of the object, virus, or cell under study. In most ways, the images resemble common light photographic images and require little interpretation, only understanding.
It has been shown that the structures of viruses observed by AFM are entirely consistent with models derived by X-ray crystallography and cryo-EM. AFM has also been proven effective in visualizing viruses in situ emerging from infected, living cells. Although there are currently no examples, there is certainly no reason why structural information derived from X-ray crystallography and/or electron microscopy cannot be combined with AFM images, just as it has been for those latter two technologies.
New techniques, reagents, and methods that will further enhance the value of AFM to structural biologists are currently under development (3). Finally, the resolution of the technology and the unique insights into virus structure that it yields will be highly influenced by advances in tip acuity and improvements in instrument design. A great deal has yet to be done in developing sample preparation techniques, methods for chemically modifying cantilever tips, and expanding the use of histological procedures appropriate to AFM.
REFERENCES
- 1. Adrian M., Dubochet J., Lepault J., McDowall A. W. 1984. Cryoelectron microscopy of viruses. Nature 308:32–36 [DOI] [PubMed] [Google Scholar]
- 2. Allen S., Davies M. C., Roberts C. J., Tendler S. J. B., Williams P. M. 1997. Atomic force microscopy in analytical biotechnology. Trends Biotechnol. 15:101–105 [Google Scholar]
- 3. Baclayon M., Wuite G. J. L., Roos W. H. 2010. Imaging and manipulation of single viruses by atomic force microscopy. Soft Matter 6:5273–5285 [Google Scholar]
- 4. Baker T. S., Olson N. H., Fuller S. D. 1999. Adding the third dimension to virus life cycles: three-dimensional reconstruction of icosahedral viruses from cryo-electron micrographs. Microbiol. Mol. Biol. Rev. 63:862–922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Binnig G., Quate C. F. 1986. Atomic force microscope. Phys. Rev. Lett. 56:930–933 [DOI] [PubMed] [Google Scholar]
- 6. Bustamante C., Keller D. 1995. Scanning force microscopy in biology. Phys. Today 48:32–38 [Google Scholar]
- 7. Carragher B., Potter C. S., Sigworth F. J. 2007. Software tools for macromolecular microscopy. J. Struct. Biol. 157:1–288 [DOI] [PubMed] [Google Scholar]
- 8. Carrillo-Tripp M., et al. 2009. VIPERdb2: an enhanced and web API enabled relational database for structural virology. Nucleic Acids Res. 37:D436–D442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Caspar D. L. D. 1965. Viral and rickettsial infections of man. Lippincott, New York, NY [Google Scholar]
- 10. Ganser B. K., Li S., Klishko V. Y., Finch J. T., Sundquist W. I. 1999. Assembly and analysis of conical models for the HIV-1 core. Science 283:80–82 [DOI] [PubMed] [Google Scholar]
- 11. Goldsbury C. S., Scheuring S., Kreplak L. 2009. Introduction to atomic force microscopy (AFM) in biology. Curr. Protoc. Protein Sci. 17.7:1–19 [DOI] [PubMed] [Google Scholar]
- 12. Hansma P. K. 1994. Tapping mode atomic force microscopy in liquids. Appl. Phys. Lett. 64:1738–1740 [Google Scholar]
- 13. Horne R. W., Wildy P. 1961. Symmetry in virus architecture. Virology 15:348–373 [DOI] [PubMed] [Google Scholar]
- 14. Horne R. W., Wildy P. 1963. Virus structure revealed by negative staining. Adv. Virus Res. 10:101–170 [DOI] [PubMed] [Google Scholar]
- 15. Jena B. P., Horber J. K. (ed.) 2002. Atomic force microscopy in cell biology. Methods Cell Biol. 68:1–41512053726 [Google Scholar]
- 16. Kaper J. M. 1975. The chemical basis of virus structure, dissociation and reassembly. Front. Biol. Ser. 39:1–185 [PubMed] [Google Scholar]
- 17. Kuznetsov Y., Gershon P. D., McPherson A. 2008. Atomic force microscopy investigation of vaccinia virus structure. J. Virol. 85:7551–7566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kuznetsov Y. G., Daijogo S., Zhou J., Semler B. L., McPherson A. 2005. Atomic force microscopy analysis of icosahedral virus RNA. J. Mol. Biol. 347:41–52 [DOI] [PubMed] [Google Scholar]
- 19. Kuznetsov Y. G., et al. 2002. Atomic force microscopy investigation of fibroblasts infected with wild-type and mutant murine leukemia virus (MuLV). Biophys. J. 83:3665–3674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kuznetsov Y. G., Dowell J. J., Gavira J. A., Ng J. D., McPherson A. 2010. Biophysical and atomic force microscopy characterization of the RNA from satellite tobacco mosaic virus. Nucleic Acids Res. 38:8284–8294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kuznetsov Y. G., Gurnon J. R., Van Etten J. L., McPherson A. 2005. Atomic force microscopy investigation of a chlorella virus, PBCV-1. J. Struct. Biol. 149:256–263 [DOI] [PubMed] [Google Scholar]
- 22. Kuznetsov Y. G., Low A., Fan H., McPherson A. 2005. Atomic force microscopy investigation of isolated virions of murine leukemia virus. J. Virol. 79:1970–1974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kuznetsov Y. G., Low A., Fan H., McPherson A. 2004. Atomic force microscopy investigation of wild-type Moloney murine leukemia virus particles and virus particles lacking the envelope protein. Virology 323:189–196 [DOI] [PubMed] [Google Scholar]
- 24. Kuznetsov Y. G., Malkin A. J., Lucas R. W., Plomp M., McPherson A. 2001. Imaging of viruses by atomic force microscopy. J. Gen. Virol. 82:2025–2034 [DOI] [PubMed] [Google Scholar]
- 25. Kuznetsov Y. G., Malkin A. J., McPherson A. 1997. Atomic force microscopy studies of living cells: visualization of motility, division, aggregation, transformation, and apoptosis. J. Struct. Biol. 120:180–191 [DOI] [PubMed] [Google Scholar]
- 26. Kuznetsov Y. G., McPherson A. 2006. Atomic force microscopy investigation of turnip yellow mosaic virus capsid disruption and RNA extrusion. Virology 352:329–337 [DOI] [PubMed] [Google Scholar]
- 27. Kuznetsov Y. G., McPherson A. 2006. Identification of DNA and RNA from retroviruses using ribonuclease A. Scanning 28:278–281 [DOI] [PubMed] [Google Scholar]
- 28. Kuznetsov Y. G., Ulbrich P., Haubova S., Ruml T., McPherson A. 2007. Atomic force microscopy investigation of Mason-Pfizer monkey virus and human immunodeficiency virus type 1 reassembled particles. Virology 360:434–446 [DOI] [PubMed] [Google Scholar]
- 29. Kuznetsov Y. G., Victoria J. G., Robinson W. E., Jr., McPherson A. 2003. Atomic force microscopy investigation of human immunodeficiency virus (HIV) and HIV-infected lymphocytes. J. Virol. 77:11896–11909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kuznetsov Y. G., et al. 2010. Atomic force microscopy investigation of the giant mimivirus. Virology 404:127–137 [DOI] [PubMed] [Google Scholar]
- 31. Kuznetsov Y. G., Zhang M., Menees T. M., McPherson A., Sandmeyer S. 2005. Investigation by atomic force microscopy of the structure of Ty3 retrotransposon particles. J. Virol. 79:8032–8045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Low A., et al. 2007. Mutation in the glycosylated gag protein of murine leukemia virus results in reduced in vivo infectivity and a novel defect in viral budding or release. J. Virol. 81:3685–3692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Lucas R. W., Kuznetsov Y. G., Larson S. B., McPherson A. 2001. Crystallization of brome mosaic virus (BMV) and T=1 brome mosaic virus particles following a structural transition. Virology 286:290–303 [DOI] [PubMed] [Google Scholar]
- 34. Malkin A. J., Kuznetsov Y. G., Lucas R. W., McPherson A. 1999. Surface processes in the crystallization of turnip yellow mosaic virus visualized by atomic force microscopy. J. Struct. Biol. 127:35–43 [DOI] [PubMed] [Google Scholar]
- 35. Malkin A. J., Land T. A., Kuznetsov Y. G., McPherson A., DeYoreo J. J. 1995. Investigation of virus crystal growth mechanisms by in situ atomic force microscopy. Phys. Rev. Lett. 75:2778–2781 [DOI] [PubMed] [Google Scholar]
- 36. Malkin A. J., McPherson A., Gershon P. D. 2003. Structure of intracellular mature vaccinia virus visualized by in situ AFM. J. Virol. 77:6332–6340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Malkin A. J., Plomp M., McPherson A. 2002. Application of atomic force microscopy to studies of surface processes in virus crystallization and structural biology. Acta Crystallogr. 58:1617–1621 [DOI] [PubMed] [Google Scholar]
- 38. McPherson A. 2005. Micelle formation and crystallization as paradigms for virus assembly. Bioessays 27:447–458 [DOI] [PubMed] [Google Scholar]
- 39. McPherson A., Kuznetsov Y. G., Malkin A. J., Plomp M. 2004. Macromolecular crystal growth investigations using atomic force microscopy. J. Synchrotron Radiat. 11:21–23 [DOI] [PubMed] [Google Scholar]
- 40. McPherson A., Malkin A. J., Kuznetsov Y. G., Plomp M. 2001. Atomic force microscopy applications in macromolecular crystallography. Acta Crystallogr. D 57:1053–1060 [DOI] [PubMed] [Google Scholar]
- 41. Morris V. J., Kirby A. R., Gunning A. P. 2010. Atomic force microscopy for biologists, 2nd ed. Imperial College Press, London, United Kingdom [Google Scholar]
- 42. Nitta T., Kuznetsov Y., McPherson A., Fan H. 2010. Murine leukemia virus glycosylated Gag (gPr80gag) facilitates interferon-sensitive virus release through lipid rafts. Proc. Natl. Acad. Sci. U. S. A. 107:1190–1195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Plomp M., Rice M. K., Wagner E. K., McPherson A., Malkin A. J. 2002. Rapid visualization at high resolution of pathogens by atomic force microscopy: structural studies of herpes simplex virus-1. Am. J. Pathol. 160:1959–1966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Rayment I. 1984. Animal virus structure, vol. 1 Wiley Interscience, New York, NY [Google Scholar]
- 45. Rossmann M. G. 2000. Fitting atomic models into electron-microscopy maps. Acta Crystallogr. 56:1341–1349 [DOI] [PubMed] [Google Scholar]
- 46. Rossmann M. G., Bernal R., Pletnev S. V. 2001. Combining electron microscopic with X-ray crystallographic structures. J. Struct. Biol. 136:190–200 [DOI] [PubMed] [Google Scholar]
- 47. Wikoff W. R., et al. 2000. Topologically linked protein rings in the bacteriophage HK97 capsid. Science 289:2129–2133 [DOI] [PubMed] [Google Scholar]
- 48. Xiao C., et al. 2009. Structural studies of the giant mimivirus. PLoS Biol. 7:958–966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Yan X., et al. 2000. Structure and assembly of large lipid-containing dsDNA viruses. Nat. Struct. Biol. 7:101–103 [DOI] [PMC free article] [PubMed] [Google Scholar]