Abstract
Naturally contaminated bovine bulk tank milk (n = 44) and feces (n = 39) were tested for the presence of viable Mycobacterium avium subsp. paratuberculosis by a novel peptide-mediated magnetic separation-phage (PMS-phage) assay. Counts of viable M. avium subsp. paratuberculosis cells ranging from 1 to 110 PFU/50 ml of milk and 6 to 41,111 PFU/g of feces were indicated by the PMS-phage assay.
TEXT
Culture is considered the definitive diagnostic test for Mycobacterium avium subsp. paratuberculosis in diagnostic specimens such as feces and bulk tank milk (BTM) (18). However, it is labor intensive and time consuming and involves chemical decontamination and several months of incubation. Recently, we optimized a peptide-mediated magnetic separation-phage (PMS-phage) assay to rapidly detect viable M. avium subsp. paratuberculosis (4, 5). PMS selectively captures and concentrates M. avium subsp. paratuberculosis cells from a sample, effectively removing the vast majority of contaminating microorganisms, and the phage amplification assay enables rapid enumeration of viable M. avium subsp. paratuberculosis cells within 24 h. When employed together, PMS and the phage amplification assay are selective for low numbers of viable M. avium subsp. paratuberculosis cells in artificially spiked milk samples (5) without the need for chemical decontamination. The objective of this study was to assess the performance of this novel PMS-phage assay when applied to naturally contaminated bovine BTM and feces samples.
Fresh BTM samples (n = 25) from dairy farms in Northern Ireland (NI) on which one or more animals were seropositive for M. avium subsp. paratuberculosis and previously frozen (−70°C for several months) BTM (n = 19) and feces samples (n = 39) from dairy herds of known Johne's disease status connected with the University of Pennsylvania School of Veterinary Medicine were tested. A 50-ml aliquot of each BTM sample was centrifuged at 2,500 × g for 15 min and the pellet was resuspended in 1 ml of Middlebrook 7H9 broth containing 10% oleic acid-albumin-dextrose-catalase (OADC) prior to the optimized PMS-phage assay. One gram of feces was mixed thoroughly with 4 ml of sterile water, and the sample was subjected to low-speed centrifugation (300 × g for 3 min). One milliliter of the resultant supernatant was processed through the optimized PMS-phage assay (5). PMS beads were resuspended in Middlebrook 7H9 broth containing a nystatin-oxacillin-aztreonam (NOA) antimicrobial supplement (final concentrations per ml, 50 IU of nystatin, 2 μg of oxacillin, and 30 μg of aztreonam; Biotec Laboratories Limited, United Kingdom) to limit the growth of background microflora during the subsequent incubation steps. Plaques were counted and expressed as PFU/50 ml of BTM and PFU/g of feces. Plaque-IS900 PCR (15) was performed on PMS-phage assay-positive samples to confirm the presence of M. avium subsp. paratuberculosis DNA.
BTM samples were also cultured. A 50-ml aliquot of each NI BTM sample was decontaminated with 0.75% hexadecylpyridinium chloride (HPC; Sigma) for 5 h at room temperature (8) before being inoculated in duplicate onto Herrold's egg yolk medium containing 2 μg/ml of mycobactin J and into 10 ml of Middlebrook 7H9 broth supplemented with 10% OADC and 2 μg/ml of mycobactin J. As limited volumes of U.S. BTM were available, after PMS 50-μl aliquots of the resuspended beads were cultured without prior chemical decontamination. All cultures were incubated for 16 weeks at 37°C, and cultures that were suspected to be positive were tested for M. avium subsp. paratuberculosis DNA by IS900 PCR (11).
Ten of the 25 (fresh) NI BTM samples and 5 of the 19 (previously frozen) U.S. BTM samples tested by PMS-phage assay yielded plaques, with counts ranging from 1 to 89 and 2 to 110 PFU/50 ml of milk, respectively. Plaques from all 10 positive NI milk samples and from 4 of the 5 positive U.S. milk samples were confirmed to contain M. avium subsp. paratuberculosis DNA, with 89.3% (60 of 67) and 35.7% (15 of 42; see Fig. S1 in the supplemental material) of the plaques, respectively, testing positive by plaque PCR. The presence of viable M. avium subsp. paratuberculosis cells was confirmed by HPC decontamination plus broth culture in just three NI milk samples and by PMS plus broth culture in two U.S. milk samples (Table 1). In the case of fresh NI BTM, there was little agreement between the results of the two assays, with the PMS-phage assay yielding more M. avium subsp. paratuberculosis-positive results than HPC decontamination and culture (10 versus 3; Table 1). In contrast, when PMS was used in place of HPC decontamination before culture, as in the case of U.S. milk samples, the deleterious effects of HPC on the viability of M. avium subsp. paratuberculosis were avoided, resulting in no significant difference in proportions positive by PMS-phage assay and PMS plus culture (4 versus 2, respectively; Table 1). The culture results confirm that chemical decontamination before culture has adverse effects on the viability of M. avium subsp. paratuberculosis in milk (6), resulting in diminished detection sensitivity.
Table 1.
Comparison of the PMS-phage assay and culture for testing bovine bulk tank milka
| Source (no.) of milk samples | Milk conditionb | No. of samples positive by both culture and PMS-phage assay | No. of samples positive by culture and negative by PMS-phage assay | No. of samples negative by culture and positive by PMS-phage assay | No. of samples negative by both culture and PMS-phage assay | Kappa value (P valuec) |
|---|---|---|---|---|---|---|
| Northern Ireland (25) | Fresh | 2 | 1 | 8 | 14 | 0.151 (0.0455) |
| United States (19) | Frozen | 2 | 0 | 2 | 15 | 0.612 (0.4795) |
HPC decontamination preceded culturing of Northern Ireland milk samples, and PMS preceded culturing of milk samples from the United States.
Fresh milk was 24 h old; frozen milk was kept at −70°C for several months and thawed for testing.
Determined by McNemar's test using GraphPad InStat 3.0 (GraphPad Software, La Jolla, CA).
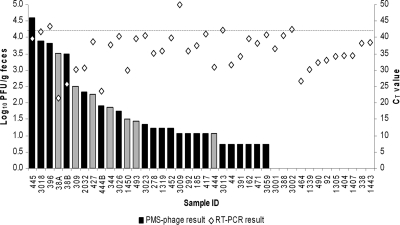
The phage amplification assay cannot be applied directly to feces; some unknown component of feces inhibits phage infection, resulting in no plaque formation (3; I. R. Grant, unpublished data). Plaques were obtained for 27 of the 39 feces samples and ranged in number from 6 to 41,111 PFU/g. Plaques from 20 of these samples tested positive by IS900 PCR (Fig. 1). Comparison of the PMS-phage assay results with the original real-time PCR results for the same fecal samples revealed limited correlation (Fig. 1). However, the two assays were performed months apart, and the viability of M. avium subsp. paratuberculosis in feces may have been adversely affected by being frozen (−70°C) and stored (10, 12). Fresh feces would be the optimal specimen to process via the PMS-phage assay.
Fig. 1.
Comparison of the PMS-phage assay (log10 PFU/g of feces) and RT-PCR (CT value) results obtained for M. avium subsp. paratuberculosis testing of 39 U.S. bovine feces samples. Black bars indicate fecal samples from which at least some plaques tested M. avium subsp. paratuberculosis positive by plaque-IS900 PCR; gray bars indicate fecal samples for which this was not the case. The dashed line indicates the threshold cycle (CT) cutoff value above which a sample was considered M. avium subsp. paratuberculosis negative by RT-PCR.
Plaque counts for viable M. avium subsp. paratuberculosis obtained for both BTM and feces were believable, in our opinion, ranging from 1 to 110 PFU/50 ml of BTM and from 6 to 41,111 PFU/g of feces. However, these counts may still be underestimates, perhaps 10-fold lower than the actual numbers of M. avium subsp. paratuberculosis PFUs present (5). The PMS-phage assay counts are in general agreement with previous estimates of the numbers of M. avium subsp. paratuberculosis cells in milk from individual infected cows (2 to 8 CFU/50 ml [16], <100 CFU/ml [7], 4 to 20 CFU/50 ml [2], and 10 to 560 cells/ml [13]), in BTM (1 to 9 cells/ml [13] and “several tens of cells/ml” [14]), and in bovine feces (9, 17). Residual background milk or fecal microflora present after PMS, particularly from the frozen samples, was not always adequately controlled by the addition of NOA (originally developed to control respiratory contaminant bacteria in sputum samples [1]), and this may have led to some false-negative results. Preliminary investigations (A. Foddai, and I. R. Grant, unpublished data) substituting one-fourth-strength MGIT PANTA (final concentrations per ml of 7H9 broth, 10 IU of polymyxin B, 1 μg of amphotericin, 4 μg of nalidixic acid, 1 μg of trimethoprim, and 1 μg of axlocillin; Becton Dickinson) for NOA suggest that this may be a solution. Further testing of fresh milk and feces employing one-fourth-strength MGIT PANTA instead of NOA is warranted in order to validate the use of the PMS-phage assay for diagnostic purposes.
Supplementary Material
Acknowledgments
Antonio Foddai was the recipient of a Masters and Back Studentship from the Sardinian government during this study.
Thanks are due to Susan McAdams of the University of Pennsylvania, who coordinated the sending of bulk milk and fecal samples from the United States to Queen's University Belfast.
Footnotes
Supplemental material for this article may be found at http://jcm.asm.org/.
Published ahead of print on 16 March 2011.
REFERENCES
- 1. Albert H., et al. 2007. Development of an antimicrobial formulation for control of specimen-related contamination in phage-based diagnostic testing for tuberculosis. J. Appl. Microbiol. 103:892–899 [DOI] [PubMed] [Google Scholar]
- 2. Ayele W. Y., Svastova P., Roubal P., Bartos M., Pavlik I. 2005. Mycobacterium avium subspecies paratuberculosis cultured from locally and commercially pasteurized cow's milk in the Czech Republic. Appl. Environ. Microbiol. 71:1210–1214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. De Buck J., Griffiths T., Barkema H., Rioux K. 2008. Detection of Mycobacterium avium subspecies paratuberculosis by modified FASTplaqueTB bacteriophage assay. In Neilsen S. S. (ed.), Proceedings of the 9th International Colloquium on Paratuberculosis International Association for Paratuberculosis, Madison, WI [Google Scholar]
- 4. Foddai A., Elliott C. T., Grant I. R. 2009. Optimization of a phage amplification assay to permit accurate enumeration of viable Mycobacterium avium subsp. paratuberculosis cells. Appl. Environ. Microbiol. 75:3896–3902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Foddai A., Elliott C. T., Grant I. R. 2010. Maximizing capture efficiency and specificity of magnetic separation for Mycobacterium avium subsp. paratuberculosis cells. Appl. Environ. Microbiol. 76:7550–7558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Gao A., Odumeru J., Raymond M., Mutharia L. 2005. Development of improved method for isolation of Mycobacterium avium subsp. paratuberculosis from bulk tank milk: effect of age of milk, centrifugation, and decontamination. Can. J. Vet. Res. 69:81–87 [PMC free article] [PubMed] [Google Scholar]
- 7. Giese S., Ahrens P. 2000. Detection of Mycobacterium avium subsp. paratuberculosis in milk from clinically affected cows by PCR and culture. Vet. Microbiol. 77:291–297 [DOI] [PubMed] [Google Scholar]
- 8. Grant I. R., Ball H. J., Rowe M. T. 2002. Incidence of Mycobacterium paratuberculosis in bulk raw and commercially pasteurized cows' milk from approved dairy processing establishments in the United Kingdom. Appl. Environ. Microbiol. 68:2428–2435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Hovingh E., et al. 2006. Identification and implications of MAP supershedders. J. Anim. Sci. 84(Suppl. 1):134 [Google Scholar]
- 10. Khare S., Adams L. G., Osterstock J., Roussel A., David L. 2008. Effects of shipping and storage conditions of fecal samples on viability of Mycobacterium paratuberculosis. J. Clin. Microbiol. 46:1561–1562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Moss M. T., et al. 1992. Polymerase chain reaction detection of Mycobacterium paratuberculosis and Mycobacterium avium subsp. silvaticum in long term cultures from Crohn's disease and control tissues. Gut 33:1209–1212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Richards W. D., Thoen C. O. 1977. Effect of freezing on the viability of Mycobacterium paratuberculosis in bovine feces. J. Clin. Microbiol. 6:392–395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Slana I., Kralik P., Kralova A., Pavlik I. 2008. On-farm spread of Mycobacterium avium subsp. paratuberculosis in raw milk studied by IS900 and F57 competitive real time quantitative PCR and culture examination. Int. J. Food Microbiol. 128:250–257 [DOI] [PubMed] [Google Scholar]
- 14. Slana I., Liapi M., Moravkova M., Kralova A., Pavlik I. 2009. Mycobacterium avium subsp. paratuberculosis in cow bulk tank milk in Cyprus detected by culture and quantitative IS900 and F57 real-time PCR. Prev. Vet. Med. 89:223–226 [DOI] [PubMed] [Google Scholar]
- 15. Stanley E. C., et al. 2007. Development of a new, combined rapid method using phage and PCR for detection and identification of viable Mycobacterium paratuberculosis bacteria within 48 hours. Appl. Environ. Microbiol. 73:1851–1857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Sweeney R. W., Whitlock R. H., Rosenberger A. E. 1992. Mycobacterium paratuberculosis cultured from milk and supramammary lymph nodes of infected asymptomatic cows. J. Clin. Microbiol. 30:166–171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. van Schaik G., Stehman S. M., Schukken Y. H., Rossiter C. R., Shin S. J. 2003. Pooled fecal culture sampling for Mycobacterium avium subsp. paratuberculosis at different herd sizes and prevalence. J. Vet. Diagn. Invest. 15:233–241 [DOI] [PubMed] [Google Scholar]
- 18. Whittington R. 2010. Cultivation of Mycobacterium avium subsp. paratuberculosis p. 244–266 In Behr M. A., Collins D. M. (ed.), Paratuberculosis: organism, disease, control. CAB International, Wallingford, UK [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.