Abstract
Summary: A substantial amount of antisense transcription is a hallmark of gene expression in eukaryotes. However, antisense transcription was first demonstrated in bacteria almost 50 years ago. The transcriptomes of bacteria as different as Helicobacter pylori, Bacillus subtilis, Escherichia coli, Synechocystis sp. strain PCC6803, Mycoplasma pneumoniae, Sinorhizobium meliloti, Geobacter sulfurreducens, Vibrio cholerae, Chlamydia trachomatis, Pseudomonas syringae, and Staphylococcus aureus have now been reported to contain antisense RNA (asRNA) transcripts for a high percentage of genes. Bacterial asRNAs share functional similarities with trans-acting regulatory RNAs, but in addition, they use their own distinct mechanisms. Among their confirmed functional roles are transcription termination, codegradation, control of translation, transcriptional interference, and enhanced stability of their respective target transcripts. Here, we review recent publications indicating that asRNAs occur as frequently in simple unicellular bacteria as they do in higher organisms, and we provide a comprehensive overview of the experimentally confirmed characteristics of asRNA actions and intimately linked quantitative aspects. Emerging functional data suggest that asRNAs in bacteria mediate a plethora of effects and are involved in far more processes than were previously anticipated. Thus, the functional impact of asRNAs should be considered when developing new strategies against pathogenic bacteria and when optimizing bacterial strains for biotechnology.
INTRODUCTION
In eukaryotes, only a small fraction of the genome codes for proteins. The majority of the transcriptional output, up to 90%, consists of RNA that does not code for proteins (6). Major developmental and evolutionary differences between humans and other primates are thought to be caused by the action of regulatory noncoding RNAs originating from intergenic and noncoding regions. In addition, substantial antisense transcription from protein-coding regions in human cells has been reported (6, 33). These transcripts are called natural antisense RNAs (asRNAs) or cis-encoded natural asRNAs. Considerable antisense transcription has also been observed for mice (21.9 to 72% of all genes) (39, 112), Saccharomyces cerevisiae (27%) (17, 109), Drosophila melanogaster (16.8%) (112), and Arabidopsis thaliana (7.4%) (34), yet the functional relevance, if any, remains to be understood.
Until very recently, the transcriptomes of bacteria appeared to be much simpler, mainly because the bulk of the genomes consist of protein-coding genes. However, this picture is currently changing dramatically because of accumulating evidence from transcriptome studies suggesting that extensive antisense transcription also occurs in bacteria. Bacterial transcriptomes seem to be unexpectedly complex, with frequent antisense as well as inter- and intragenic transcription (18, 60, 72, 76). For some bacteria, even the majority of the genomic output on the RNA level seems to be noncoding (e.g., see references 60 and 76). In Synechocystis sp. strain PCC6803, ∼65% of all different individual transcripts are noncoding RNAs (ncRNAs), whereas the genome is 87% coding (60).
Naturally occurring asRNAs were first observed in bacteria more than 30 years ago (38, 47) and were postulated even earlier for bacteriophage λ (85). In archaea, the first case of the antisense control of gene expression was reported in 1993 for the extremely halophilic Halobacterium salinarium, lysogenic for phage φH, with an asRNA complementary to the first 151 nucleotides (nt) of the transcript T1 (88). Initially, the few examples of well-studied prokaryotic chromosome-, plasmid-, and phage-encoded asRNAs were considered to be exceptions rather than the rule. However, these examples illustrated that natural antisense transcripts are present in all three kingdoms of life, although in recent years, they have been investigated largely for eukaryotes.
New evidence suggests that the regulation of gene expression through cis-encoded asRNAs constitutes a distinct layer of control in bacteria. Here, we review this recent evidence within the context of this rapidly advancing field of molecular microbiology and aim to present a comprehensive view. Computational and experimental approaches for the discovery and characterization of bacterial regulatory noncoding RNAs and their targets have been reviewed recently (2, 4, 77, 84, 92) and are outside the scope of this review.
NEW EVIDENCE FOR SUBSTANTIAL NUMBERS OF DISTINCT asRNAs IN BACTERIA
Antisense transcription in bacteria did not become the focus of systematic genome-wide analyses until very recently, mainly because of three technical problems. Two of these problems are the lack of robust bioinformatic algorithms to specifically predict asRNAs and the fact that the measurement of antisense transcription in microarray analyses was thought to be an experimental artifact generated during cDNA synthesis (68). The third problem was the interpretation of experimental data. The earliest systematic study suggested that antisense transcription occurred in 3,000 to 4,000 open reading frames (ORFs) in Escherichia coli (75). Because such a low level of transcription was reported to occur virtually throughout the genome (75), it was considered difficult to differentiate asRNAs with regulatory functions from transcriptional noise. Georg et al. (27) demonstrated that it is possible to overcome all three obstacles by (i) the direct labeling of RNA instead of cDNA prior to hybridization on tiled microarrays to avoid unintended second-strand synthesis, (ii) rigorously comparing the results to computational predictions, and (iii) focusing on very highly expressed asRNAs. As a result, the experimentally confirmed number of highly expressed asRNAs in the model cyanobacterium Synechocystis PCC6803 was raised from 1 (21) to 73; an extrapolation suggested that at least 10% of all protein-coding genes are associated with an asRNA (27). Thus, the view on how to study antisense transcription in bacteria is changing due to progress in high-throughput RNomics methods such as tiling microarrays, direct RNA-labeling methods and, in particular, RNA deep sequencing.
A recent transcriptome analysis based on Illumina sequencing confirmed that widespread antisense transcription also occurs in E. coli by identifying about 1,000 different asRNAs (18). Using Rho factor chromatin immunoprecipitation and microarray (ChIP-chip) analysis, it was established that 25 E. coli asRNAs rely on Rho-dependent termination (69). The pausing of the RNA polymerase during Rho-dependent termination could be important for the mechanism of transcriptional interference (67), as we discuss further below.
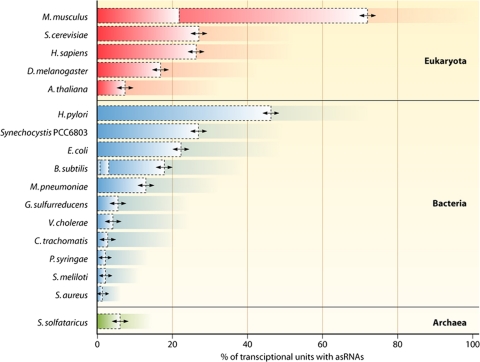
Using microarrays, the transcriptome of Bacillus subtilis was found to encompass asRNAs for 18% of a selected set of 506 genes (49) and 2.9% of all genes (72), and a recent differential RNA-Seq (dRNA-Seq) analysis revealed 29 asRNAs (37) that partially overlap with those found previously with high-density microarrays (72). For Synechocystis PCC6803, Mycoplasma pneumoniae, Sinorhizobium meliloti, Geobacter sulfurreducens, Vibrio cholerae, Chlamydia trachomatis, Pseudomonas syringae, and Staphylococcus aureus, antisense transcription rates were reported to be approximately 26.8%, 13%, 2 to 11%, 5.6%, 4.7%, 2.7%, 2.2%, and 1.3%, respectively (1, 5, 23, 27, 31, 54, 60, 71, 74). Additionally, 6.1% of all genes in the archaeon Sulfolobus solfataricus have asRNAs (Fig. 1) (107). The compact genome of the human pathogen Helicobacter pylori was, until recently, considered to be virtually free of noncoding RNA, including asRNAs. However, in studying the composition of its transcriptome by a combination of pyrosequencing and Illumina deep sequencing, along with introducing a novel approach to select for the 5′ end of primary transcripts (dRNA-Seq), Sharma et al. (76) found asRNAs for 46% of all annotated open reading frames. This observation is important because Helicobacter pylori has a very compact genome (1,667,867 bp [96]) and because this study was the first in which a bacterial transcriptome was analyzed exhaustively. Moreover, these data revealed that antisense transcription is an active, nonrandom process resulting from the genome-wide initiation of transcription rather than from read-through at leaky transcriptional terminators (76).
Fig. 1.
Antisense transcription is a hallmark of gene expression in all three domains of life. The number of reported antisense transcripts is plotted as a percentage of the total number of genes in the selected bacteria (1, 5, 23, 27, 31, 41, 49, 54, 60, 71, 72, 74–76, 81, 99, 110), various eukaryotes (17, 34, 39, 112), and archaea (91, 107). Dashed boxes indicate that publications with different numbers of asRNAs exist for a distinct organism (e.g., B. subtilis and Mus musculus). In the case of E. coli, the authors speculated that the high incidence of antisense transcription could be representing background transcription throughout the genome (75). The percentages are either directly stated in the respective publications or roughly calculated using the asRNA number and the number of annotated genes. H. sapiens, Homo sapiens.
Thus, a closer look at the bacterial kingdom shows that asRNAs are present in a wide range of individual species and families, and the likely ubiquitous distribution of asRNAs throughout the bacterial kingdom becomes obvious. It should be noted that the reported percentages of antisense transcripts in various bacteria (Fig. 1) indicate only current knowledge and will most likely change with further high-resolution transcriptome analyses. Observed differences in the percentages of antisense-associated genes might be strain specific or due to differences in experimental designs. In the case of RNA-Seq studies, the coverage should be high enough to saturate the whole transcriptome; otherwise, the number of asRNAs detected scales with the total number of reads rather than with genome size or actual numbers. Conversely, a subset of the reported RNAs could well be artifacts of cross-hybridization, second-strand synthesis, or other issues, which necessitates verification by complementary methods. Hence, the above-mentioned numbers are not perfectly comparable, but they give an overview and are the basis for further discussions. Compared to eukaryotes and archaea, it turns out that antisense transcription is as common in bacteria as it is in the other two domains of life (Fig. 1).
VARIOUS TYPES OF ANTISENSE TRANSCRIPTS IN BACTERIA
Bacterial asRNAs are rather diverse, and there is no generally shared feature other than the fact that transcription occurs from the antisense strand of a known transcriptional unit. However, they can be roughly classified based on their location as 5′-overlapping (divergent, head to head), 3′-overlapping (convergent, tail to tail), or internally located asRNAs. Transcripts from protein-coding genes with long 5′ or 3′ UTRs (untranslated regions), which overlap substantially with the mRNAs originating from other genes (e.g., in Listeria [62, 94], Synechocystis PCC6803 [27], and Anabaena sp. strain PCC7120 [36]), constitute an elegant way to achieve a regulatory connection between neighboring genes. In Anabaena PCC7120, the alr1690 gene has an extremely long 3′ UTR that overlaps the full length of the gene for the ferric uptake regulator furA. Indeed, in an Δalr1690 strain, increased levels of the Fur protein and an iron deficiency phenotype were observed (35, 36). For other asRNAs, the longer part of the transcript lies antisense to a gene, but the nonoverlapping part may contain a small open reading frame, as in the case of as_slr0882 in Synechocystis PCC6803 (27). Another example is the Pseudomonas fluorescens PF0-1 gene iiv14 (cosA), which overlaps the Pfl_0939 reading frame with 987 nucleotides of its 1,020-nucleotide total length. Both of these mRNAs are translated into proteins, and the product of cosA was demonstrated to be important for the efficient colonization of soil (80). Finally, there are also short asRNAs which overlap only the 5′ UTR of a gene (SyR7 in Synechocystis PCC6803 [27] and SymR in E. coli [40]) and that have some hallmarks of trans-acting noncoding RNAs. These observations demonstrate the extensive functional overlap with respect to the biological mechanisms involved with mRNAs, trans-acting noncoding RNAs, and asRNAs.
The sizes of asRNAs are very diverse. There are examples of rather short asRNAs of only 100 to 300 nt (e.g., SymR [40], GadY [66], and SyR7 [27]), but many asRNAs are substantially longer, ranging from 700 to 3,500 nt (3, 22, 27, 48, 86, 94). At least one example, in Prochlorococcus sp. strain MED4, is as long as 7,000 nt, overlapping 14 genes of a ribosomal protein operon (86). The steady-state levels of asRNAs range from being barely detectable to being in high abundance. For Synechocystis PCC6803, some asRNAs were reported to accumulate to levels comparable to those of strongly expressed protein-coding genes such as amt1, a gene that encodes an ammonium transporter (27).
MECHANISMS OF asRNA ACTION IN BACTERIA
Whereas knowledge of the number of chromosomally encoded cis-antisense RNAs in individual bacteria is rapidly advancing, information on the molecular mechanisms of individual asRNAs is growing at a much slower pace. In spite of this, functional characteristics have been well established for several phage- and plasmid-encoded asRNAs (for reviews, see references 8 and 102) and many trans-acting noncoding RNAs. Experimental analyses of a small number of chromosomally encoded asRNAs demonstrated that some of these functional mechanisms also apply to asRNAs. On the other hand, there are mechanisms that can uniquely be employed by asRNAs only. An overview of functionally characterized asRNAs is discussed below in detail and is summarized in Table 1.
Table 1.
Characteristic features of the functionally characterized bacterial antisense RNAs discussed in the texta
| asRNA | Host | Phylum | Length(s) | Functional mechanism(s) of asRNA action | Outcome | Reference(s) |
|---|---|---|---|---|---|---|
| AmgR | Salmonella enterica | Gammaproteobacteria | 1.2 kb | Codegradation; complementary to mgtC of the mgtCBR operon; likely RNase E dependent | Dynamic adjustment of MgtC and MgtB protein levels | 48 |
| GadY | Escherichia coli | Gammaproteobacteria | 105 nt, 90 nt, 59 nt | Cleavage of the bicistronic gadXW mRNA, likely by RNases III and E and other unknown factors | Enhanced stability of gadX mRNA | 65, 66, 90, 97 |
| SibABCDE | Escherichia coli | Gammaproteobacteria | ∼140 nt | Interference with translation and/or stability of mRNA suggested | Repression of IbsABCDE toxin synthesis | 25 |
| IsrR | Synechocystis PCC6803 | Cyanobacteria | 177 nt | Codegradation with isiA mRNA | Threshold linear response; delayed induction of isiA under stress and faster recovery | 21, 50 |
| mccA asRNA | Clostridium acetobutylicum | Firmicutes | 1,000 nt, 700 nt, 400 nt, 200 nt | Transcriptional interference with the convergent ubiG-mccBA promoter | Sulfur-dependent control of the ubiG-mccBA operon | 3 |
| OOP asRNA | Bacteriophage λ | Siphoviridae | 77 nt | Codegradation; RNase III-dependent cleavage of the mRNA cII | Repression of cII; involved in lysis-lysogeny decision; favors lysis | 44–46, 106 |
| CfrI | Cyanomyovirus S-PM2 | Myoviridae | 225 nt | Unknown | Unknown | 59 |
| Ribosomal protein operon/genomic island asRNAs | Prochlorococcus MED4 | Cyanobacteria | 3,500 nt, 7,000 nt | Masking of RNase E sites | Enhanced half-life upon phage infection | 86 |
| RNAα | Vibrio anguillarum | Gammaproteobacteria | 650 nt | Complementary to the fatB gene of the iron transport-biosynthesis operon | Repression of fatA and fatB under iron-rich conditions | 12, 73, 95, 103, 104 |
| RNAβ | Vibrio anguillarum | Gammaproteobacteria | 427 nt | Complementary to the 3′ region of fatA and the 5′ end of angR | Termination of transcription | 12, 73, 89 |
| RnaG | Shigella flexneri | Gammaproteobacteria | 450 nt | Complementary to the 5′ UTR of the icsA (virG) virulence gene; transcriptional interference with the convergent icsA promoter and transcription attenuation of icsA mRNA | Premature termination of transcription and direct reduction of icsA transcription | 28 |
| SymR | Escherichia coli | Gammaproteobacteria | 77 nt | Overlaps the symE mRNA 5′ end and RBS, preventing the initiation of translation | Control of SymE translation | 40 |
| ureAB asRNAs | Helicobacter pylori | Epsilonproteobacteria | 290 nt | Processing or termination | pH-dependent regulation of ureAB | 105 |
RBS, ribosome binding site.
Alteration of Target RNA Stability
The interaction of an asRNA with its target RNA alters the secondary structure of both interacting molecules and results in duplex (double-stranded RNA [dsRNA]) formation. Those structural changes influence the stability and half-life of the RNAs, with various outcomes. In some cases, duplex formation results in the rapid and complete degradation of both RNAs. A prominent example is the 77-nt OOP asRNA of the λ phage. The RNA is complementary to the 3′ end of the λ cII-repressor mRNA. The overexpression of the OOP asRNA from a plasmid vector results in an RNase III-dependent cleavage of the cII mRNA, initially at two sites, one in the 3′ end of the coding region and one in the cII and O gene intergenic region (44–46).
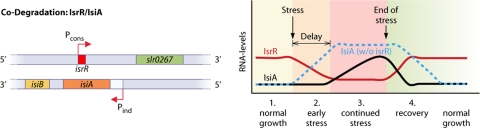
The isiA/IsrR sense/antisense pair of Synechocystis PCC6803.
A well-studied example of codegradation is the isiA/IsrR sense/antisense pair in Synechocystis PCC6803 (21) (Fig. 2). The IsiA protein belongs to the iron stress response regulon, and the expression of IsiA results in a massive reorganization of the photosynthesis apparatus (21) by the formation of ring-like structures around photosystem I. Thus, the expression of isiA requires tight regulation, which is in part achieved by IsrR. The transcript accumulations are strictly inversely related to each other, and therefore, both RNAs exist as almost mutually exclusive species. The asRNA IsrR is transcribed from a constitutive promoter, whereas the transcription of isiA is induced upon iron, redox, or light stress. When both species are expressed simultaneously, they form an RNA duplex that is immediately degraded by an unknown mechanism. Therefore, the message cannot accumulate until the number of isiA mRNA molecules titrates out the number of asRNA molecules. The overexpression of both the asRNA as well as the isiA fragment complementary to IsrR in trans also results in a knockdown of its respective counterpart. Interestingly, the single-stranded molecules alone are very stable, with a reported half-life of more than 45 min for IsrR (50). As a result, IsrR causes not only a delay of isiA expression in the early stress phase but also a faster depletion of isiA during recovery from stress, when the transcription of isiA ceases. This mode of regulation is known as the “threshold linear response” (50) and is discussed below. The extraordinarily long half-life of IsrR under standard conditions has the implication that a high steady-state level of the asRNA could be maintained with a moderate or low rate of de novo transcription. Consequently, threshold regulation by IsrR is very efficient from an energetic point of view.
Fig. 2.
Mechanisms of bacterial asRNAs: codegradation of IsrR together with its target, the mRNA isiA, in the cyanobacterium Synechocystis PCC6803. The asRNA IsrR originates from the central part of the isiA gene from a constitutive promoter (Pcons). The isiA gene is under the control of the inducible promoter Pind. Under early-stress conditions, isiA transcription becomes activated. Both transcripts are codegraded. The mRNA cannot accumulate as long as IsrR > isiA, and no protein is made. When stressful conditions persist, IsrR is still transcribed, but its turnover is very high, and consequently, it becomes titrated out. The mRNA accumulates, translation occurs, and supercomplexes between IsiA and photosystem I are formed. As a result, IsrR causes a threshold linear response of gene activation and inactivation, with an initial delay and a fast degradation after the stress compared to the expected kinetics of gene expression in the absence of the asRNA (blue dashed line).
E. coli GadY asRNA.
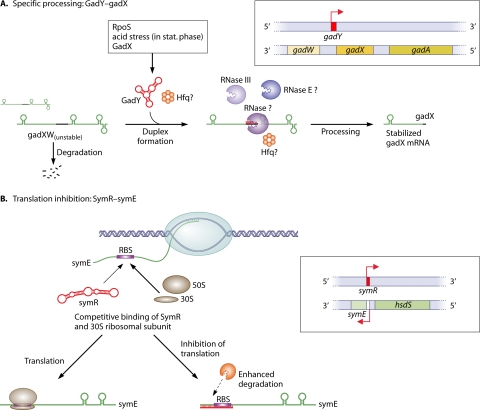
The interaction of mRNA and asRNA does not necessarily result in rapid degradation. An RNA duplex can also generate a specific processing site, which may lead to a translationally inactive mRNA or yield a mature or stabilized form of the mRNA. The E. coli GadY asRNA is involved in the response to acid stress. Its gene lies antisense to the 3′ end of gadX, which is an activator of the glutamate-dependent acid resistance system (gad system) (66) and was originally named IS183 (93). GadY posttranscriptionally controls gadX levels by inducing the cleavage of the bicistronic gadXW message (Fig. 3A). Processed gadX and gadW mRNAs alone have an enhanced stability (65, 97). The processing machinery has been investigated in more detail. Of the five known E. coli RNA endonucleases, only RNase III, and not RNase E, G, BN, or P, seems to be involved. The factor inferring the main activity could not be detected (65), raising the possibility of thus-far-unknown proteins being involved in regulatory RNA mechanisms. However, in another study the levels of GadY and gadX mRNA were significantly decreased in an RNase E knockout strain in comparison to controls, and the survival of the cell under acid stress conditions was reduced, indicating that RNase E is somehow necessary for processing (90).
Fig. 3.
Two mechanisms for bacterial asRNAs that overlap at the 3′ or the 5′ end of a protein-coding gene. (A) Specific processing of the gadXW transcript through the asRNA GadY. GadY originates from a gene overlapping the 3′ end of the gadX gene that is highly induced during stationary phase and is dependent on the alternative sigma factor RpoS (66). GadY base pairs with the 3′ untranslated region of the gadX mRNA and confers increased stability through posttranscriptional processing, which allows gadX and gadW mRNA accumulation and increased expression levels of the downstream acid resistance genes (97). Protein factors involved include RNase III, other so-far-unknown RNases (65), probably RNase E (90), and possibly Hfq. (B) Inhibition of translation through SymR (40, 41). SymR is complementary over its full length to the symE 5′ UTR, including the ribosome binding site (RBS), and probably causes a block in ribosome binding and, to a lesser extent, enhanced degradation of the untranslated mRNA. GadY and SymR are drawn according to their RNAfold maximum free energy (mfe) secondary structures.
Despite the lack of a larger number of well-characterized examples, there are likely to be more asRNAs that contribute directly to target stabilization or positively influence target expression.
Interplay between bacterial asRNAs and RNase E.
Evidence for an interplay between asRNAs and RNase E was found, with asRNAs overlapping the 5′ region of an mRNA that inhibits RNase E-dependent decay, at least in vitro (56). asRNA-dependent protection from RNase E was also suggested to be involved in the phage-host interplay. In the cyanobacterium Prochlorococcus MED4, infection by the lytic phage P-SSP7 triggers a shortened half-life of most host mRNAs. This process is likely supported by the upregulation of host RNase E, as evidenced by its increased mRNA level (53). In addition, the levels of 40 other mRNAs increased upon phage infection, raising the question of how these mRNAs were selectively upregulated or stabilized. Interestingly, some of these mRNAs have extraordinarily long asRNAs (up to 7 kb) that are coinduced (86). The interaction of these long asRNAs with their complementary polycistronic mRNAs masks RNase E cleavage sites and prevents RNase E-dependent degradation (86). It is so far unclear if the asRNAs are a host defense mechanism or if they are hijacked by the phage to ensure the expression of important host factors (86).
RNase E is also involved in the asRNA-dependent degradation of mRNAs, as shown for the AmgR/mgtC system in Salmonella enterica (48). A proposed endonucleolytic mRNA destabilization pathway by RNase E or other RNases would be consistent with the function of further bacterial asRNAs.
Modulation of Translation
The enterobacterial symR/SymE sense/antisense pair.
In some cases, the degradation of the RNA is of secondary importance for the suppression of gene expression. An example is the regulation of the SOS response-induced protein SymE in enterobacteria. It is thought to be a toxin-like RNA endonuclease that is tightly controlled under standard conditions. One of at least three repression mechanisms requires the asRNA SymR (40). SymR overlaps the 5′ end of the symE mRNA, including the ribosome binding site and the AUG start codon (Fig. 3B). Therefore, the formation of a symE mRNA/SymR duplex is incompatible with the binding of the 30S ribosomal subunit and prevents the initiation of SymE translation. Indeed, in an E. coli ΔsymR strain, protein levels increased more than 7-fold, whereas the mRNA amounts increased only 3-fold. It is not clear if the binding of the asRNA directly triggers the enhanced degradation of symE mRNA or, rather, if it is a secondary effect due to the absence of translating ribosomes on the mRNA. It was shown that RNA decay was not RNase III dependent (40). Interestingly, the SymR-to-symE mRNA ratio is 10:1, even after symE induction (40). This indicates that the repression of symE is not complete and that SymR may be sequestered by additional targets or that the binding of the ribosome to the symE mRNA is favored in comparison to SymR. This example shows that both RNA degradation and the inhibition of translation can act simultaneously and can contribute jointly to the repression of a target gene.
Regulation of translation may not necessarily include the ribosome binding site.
A similarly negative regulation of translation is likely for many asRNAs that overlap the ribosome binding site or the start codon of their target. Furthermore, recent findings for trans-acting noncoding RNAs showed that the region sufficient to achieve translation inhibition may not necessarily include the ribosome binding site. The binding of a regulatory RNA in a window 5 codons after the start was also found to inhibit ribosome binding (7). Conversely, binding far upstream of the ribosome binding site was able to repress (istR) (16) or induce (dsrA) (57) translation. Extended sense/asRNA duplexes may also be stable enough to inhibit or slow down the elongation of the 70S ribosome far downstream of the translation start site.
Transcription Termination
The Vibrio anguillarum iron transport-biosynthesis operon.
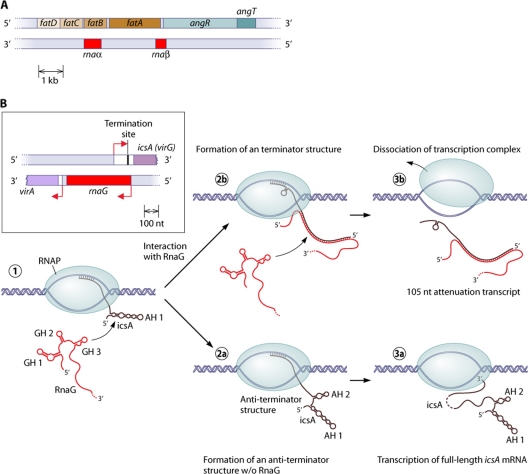
In addition to posttranscriptional mechanisms, there are also mechanisms that directly influence the transcription of target genes. The Vibrio anguillarum iron transport-biosynthesis operon, which includes four ferric siderophore transport genes (fatDCBA) and two siderophore biosynthesis genes (angR and angT), also gives rise to two asRNAs (RNAα and RNAβ) (12, 73, 95, 103, 104) (Fig. 4A). Whereas RNAα represses fatA and fatB expressions under iron-rich conditions, RNAβ causes the differential transcription of the full-length operon fatDCBA-angRT and a shortened fatDCBA message (89). The short form is about 17 times more abundant than the long form. RNAβ is complementary to the 3′ region of fatA and the 5′ end of angR. The binding of the asRNA to the growing polycistronic fatDCBA message leads to transcription termination at a potential hairpin close to the fatA stop codon. This also occurred in an in vitro transcription assay with the external addition of RNAβ, suggesting that other mechanisms, such as codegradation or transcriptional interference, could be excluded (89).
Fig. 4.
Mechanisms of transcription termination by bacterial asRNAs. (A) Organization of the Vibrio anguillarum iron transport-biosynthesis operon. The asRNA RNAβ induces transcription termination at a predicted stem-loop after the fatABCD part of the mRNA (89). (B) Schematic representation of the virA-rnaG-icsA (virG) locus of the Shigella flexneri pVIN plasmid and visualization of the proposed transcription termination mechanism (28). The 5′ region of the newly transcribed icsA message forms the stem-loop structure AH1 (1), and, without binding to RnaG, another stem-loop (AH2) is formed. This 5′ structure resembles an antiterminator structure (2a), and the full-length mRNA could be transcribed (3a). When RnaG is present, it forms a heteroduplex with the growing icsA message. This inhibits the formation of the antiterminator structure, and a terminator hairpin is formed (2b). Subsequently, transcription is attenuated, and a ∼100-nt abortion RNA is released (3b).
RnaG controls the Shigella flexneri virulence gene icsA.
Another example is the Shigella flexneri virulence gene icsA (virG), which is controlled by the asRNA RnaG. The asRNA represses the transcription of its target by transcriptional interference (described below) and by transcription attenuation (28). The 5′ part of the icsA RNA forms two long hairpin motifs that presumably resemble an antiterminator structure. The binding of the asRNA to the actively transcribed mRNA inhibits the formation of the antiterminator and favors the formation of a terminator hairpin (Fig. 4B). This proposed mechanism is supported by structural probing experiments. Furthermore, mutations that avoid base pairing in the terminator stem significantly reduce asRNA-induced transcription attenuation (28).
Transcriptional Interference
In the fascinating regulatory mechanism of transcriptional interference, divergently or tandemly transcribed promoters influence each other. Therefore, the process of transcription is the point at which regulation takes place, and the resulting RNA could be a side effect. The recent discovery of a huge number of asRNAs and other noncoding RNAs in bacteria also means that there are unexpectedly large numbers of closely spaced promoters, dramatically increasing the potential for transcriptional interference.
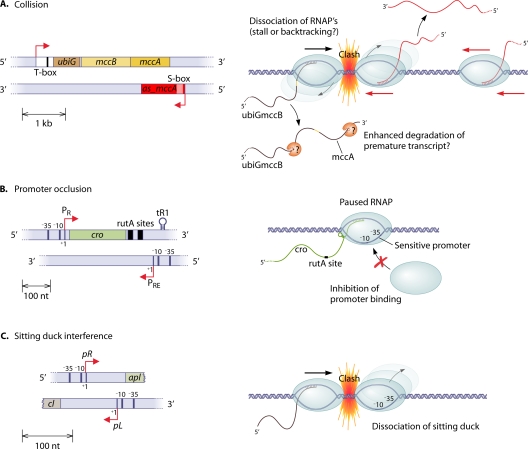
Three mechanisms (78, 83) that contribute in various amounts to total interference are discussed below (Fig. 5).
Fig. 5.
Transcriptional interference: asRNAs as a by-product of interfering promoters. (A) Proposed collision mechanism for the ubiG-mccAB-as_mccA system (as_mccA stands for mccA antisense RNA). The two divergently elongating RNA polymerases, transcribing the asRNA and the ubiG-mccAB operon, collide and give rise to the 1,000-nt fragment for as_mccA, which represents the sole known mechanism of termination. Short fragments for the mRNA were not detected, indicating a rapid degradation of the prematurely terminated transcript. (B) Promoter occlusion. (C) The sitting-duck mechanism of transcriptional interference.
Collision.
The collision of two divergently elongating RNA polymerase complexes results in the premature termination of one or both transcription events. The clash is not likely to be a direct steric interaction but rather a longer-distance interaction (e.g., electrostatic) or an effect of the bow wave of positively supercoiled DNA in front of an elongating RNA polymerase (14). There are several possible fates for the RNA polymerase after the clash, including the dissociation of one or both complexes, the backtracking of one complex, or a stall of the polymerases (14, 83). A collision of this type was shown for the elongating polymerase complexes involved in the transcription of the gal7 and gal10 genes in yeast (70). The model shows that interference by collisions is most effective when the distance between the divergent promoters, and, hence, the overlapping region, is long. Mathematical modeling suggests that relatively small differences in promoter strengths can result in a strong asymmetric interference of a stronger “aggressive” promoter on the weaker “sensitive” promoter, especially in cases where there is a long sequence overlap (83). This fact is especially interesting given that some asRNAs tend to be rather long (3, 27, 86, 94). A likely example of the collision mechanism in eubacteria was found for Clostridium acetobutylicum (3). The ubiG-mccBA operon contains genes necessary for the conversion from methionine to cysteine (Fig. 5A). Consequently, its expression is upregulated in the presence of methionine and downregulated in the presence of cysteine. The main player mediating this regulation is an asRNA that is regulated in response to sulfur availability and shows an inverse relationship to the ubiG-mccBA message. This asRNA is up to 1,000 nt long, with three major shorter fragments with lengths of 700 nt, 400 nt, and 200 nt. Interestingly, the transcript ends of the longer fragments do not correlate with obvious terminator structures, nor does the fragment pattern change in RNase III or RNase J1/J2 mutants, indicating an alternative termination mechanism and ruling out codegradation. Nevertheless, destroying the asRNA promoter region also corrupts the sulfur-specific regulation of ubiG-mccBA, and the regulation cannot be reestablished by expressing the asRNA in trans (3). These observations clearly indicate regulation by transcriptional interference. Interestingly, the transcription of as_mccA appears to be constitutive, but the accumulation of the full-length asRNA is controlled by an S-box riboswitch. The binding of S-adenosylmethionine stabilizes the terminator conformation of the riboswitch. Without methionine as a sulfur source, no termination occurs, and full-length as_mccA is expressed (3). This example strikingly demonstrates the complexity and flexibility of RNA-based gene regulation.
Promoter occlusion.
Promoter occlusion occurs when an elongating RNA polymerase coming from an “aggressive” promoter passes over a “sensitive” promoter element so that it prevents the formation of an initiation complex at that “sensitive” promoter (Fig. 5B). Mathematical modeling showed that efficient interference by occlusion normally requires a very strong “aggressive” promoter, due to the short time window in which the elongating RNA polymerase blocks the “sensitive” promoter region by its passage (83). Nevertheless, a study of the interference of the divergent λ phage PR and PRE promoters (67) showed that the pausing of RNA polymerase at a tR1 site (because of Rho-dependent termination) opposite the “sensitive” promoter strongly enhances interference. That study also showed that promoter occlusion in combination with pausing can lead to strong asymmetric interference between two promoters of an otherwise relatively similar strength.
Sitting-duck interference.
With sitting-duck interference, RNA polymerase bound at an open complex of the “sensitive” promoter is removed by the collision of another elongating RNA polymerase complex before the first polymerase can proceed to elongation (Fig. 5C). Sitting-duck interference seems to be the major type of interference between the lytic-phase promoter (pR) and the lysogenic promoter (pL) in bacteriophage 186 (10, 83). Modeling studies proposed this mechanism to be the strongest interference mechanism when promoters are closely spaced and of moderate strength; the effect is maximized when the on-rate of RNA polymerase and the initiation rate of elongation at the “sensitive” promoter contribute equally to promoter strength (83).
Environmental Cues and asRNA-Mediated Regulatory Functions
If there is a regulatory relationship, it appears likely that the expression level of an asRNA is somehow coupled to the expression level of the corresponding mRNA. Moreover, as expected for a regulatory factor, some asRNAs respond to changes in the environment and hence become differentially expressed under certain conditions. There are many examples of environmental conditions impacting the expression of asRNAs in various bacteria. Prochlorococcus MED4 asRNA levels respond to phage infection and light conditions (86, 87); B. subtilis asRNA levels respond to sporulation (81); Listeria monocytogenes asRNA levels respond to O2 concentrations, temperature (94), or intracellular conditions (62); Synechocystis PCC6803 asRNA levels respond to variations in CO2 supply, light conditions, and iron availability (21, 27, 60); and S. enterica serovar Typhi asRNAs were shown to have growth state-dependent expression patterns (13).
Mineral homeostasis and pathogenicity.
In Clostridium acetobutylicum, the transcription of the as_mccA asRNA is controlled by sulfur availability, and as_mccA in turn controls a sulfur metabolic operon, as discussed above (3). In addition, the Synechocystis PCC6803 IsrR asRNA controlling isiA abundance is an example of the regulation of iron homeostasis, and RNAα and RNAβ of V. anguillarum represent another. There are now several reports of the role of asRNA-mediated regulatory effects relevant for the pathogenicity of the corresponding bacteria. The asRNA AmgR of S. enterica is complementary to the mgtC part of the mgtCBR operon, which is involved in pathogenicity and Mg2+ homeostasis. AmgR acts predominantly on the mgtC part of the message and is likely a timing device that dynamically adjusts the levels of MgtC and MgtB, which may ensure different expression modes of mgtCBR for host interactions and low-Mg2+ conditions (48). The expressions of both AmgR and mgtCBR are regulated by the response regulator PhoP, but the mgtCBR promoter has a higher affinity for the regulator. Thus, low levels of activated PhoP result in the expression of the mgtCBR message with an MgtC/MgtB ratio needed for growth under low-Mg2+ conditions and survival in macrophages. High levels of active PhoP also induce AmgR expression, leading to the degradation of mgtC and the realization of a different MgtC/MgtB ratio (48).
In H. pylori, the ureAB operon is posttranscriptionally controlled by a thus-far-unnamed asRNA (105). UreA and UreB are the enzymatically active subunits of the urease enzyme necessary for the cytoplasmic pH homeostasis of H. pylori in its acidic environment. However, the activity of the urease could be lethal at a neutral pH. Thus, the expression is regulated by the HP0165-HP0166 two-component system. Under acidic conditions, HP0166 is phosphorylated and activates the transcription of ureAB into a 2.7-kb mRNA. In contrast, a neutral pH results in unphosphorylated HP0166, which activates the expression of the asRNA, leading to the subsequent accumulation of a truncated 1.4-kb ureAB transcript. The asRNA ensures a rapid shutdown of ureAB expression under neutral-pH conditions (105). The underlying mechanism, e.g., asRNA-dependent processing or termination, is so far unknown.
RNA-RNA INTERACTION AS A PREREQUISTE FOR asRNA-BASED REGULATION
Except for transcriptional interference, the physical interaction of the asRNA with its target is an essential prerequisite for any asRNA-based regulatory mechanism. The affinity of two complementary RNAs and the free energy of the duplex are given by the length of the RNA-RNA interaction and the nucleotide composition. At first glance, one would expect that perfectly complementary RNAs easily form the thermodynamically favored full-length double-stranded RNA helix. However, the real situation is more complex. Both RNAs have their individual secondary structures; all nucleotides buried within a stable intramolecular helix cannot interact with complementary bases of a second RNA. Single-stranded or weakly structured complementary regions are necessary for the first interaction step (9, 42, 63). Furthermore, the coding regions of bacterial mRNAs are frequently covered by elongating ribosomes, and finally, both RNAs could interact with RNA binding proteins, which interfere with asRNA-mRNA interactions. The importance of structural accessibility is exemplarily illustrated by the E. coli Ibs/Sib type I toxin-antitoxin system. Sib stands for short intergenic abundant sequences, and Ibs stands for induction brings stasis, and there are five such loci in E. coli K-12 (25), called IbsA/SibA to IbsE/SibE. The IbsABCDE toxins are conserved, very hydrophobic, 18- to 19-amino-acid proteins. Despite the high sequence similarity between the ibs toxin mRNA sequences, each asRNA (except SibA) recognizes and represses only its cognate mRNA target. SibC contains two, likely structurally favored, target recognition domains, which are responsible for the initial interaction and target recognition. These domains target highly variable regions in ibsC and are responsible for the specificity (32).
When a first interaction is achieved, the RNAs likely form a full-length duplex over a certain time scale, which is then a substrate for the dsRNA processing RNase III (19). However, on a relevant time scale, the biologically active asRNA/mRNA complex is not necessarily the full-length heteroduplex. Several plasmid-borne asRNAs form functionally relevant four-way junction structures with their targets (43). Indeed, several asRNAs discussed in this review do not require RNase III but rather require RNase E or an unknown RNase for their mechanism (90) (SymR [40] and AmgR [48[), which indicates that there are functional complexes that are not completely paired.
BIOLOGICAL RELEVANCE OF BACTERIAL asRNAs
The examples presented here provide an overview of the functions of cis-asRNAs in bacteria. In light of the large number of asRNAs in diverse bacteria, the question arises regarding what the benefit of asRNA-based gene regulation is compared with regulatory proteins on the one hand and trans-acting noncoding RNAs on the other hand.
Quantitative Aspects of Regulation through RNA
Deterministic view.
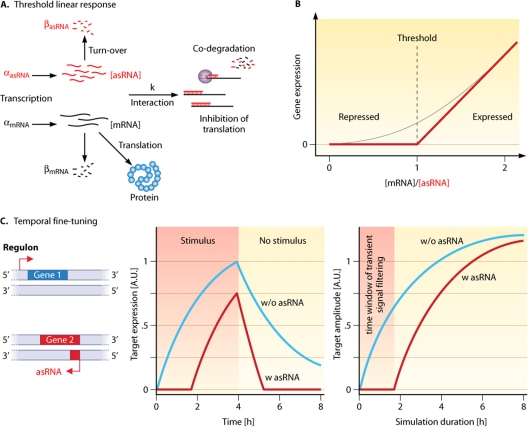
A theoretical framework for the comparison of the quantitative aspects of RNA-based (cis-asRNAs and trans-acting noncoding RNAs) and protein-based gene regulation has been developed in several studies. Regulatory RNAs that interact stoichiometrically with their target and are noncatalytic (i.e., they are codegraded with or sequestered by their target, e.g., IsrR [21, 50]) can be described by a threshold linear response model (52) (Fig. 6A and B). For the protein-based regulation of transcription, target mRNA expression is always proportional to promoter activity, resulting in a linear dependency on the protein regulator (52). For RNA-based regulation, target expression is dependent on the target promoter activity and the number of regulatory RNA molecules available for posttranscriptional regulation (52). The expression of the target is, roughly said, repressed as long as the transcription of the regulator exceeds the transcription of the target (repressed regime), and its expression increases linearly when the mRNA concentration outruns that of the regulator (expressed regime). The threshold is defined by the transcription (and degradation) rate of the regulatory RNA and could be dynamically adjusted by changing the transcription rate (52). This linear threshold behavior was experimentally verified for the trans-acting noncoding RNA RyhB and the cis-asRNA RNA-OUT (52). The quantitative impact of the IsrR asRNA in the IsrR/isiA system was also investigated theoretically and experimentally (21, 50). In this case, and in addition to the threshold linear response, the asRNA causes an expression delay of the mRNA compared to the other genes in the same regulon (Fig. 6C). Because of the great impact of IsiA on the photosynthesis apparatus (21), it is advantageous for the organism to express isiA later than other iron-stress-inducible genes. This delay of approximately 17 h (21) is controlled by a fixed steady-state amount of IsrR, as illustrated in Fig. 6C. In the case of only a short stress period, the protein is not expressed at all; in other words, transient input signals are filtered out (Fig. 6C). Furthermore, recovery to the prestress state is faster due to the rapid depletion of the target by IsrR and other regulatory RNAs (50, 79).
Fig. 6.
Quantitative effects of asRNAs on the modulation of gene expression in the threshold linear response model. (A) In the threshold linear response, the mRNA and its asRNA regulator interact in a stoichiometric way with each other. The transcription rates (α) and the individual degradation rates (β) define the concentrations of mRNA ([mRNA]) and asRNA ([asRNA]) that are available for interaction with the rate (k) to be sequestered or codegraded (also see Fig. 2 for the special case of IsrR/isiA codegradation that follows this model) (50). (B) Graphical representation of the threshold linear response caused by the presence of an asRNA. The threshold is set by the ratio of mRNA/asRNA (52). The black line illustrates the theoretical behavior if the asRNA and target interact completely and instantly. In reality, there is no sharp transition point, and the actual response depends on the interaction rate between the regulator and target (illustrated by the gray line). (C) The expression of genes belonging to one and the same regulon is temporally shifted by the presence of an asRNA. Without an asRNA regulator, a gene is expressed directly upon the onset of a stimulus, and when the stimulus disappears, the mRNA level declines relatively slowly via its intrinsic turnover rate (blue line, middle). The asRNA facilitates a temporal delay of gene expression and a faster expression cutoff due to codegradation with its target (red line, middle). If a stimulus is only of a transient nature, e.g., because of short-term environmental changes or noise in upstream regulatory loops, it is filtered by the function of the asRNA (right). A.U., arbitrary units. (The two plots are adapted from reference 50 with permission of Elsevier.)
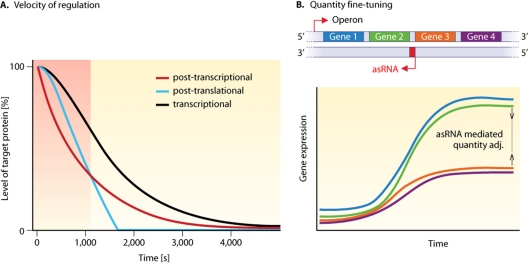
Shimoni et al. (79) compared the dynamics of regulation by posttranslational protein-protein interactions, transcriptional regulation, and posttranscriptional regulation by regulatory RNAs. If the respective regulators are already present in the cell when regulation is turned on, RNA-based regulation is faster than transcriptional regulation but slower than the protein-protein interaction. However, if the regulators have to be newly synthesized upon a sudden stress, RNA-based regulation outperforms the other two mechanisms (Fig. 7A). Hence, this kind of regulation is advantageous when fast responses are needed (79). Common to all these models is that the quantitative properties of RNA-based regulation are discussed for a bacterial population as a whole, in which the measured or modeled expression values are steady-state levels averaged over a large number of cells (deterministic view).
Fig. 7.
Dynamic effects of asRNAs on the regulation of gene expression. (A) The speed of a regulatory response to environmental stress depends on the level at which control is exerted. The plot illustrates an example where the regulators have to be newly synthesized upon stress. (Adapted from reference 79 with permission of Macmillan Publishers Ltd. Copyright 2007.) In the time window soon after the onset of the downregulation, posttranscriptional regulation by RNAs is the fastest (gray region). (B) Schematic visualization of an asRNA-regulated, uncoordinated expression of genes within an operon, thereby tuning the amounts of gene products made from the different genes. Thus far, this mechanism has been described only qualitatively (66, 89, 97).
Stochastic view of RNA-based regulation.
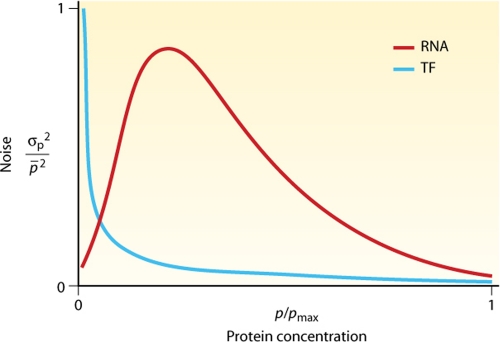
When the focus is shifted from the deterministic view to the individual consideration of single cells in a population, stochastic effects come into play. Biological processes like transcription are affected by stochastic fluctuations, such as transcriptional bursts (29). This necessarily results in gene expression noise, which describes how far the expression value for each single cell differs from the mean gene expression level of the population. The noise of transcriptional regulation was compared with the noise resulting from posttranscriptional regulation by regulatory RNAs (58). It was shown that regulatory RNA can suppress the expression of a gene more reliably and with more resistance to noise than what is possible by transcriptional regulation in the repressed regime (51, 58) (Fig. 8). This is especially important when the expressed gene has a great impact on the organism, such as the cases for isiA (21), symE (SOS response) (40), and potentially harmful genetic elements such as the toxins in type I toxin/antitoxin systems (24) or transposons (5, 64, 82). In contrast, the crossover regime, in which the mRNA and regulator have similar concentrations (near the threshold), is extremely vulnerable to noise (58) (Fig. 8). This leads to high levels of intrinsic noise for intermediate-to-high protein output levels compared with protein-based transcription regulation. A systematic investigation of genes with asRNA in yeast showed that they do indeed have a higher level of gene expression variability (108). These noise effects should make regulatory RNAs disadvantageous for the accurate adjustment of protein concentrations. However, in some cases, the enhanced noise might be of biological relevance, as it may lead to an enhanced phenotypic heterogeneity of cells within a genetically uniform microbial population and probably makes the population as a whole more resistant to rapid environmental changes. Furthermore, several bacterial cell fate decisions (e.g., persistence and many bet-hedging strategies) require noise at the decision-making point (20, 98).
Fig. 8.
Predicted effects of asRNAs on the level of biological noise in gene expression. The noise properties of transcription factor (TF)- and asRNA (RNA)-based regulation depend on the relative output protein level. For a low protein output (repressed regime), the noise of RNA-based posttranscriptional regulation is lower (gray region). For an intermediate-to-high protein output, the noise of transcriptional regulation is clearly lower. (Adapted from reference 58 with permission of Macmillan Publishers Ltd., copyright 2008.)
Other Aspects of asRNA-Based Regulation
Previously, other mechanisms of asRNA function such as transcription termination (28, 89) or transcript stabilization (66, 90, 97) were investigated only qualitatively. Because the transcription termination mechanism is not dependent on codegradation, this kind of regulation of the catalytic activity of the asRNA is possible if the asRNA dissociates from its target after action. In the case of a stoichiometric interaction, quantitative parameters can also be described by the threshold linear response model. Quantitative aspects of transcriptional interference have been modeled (83), but the focus lies on factors that influence the interference, whereas aspects of gene regulation with and without interference have not been discussed.
Protein regulators such as transcription factors often control regulons consisting of many genes, and operon structures are generally transcribed as one unit. asRNAs can be utilized to fine-tune this rather rough and more global gene regulation. They can set temporal gene expression thresholds (21, 50) (Fig. 6C) and implement a quantitative adjustment of the mRNA amounts from genes within an operon (Fig. 7B). This could be achieved by the specific processing of the polycistronic message (in the case of gadY [Fig. 3A] [66, 90, 97] or AmgR [48]) or by alternative termination (in the case of RNAβ [Fig. 4A] [89]).
Antisense transcription appears to also be important for the linkage of neighboring gene expression. This could be achieved by asRNAs that overlap more than one gene, by overlapping mRNAs, or by bidirectional promoters (as shown for yeast [108]). This linkage may also extend to the conservation of gene arrangements on evolutionary time scales, as discussed previously for the lytic cyanophage S-PM2 that infects marine Synechococcus strains (59). In this cyanomyovirus, an asRNA named cyanophage functional RNA I (CfrI) was found linking two separate genetic elements. One element is the psbA gene for the viral form of the D1 protein of photosystem II, and the other is the gene encoding the homing endonuclease F-CphI. This enzyme serves as the active homing endonuclease for the self-splicing group I intron located within the phage psbA gene, as was demonstrated experimentally (111). Therefore, a possible function was discussed for CfrI as physically connecting the F-CphI gene to its target intron, driven by a possible protective function for the psbA 3′ exon (59), which might remain ribosome-free for some time due to the slow splicing of the intron. Analysis of data sets from the global ocean survey predicted the presence of CfrI in 9 of the 11 scaffolds tested, implying an evolutionary conservation of this situation (59).
Finally, asRNAs can suppress gene expression, and they are part of regulatory circuits that respond to external and internal signals. There are several examples of genes with more than a single asRNA (15, 27, 60, 76), and together with regulatory proteins and other ncRNAs, several input signals can be integrated to adjust the expression of the target gene to the actual need.
Comparison to trans-Acting Noncoding RNAs
Examples for the various mechanisms used by experimentally characterized asRNAs are summarized in Table 1. Bacterial asRNAs show substantial similarities to trans-acting noncoding RNAs, particularly with regard to mechanisms that inhibit translation or decrease target stability. Both types of regulatory RNAs (i) form Watson-Crick hydrogen bonds with complementary bases in their RNA target, (ii) interact stoichiometrically with their target, and (iii) can be codegraded or sequestered by interactions with their target.
However, there are also several important differences. (i) asRNAs have complete or extended complementarity with their target and can form more stable RNA duplexes. This may have implications for binding kinetics, and the interaction may be independent of proteins such as Hfq. (ii) asRNAs originate from the same genomic region as their target. This could be a key advantage because the space requirements are negligible, and an organism can theoretically afford one or more RNA regulators for every single gene without the need for an increase of the genome size. Recent transcriptome studies with high-throughput methods showed that this situation is not too far from reality for some bacteria (76). Furthermore, compared to other regulators, new asRNAs should easily originate during evolution because a small number of point mutations would yield an active promoter. The sequence complementarity would be automatic. (iii) The cis position has the additional effect that the asRNA and target are transcribed in close steric proximity. This should result in an enhanced local concentration and facilitate interactions. Furthermore, there is evidence that the spatial pattern of transcription is more important than was anticipated until very recently. Some mRNAs in E. coli and Caulobacter crescentus show only limited diffusion from their sites of transcription, indicating some kind of spatial organization (61). (iv) Transcriptional interference relies on transcription from closely spaced promoters, and therefore, this kind of regulation is restricted to asRNAs. Transcriptional interference and other asRNA mechanisms can act additively to enhance the regulatory effect on the target (28). (v) The remarkable length of some asRNAs (up to more than 7,000 nt [86]) is another major difference in comparison to trans-acting noncoding RNAs. Length could be advantageous for the transcriptional interference collision mechanism (83) or other unknown mechanisms.
CONCLUDING REMARKS AND FURTHER DIRECTIONS
Recently used techniques such as next-generation sequencing, dRNA-Seq, and tiling microarrays have made it possible to determine the complete transcriptome of an organism. Applied to bacteria, these methods have dramatically changed the view of antisense transcription to being a common and widespread phenomenon rather than being exceptional. In fact, the percentage of sense/antisense pairs can be as high as that in eukaryotes. Surely, new transcriptome projects will further increase our knowledge of the prevalence of asRNAs in bacteria. Among other observations, it is very interesting that bacteria with streamlined genomes and reduced sets of regulatory proteins (such as M. pneumoniae [31], H. pylori, and Prochlorococcus MED4 [87]) have substantial numbers of asRNAs. Nevertheless, there are many open questions. (i) Is there a more frequent association of asRNAs with genes of a particular functional class, e.g., the class of outer membrane proteins for trans-acting noncoding RNAs (100)? (ii) The ndhB gene, coding for a subunit of the NADH dehydrogenase, is associated with an asRNA in cyanobacteria (60) as well as in chloroplasts (26), their remote evolutionary descendants. However, these asRNAs target different regions in the mRNA, and the question arises, Are some asRNAs conserved among different organisms, such as the Yfr1 noncoding RNA in cyanobacteria (101) or 6S RNA and tmRNA, which are distributed ubiquitously throughout the bacterial kingdom? Or is the evolution of asRNAs too rapid to establish such conservation? Possibly, regulation by asRNAs is an evolutionarily fast strategy to cope with the regulatory needs of individual genes in a distinct host. (iii) Are there more asRNAs that are translated into proteins, such as cosA in P. fluorescens, or that form hybrid RNAs, which are both translated and act as asRNAs? High-coverage proteomic studies will help to answer this question. (iv) In a more global sense, what are the roles of asRNAs in regulatory networks? Are asRNAs (and trans-acting noncoding RNAs) able to replace lost regulatory proteins in a streamlined genome or perhaps act in combination with other cis elements like riboswitches? (v) There are likely other mechanisms still to be found. Direct targeting of DNA modifications would be one possibility. (vi) Which proteins are involved in regulation by asRNAs? It was shown that a dsRNA duplex is not necessarily degraded by RNase III (3, 40, 97), and for some mechanisms, the corresponding RNase is unknown (21, 40). In enterobacteria, the RNA chaperone Hfq is necessary for a wide range of mechanisms employed by trans-acting noncoding RNAs (30). Some asRNAs also bind to Hfq (55, 82, 97), but does this result from a general affinity of Hfq for RNA or from a specific interaction? What other kinds of proteins are involved? Another interesting class of proteins is RNA helicases. In Synechocystis PCC6803, the helicase CrhR can catalyze the melting and the formation of RNA duplexes (11).
Emerging functional data suggest that asRNAs in bacteria mediate a plethora of effects and are involved in far more processes than were previously anticipated. Thus, the functional impact of asRNAs should be considered in developing new strategies against pathogenic bacteria or when optimizing bacterial strains for biotechnology.
ACKNOWLEDGMENTS
Work presented in this review was supported by the Freiburg Initiative in Systems Biology BMBF (grant 0313921) and by the German Research Foundation (DFG) focus program SPP1258, Sensory and Regulatory RNAs in Prokaryotes (grant HE 2544/4-2).
REFERENCES
- 1. Albrecht M., Sharma C. M., Reinhardt R., Vogel J., Rudel T. 2010. Deep sequencing-based discovery of the Chlamydia trachomatis transcriptome. Nucleic Acids Res. 38:868–877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Altuvia S. 2007. Identification of bacterial small non-coding RNAs: experimental approaches. Curr. Opin. Microbiol. 10:257–261 [DOI] [PubMed] [Google Scholar]
- 3. Andre G., et al. 2008. S-box and T-box riboswitches and antisense RNA control a sulfur metabolic operon of Clostridium acetobutylicum. Nucleic Acids Res. 36:5955–5969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Backofen R., Hess W. R. 2010. Computational prediction of sRNAs and their targets in bacteria. RNA Biol. 7:1–10 [DOI] [PubMed] [Google Scholar]
- 5. Beaume M., et al. 2010. Cartography of methicillin-resistant S. aureus transcripts: detection, orientation and temporal expression during growth phase and stress conditions. PLoS One 5:e10725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Beiter T., Reich E., Williams R. W., Simon P. 2009. Antisense transcription: a critical look in both directions. Cell. Mol. Life Sci. 66:94–112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bouvier M., Sharma C. M., Mika F., Nierhaus K. H., Vogel J. 2008. Small RNA binding to 5′ mRNA coding region inhibits translational initiation. Mol. Cell 32:827–837 [DOI] [PubMed] [Google Scholar]
- 8. Brantl S. 2007. Regulatory mechanisms employed by cis-encoded antisense RNAs. Curr. Opin. Microbiol. 10:102–109 [DOI] [PubMed] [Google Scholar]
- 9. Busch A., Richter A. S., Backofen R. 2008. IntaRNA: efficient prediction of bacterial sRNA targets incorporating target site accessibility and seed regions. Bioinformatics 24:2849–2856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Callen B. P., Shearwin K. E., Egan J. B. 2004. Transcriptional interference between convergent promoters caused by elongation over the promoter. Mol. Cell 14:647–656 [DOI] [PubMed] [Google Scholar]
- 11. Chamot D., Colvin K. R., Kujat-Choy S. L., Owttrim G. W. 2005. RNA structural rearrangement via unwinding and annealing by the cyanobacterial RNA helicase, CrhR. J. Biol. Chem. 280:2036–2044 [DOI] [PubMed] [Google Scholar]
- 12. Chen Q., Crosa J. H. 1996. Antisense RNA, fur, iron, and the regulation of iron transport genes in Vibrio anguillarum. J. Biol. Chem. 271:18885–18891 [DOI] [PubMed] [Google Scholar]
- 13. Chinni S. V., et al. 2010. Experimental identification and characterization of 97 novel npcRNA candidates in Salmonella enterica serovar Typhi. Nucleic Acids Res. 38:5893–5908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Crampton N., Bonass W. A., Kirkham J., Rivetti C., Thomson N. H. 2006. Collision events between RNA polymerases in convergent transcription studied by atomic force microscopy. Nucleic Acids Res. 34:5416–5425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. D'Alia D., et al. 2010. Noncoding RNA of glutamine synthetase I modulates antibiotic production in Streptomyces coelicolor A3(2). J. Bacteriol. 192:1160–1164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Darfeuille F., Unoson C., Vogel J., Wagner E. G. 2007. An antisense RNA inhibits translation by competing with standby ribosomes. Mol. Cell 26:381–392 [DOI] [PubMed] [Google Scholar]
- 17. David L., et al. 2006. A high-resolution map of transcription in the yeast genome. Proc. Natl. Acad. Sci. U. S. A. 103:5320–5325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Dornenburg J. E., DeVita A. M., Palumbo M. J., Wade J. T. 2010. Widespread antisense transcription in Escherichia coli. mBio 1:e00024–e10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Drider D., Condon C. 2004. The continuing story of endoribonuclease III. J. Mol. Microbiol. Biotechnol. 8:195–200 [DOI] [PubMed] [Google Scholar]
- 20. Dubnau D., Losick R. 2006. Bistability in bacteria. Mol. Microbiol. 61:564–572 [DOI] [PubMed] [Google Scholar]
- 21. Dühring U., Axmann I. M., Hess W. R., Wilde A. 2006. An internal antisense RNA regulates expression of the photosynthesis gene isiA. Proc. Natl. Acad. Sci. U. S. A. 103:7054–7058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Eiamphungporn W., Helmann J. D. 2009. Extracytoplasmic function sigma factors regulate expression of the Bacillus subtilis yabE gene via a cis-acting antisense RNA. J. Bacteriol. 191:1101–1105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Filiatrault M. J., et al. 2010. Transcriptome analysis of Pseudomonas syringae identifies new genes, noncoding RNAs, and antisense activity. J. Bacteriol. 192:2359–2372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Fozo E. M., Hemm M. R., Storz G. 2008. Small toxic proteins and the antisense RNAs that repress them. Microbiol. Mol. Biol. Rev. 72:579–589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Fozo E. M., et al. 2008. Repression of small toxic protein synthesis by the Sib and OhsC small RNAs. Mol. Microbiol. 70:1076–1093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Georg J., Honsel A., Voss B., Rennenberg H., Hess W. R. 2010. A long antisense RNA in plant chloroplasts. New Phytol. 186:615–622 [DOI] [PubMed] [Google Scholar]
- 27. Georg J., et al. 2009. Evidence for a major role of antisense RNAs in cyanobacterial gene regulation. Mol. Syst. Biol. 5:305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Giangrossi M., et al. 2010. A novel antisense RNA regulates at transcriptional level the virulence gene icsA of Shigella flexneri. Nucleic Acids Res. 38:3362–3375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Golding I., Paulsson J., Zawilski S. M., Cox E. C. 2005. Real-time kinetics of gene activity in individual bacteria. Cell 123:1025–1036 [DOI] [PubMed] [Google Scholar]
- 30. Gottesman S. 2004. The small RNA regulators of Escherichia coli: roles and mechanisms. Annu. Rev. Microbiol. 58:303–328 [DOI] [PubMed] [Google Scholar]
- 31. Guell M., et al. 2009. Transcriptome complexity in a genome-reduced bacterium. Science 326:1268–1271 [DOI] [PubMed] [Google Scholar]
- 32. Han K., Kim K.-S., Bak G., Park H., Lee Y. 2010. Recognition and discrimination of target mRNAs by Sib RNAs, a cis-encoded sRNA family. Nucleic Acids Res. 38:5851–5866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. He Y., Vogelstein B., Velculescu V. E., Papadopoulos N., Kinzler K. W. 2008. The antisense transcriptomes of human cells. Science 322:1855–1857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Henz S. R., et al. 2007. Distinct expression patterns of natural antisense transcripts in Arabidopsis. Plant Physiol. 144:1247–1255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Hernandez J. A., et al. 2010. Mutants of Anabaena sp. PCC 7120 lacking alr1690 and alpha-furA antisense RNA show a pleiotropic phenotype and altered photosynthetic machinery. J. Plant Physiol. 167:430–437 [DOI] [PubMed] [Google Scholar]
- 36. Hernandez J. A., et al. 2005. Identification of a furA cis antisense RNA in the cyanobacterium Anabaena sp. PCC 7120. J. Mol. Biol. 355:325–334 [DOI] [PubMed] [Google Scholar]
- 37. Irnov I., Sharma C. M., Vogel J. R., Winkler W. C. 2010. Identification of regulatory RNAs in Bacillus subtilis. Nucleic Acids Res. 38:6637–6651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Itoh T., Tomizawa J. 1980. Formation of an RNA primer for initiation of replication of ColE1 DNA by ribonuclease H. Proc. Natl. Acad. Sci. U. S. A. 77:2450–2454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Katayama S., et al. 2005. Antisense transcription in the mammalian transcriptome. Science 309:1564–1566 [DOI] [PubMed] [Google Scholar]
- 40. Kawano M., Aravind L., Storz G. 2007. An antisense RNA controls synthesis of an SOS-induced toxin evolved from an antitoxin. Mol. Microbiol. 64:738–754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Kawano M., Reynolds A. A., Miranda-Rios J., Storz G. 2005. Detection of 5′- and 3′-UTR-derived small RNAs and cis-encoded antisense RNAs in Escherichia coli. Nucleic Acids Res. 33:1040–1050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Kertesz M., Iovino N., Unnerstall U., Gaul U., Segal E. 2007. The role of site accessibility in microRNA target recognition. Nat. Genet. 39:1278–1284 [DOI] [PubMed] [Google Scholar]
- 43. Kolb F. A., et al. 2001. Four-way junctions in antisense RNA-mRNA complexes involved in plasmid replication control: a common theme? J. Mol. Biol. 309:605–614 [DOI] [PubMed] [Google Scholar]
- 44. Krinke L., Mahoney M., Wulff D. L. 1991. The role of the OOP antisense RNA in coliphage λ development. Mol. Microbiol. 5:1265–1272 [DOI] [PubMed] [Google Scholar]
- 45. Krinke L., Wulff D. L. 1987. OOP RNA, produced from multicopy plasmids, inhibits lambda cII gene expression through an RNase III-dependent mechanism. Genes Dev. 1:1005–1013 [DOI] [PubMed] [Google Scholar]
- 46. Krinke L., Wulff D. L. 1990. RNase III-dependent hydrolysis of lambda cII-O gene mRNA mediated by lambda OOP antisense RNA. Genes Dev. 4:2223–2233 [DOI] [PubMed] [Google Scholar]
- 47. Lacatena R. M., Cesareni G. 1981. Base pairing of RNA I with its complementary sequence in the primer precursor inhibits ColE1 replication. Nature 294:623–626 [DOI] [PubMed] [Google Scholar]
- 48. Lee E. J., Groisman E. A. 2010. An antisense RNA that governs the expression kinetics of a multifunctional virulence gene. Mol. Microbiol. 76:1020–1033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Lee J. M., et al. 2001. RNA expression analysis using an antisense Bacillus subtilis genome array. J. Bacteriol. 183:7371–7380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Legewie S., Dienst D., Wilde A., Herzel H., Axmann I. M. 2008. Small RNAs establish delays and temporal thresholds in gene expression. Biophys. J. 95:3232–3238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Levine E., Hwa T. 2008. Small RNAs establish gene expression thresholds. Curr. Opin. Microbiol. 11:574–579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Levine E., Zhang Z., Kuhlman T., Hwa T. 2007. Quantitative characteristics of gene regulation by small RNA. PLoS Biol. 5:e229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Lindell D., et al. 2007. Genome-wide expression dynamics of a marine virus and host reveal features of co-evolution. Nature 449:83–86 [DOI] [PubMed] [Google Scholar]
- 54. Liu J. M., et al. 2009. Experimental discovery of sRNAs in Vibrio cholerae by direct cloning, 5S/tRNA depletion and parallel sequencing. Nucleic Acids Res. 37:e46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Lorenz C., et al. 2010. Genomic SELEX for Hfq-binding RNAs identifies genomic aptamers predominantly in antisense transcripts. Nucleic Acids Res. 38:3794–3808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Mackie G. A. 1998. Ribonuclease E is a 5′-end-dependent endonuclease. Nature 395:720–723 [DOI] [PubMed] [Google Scholar]
- 57. Majdalani N., Cunning C., Sledjeski D., Elliott T., Gottesman S. 1998. DsrA RNA regulates translation of RpoS message by an anti-antisense mechanism, independent of its action as an antisilencer of transcription. Proc. Natl. Acad. Sci. U. S. A. 95:12462–12467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Mehta P., Goyal S., Wingreen N. S. 2008. A quantitative comparison of sRNA-based and protein-based gene regulation. Mol. Syst. Biol. 4:221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Millard A. D., et al. 2010. An antisense RNA in a lytic cyanophage links psbA to a gene encoding a homing endonuclease. ISME J. 4:1121–1135 [DOI] [PubMed] [Google Scholar]
- 60. Mitschke J., et al. 2011. An experimentally anchored map of transcriptional start sites in the model cyanobacterium Synechocystis sp. PCC 6803. Proc. Natl. Acad. Sci. U. S. A. 108:2124–2129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Montero Llopis P., et al. 2010. Spatial organization of the flow of genetic information in bacteria. Nature 466:77–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Mraheil M. A., et al. 29 January 2011, posting date. The intracellular sRNA transcriptome of Listeria monocytogenes during growth in macrophages. Nucleic Acids Res. doi:10.1093/nar/gkr033 [Epub ahead of print.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Muckstein U., et al. 2006. Thermodynamics of RNA-RNA binding. Bioinformatics 22:1177–1182 [DOI] [PubMed] [Google Scholar]
- 64. Nagy Z., Chandler M. 2004. Regulation of transposition in bacteria. Res. Microbiol. 155:387–398 [DOI] [PubMed] [Google Scholar]
- 65. Opdyke J. A., Fozo E. M., Hemm M. R., Storz G. 2010. RNase III participates in GadY-dependent cleavage of the gadX-gadW mRNA. J. Mol. Biol. 406:29–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Opdyke J. A., Kang J. G., Storz G. 2004. GadY, a small-RNA regulator of acid response genes in Escherichia coli. J. Bacteriol. 186:6698–6705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Palmer A. C., Ahlgren-Berg A., Egan J. B., Dodd I. B., Shearwin K. E. 2009. Potent transcriptional interference by pausing of RNA polymerases over a downstream promoter. Mol. Cell 34:545–555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Perocchi F., Xu Z., Clauder-Münster S., Steinmetz L. M. 2007. Antisense artifacts in transcriptome microarray experiments are resolved by actinomycin D. Nucleic Acids Res. 35:e128.1–e1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Peters J. M., et al. 2009. Rho directs widespread termination of intragenic and stable RNA transcription. Proc. Natl. Acad. Sci. U. S. A. 106:15406–15411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Prescott E. M., Proudfoot N. J. 2002. Transcriptional collision between convergent genes in budding yeast. Proc. Natl. Acad. Sci. U. S. A. 99:8796–8801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Qiu Y., et al. 2010. Structural and operational complexity of the Geobacter sulfurreducens genome. Genome Res. 9:1304–1311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Rasmussen S., Nielsen H. B., Jarmer H. 2009. The transcriptionally active regions in the genome of Bacillus subtilis. Mol. Microbiol. 73:1043–1057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Salinas P. C., Waldbeser L. S., Crosa J. H. 1993. Regulation of the expression of bacterial iron transport genes: possible role of an antisense RNA as a repressor. Gene 123:33–38 [DOI] [PubMed] [Google Scholar]
- 74. Schluter J. P., et al. 2010. A genome-wide survey of sRNAs in the symbiotic nitrogen-fixing alpha-proteobacterium Sinorhizobium meliloti. BMC Genomics 11:245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Selinger D. W., et al. 2000. RNA expression analysis using a 30 base pair resolution Escherichia coli genome array. Nat. Biotechnol. 18:1262–1268 [DOI] [PubMed] [Google Scholar]
- 76. Sharma C. M., et al. 2010. The primary transcriptome of the major human pathogen Helicobacter pylori. Nature 464:250–255 [DOI] [PubMed] [Google Scholar]
- 77. Sharma C. M., Vogel J. 2009. Experimental approaches for the discovery and characterization of regulatory small RNA. Curr. Opin. Microbiol. 12:536–546 [DOI] [PubMed] [Google Scholar]
- 78. Shearwin K. E., Callen B. P., Egan J. B. 2005. Transcriptional interference—a crash course. Trends Genet. 21:339–345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Shimoni Y., et al. 2007. Regulation of gene expression by small non-coding RNAs: a quantitative view. Mol. Syst. Biol. 3:138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Silby M. W., Levy S. B. 2008. Overlapping protein-encoding genes in Pseudomonas fluorescens Pf0-1. PLoS Genet. 4:e1000094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Silvaggi J. M., Perkins J. B., Losick R. 2006. Genes for small, noncoding RNAs under sporulation control in Bacillus subtilis. J. Bacteriol. 188:532–541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Sittka A., et al. 2008. Deep sequencing analysis of small noncoding RNA and mRNA targets of the global post-transcriptional regulator, Hfq. PLoS Genet. 4:e1000163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Sneppen K., et al. 2005. A mathematical model for transcriptional interference by RNA polymerase traffic in Escherichia coli. J. Mol. Biol. 346:399–409 [DOI] [PubMed] [Google Scholar]
- 84. Sorek R., Cossart P. 2010. Prokaryotic transcriptomics: a new view on regulation, physiology and pathogenicity. Nat. Rev. Genet. 11:9–16 [DOI] [PubMed] [Google Scholar]
- 85. Spiegelman W. G., et al. 1972. Bidirectional transcription and the regulation of phage lambda repressor synthesis. Proc. Natl. Acad. Sci. U. S. A. 69:3156–3160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Stazic D., Lindell D., Steglich C. 2011. Antisense RNA protects from RNase E degradation by RNA-RNA duplex formation during phage infection. Nucleic Acids Res. [Epub ahead of print.] doi:10.1093/nar/gkr037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Steglich C., et al. 2008. The challenge of regulation in a minimal phototroph: non-coding RNAs in Prochlorococcus. PLoS Genet. 4:e1000173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Stolt P., Zillig W. 1993. Antisense RNA mediates transcriptional processing in an archaebacterium, indicating a novel kind of RNase activity. Mol. Microbiol. 7:875–882 [DOI] [PubMed] [Google Scholar]
- 89. Stork M., Di Lorenzo M., Welch T. J., Crosa J. H. 2007. Transcription termination within the iron transport-biosynthesis operon of Vibrio anguillarum requires an antisense RNA. J. Bacteriol. 189:3479–3488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Takada A., Umitsuki G., Nagai K., Wachi M. 2007. RNase E is required for induction of the glutamate-dependent acid resistance system in Escherichia coli. Biosci. Biotechnol. Biochem. 71:158–164 [DOI] [PubMed] [Google Scholar]
- 91. Tang T. H., et al. 2005. Identification of novel non-coding RNAs as potential antisense regulators in the archaeon Sulfolobus solfataricus. Mol. Microbiol. 55:469–481 [DOI] [PubMed] [Google Scholar]
- 92. Thomason M. K., Storz G. 2010. Bacterial antisense RNAs: how many are there and what are they doing? Annu. Rev. Genet. 44:167–188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Tjaden B., et al. 2002. Transcriptome analysis of Escherichia coli using high density oligonucleotide probe arrays. Nucleic Acids Res. 30:3732–3738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Toledo-Arana A., et al. 2009. The Listeria transcriptional landscape from saprophytism to virulence. Nature 459:950–956 [DOI] [PubMed] [Google Scholar]
- 95. Tolmasky M. E., Crosa J. H. 1995. Iron transport genes of the pJM1-mediated iron uptake system of Vibrio anguillarum are included in a transposon like structure. Plasmid 33:180–190 [DOI] [PubMed] [Google Scholar]
- 96. Tomb J.-F., et al. 1997. The complete genome sequence of the gastric pathogen Helicobacter pylori. Nature 388:539–547 [DOI] [PubMed] [Google Scholar]
- 97. Tramonti A., De Canio M., De Biase D. 2008. GadX/GadW-dependent regulation of the Escherichia coli acid fitness island: transcriptional control at the gadY-gadW divergent promoters and identification of four novel 42 bp GadX/GadW-specific binding sites. Mol. Microbiol. 70:965–982 [DOI] [PubMed] [Google Scholar]
- 98. Veening J. W., Smits W. K., Kuipers O. P. 2008. Bistability, epigenetics, and bet-hedging in bacteria. Annu. Rev. Microbiol. 62:193–210 [DOI] [PubMed] [Google Scholar]
- 99. Vogel J., et al. 2003. RNomics in Escherichia coli detects new sRNA species and indicates parallel transcriptional output in bacteria. Nucleic Acids Res. 31:6435–6443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Vogel J., Papenfort K. 2006. Small non-coding RNAs and the bacterial outer membrane. Curr. Opin. Microbiol. 9:605–611 [DOI] [PubMed] [Google Scholar]
- 101. Voss B., Gierga G., Axmann I. M., Hess W. R. 2007. A motif-based search identifies the ortholog of the small RNA Yfr1 in all major lineages of cyanobacteria. BMC Genomics 8:375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Wagner E. G., Simons R. W. 1994. Antisense RNA control in bacteria, phages, and plasmids. Annu. Rev. Microbiol. 48:713–742 [DOI] [PubMed] [Google Scholar]
- 103. Waldbeser L. S., Chen Q., Crosa J. H. 1995. Antisense RNA regulation of the fatB iron transport protein gene in Vibrio anguillarum. Mol. Microbiol. 17:747–756 [DOI] [PubMed] [Google Scholar]
- 104. Waldbeser L. S., Tolmasky M. E., Actis L. A., Crosa J. H. 1993. Mechanisms for negative regulation by iron of the fatA outer membrane protein gene expression in Vibrio anguillarum 775. J. Biol. Chem. 268:10433–10439 [PubMed] [Google Scholar]
- 105. Wen Y., Feng J., Scott D. R., Marcus E. A., Sachs G. 2011. A cis-encoded antisense small RNA regulated by the HP0165-HP0166 two-component system controls expression of ureB in Helicobacter pylori. J. Bacteriol. 193:40–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Wrobel B., Herman-Antosiewicz A., Szalewska-Palasz S., Wegrzyn G. 1998. Polyadenylation of oop RNA in the regulation of bacteriophage lambda development. Gene 212:57–65 [DOI] [PubMed] [Google Scholar]
- 107. Wurtzel O., et al. 2010. A single-base resolution map of an archaeal transcriptome. Genome Res. 20:133–141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Xu Z., et al. 2011. Antisense expression increases gene expression variability and locus interdependency. Mol. Syst. Biol. 7:468.1–46810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Xu Z., et al. 2009. Bidirectional promoters generate pervasive transcription in yeast. Nature 457:1033–1037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Yachie N., Numata K., Saito R., Kanai A., Tomita M. 2006. Prediction of non-coding and antisense RNA genes in Escherichia coli with gapped Markov model. Gene 372:171–181 [DOI] [PubMed] [Google Scholar]
- 111. Zeng Q., Bonocora R. P., Shub D. A. 2009. A free-standing homing endonuclease targets an intron insertion site in the psbA gene of cyanophages. Curr. Biol. 19:218–222 [DOI] [PubMed] [Google Scholar]
- 112. Zhang Y., Liu X. S., Liu Q. R., Wei L. 2006. Genome-wide in silico identification and analysis of cis natural antisense transcripts (cis-NATs) in ten species. Nucleic Acids Res. 34:3465–3475 [DOI] [PMC free article] [PubMed] [Google Scholar]