Abstract
CTX-M-15-producing Escherichia coli has emerged worldwide as an important pathogen associated with community-onset infections, but in South America reports are scarce. We document the presence of CTX-M-15-producing E. coli of the international ST131 and ST405 clones in Colombia and present the first molecular characterization of these isolates in South America.
TEXT
Since the end of the 1990s, a new family of extended-spectrum β-lactamases (ESBL), the CTX-M type, has spread among continents, becoming the most prevalent in the world (1). Within this family, Escherichia coli strains producing CTX-M-15 have emerged as an important causative agent of community-onset infections (COI), predominantly urinary tract infections (UTI) (1). Moreover, the successful dispersion of CTX-M-15 has been associated with specific clones, such as ST131 and ST405, which belong to virulent phylogenetic groups B2 and D, respectively (4, 14). Other genes normally present on the same plasmid carrying blaCTX-M-15, like blaTEM and blaOXA, as well as aac-(6′)-Ib-cr and qnr determinants, explain the multiresistant phenotype of CTX-M-15-producing bacteria (6, 14).
In South America, CTX-M-2, CTX-M-9, and CTX-M-12 have been reported previously among Enterobacteriaceae (15). However, reports of CTX-M-15-producing Enterobacteriaceae remain scarce in this region, and only recently a single report from Brazil of the clone ST131 was presented in an abstract lacking clinical data or further molecular characterization (9).
In this study, we present the first molecular characterization of CTX-M-15-producing Escherichia coli isolates belonging to the clones ST131 and ST405 associated with COI in Colombia and South America, describing demographic and clinical data of patients from which these isolates were cultured.
(Part of this work was presented at the 50th Annual Interscience Conference on Antimicrobial Agents and Chemotherapy [ICAAC], Boston, MA, 12 to 15 September 2010).
From January 2010 to May 2010, ESBL-positive E. coli isolates were sent from three Colombian hospitals to our reference laboratory at CIDEIM in Cali, Colombia. All isolates were obtained from patients who presented to the emergency room with community-onset infections, defined by the patient's having no history of being in a hospital, long-term-care facility, or nursing home, receiving home care, or having an invasive procedure done in the last 3 months. Bacterial identification was confirmed using the Vitek 2 automatic system (bioMérieux, Marcy l'Etoile, France). Detection of blaTEM, blaSHV, blaCTX-M, and β-lactamase genes was performed by PCR, and all isolates in which cluster 1 blaCTX-M was detected were sequenced.
CTX-M-15-producing E. coli isolates were further characterized. Susceptibility testing was performed using the broth microdilution method (Sensititre panels; TREK Diagnostic Systems, Westlake, OH), and the results were interpreted according to the CLSI breakpoints (3). Multiplex PCR for qnr determinants (qnrA, qnrB, qnrS) was carried out as previously described (13). PCR for aac(6′)-Ib was performed in all isolates, and those that tested positive were further analyzed by digestion with BstF51 to identify strains harboring aac(6′)-Ib-cr (8). Detection of virulence-associated genes was performed based on published methods (5); virulence markers screened included usp (uropathogenic-specific protein), ompT (outer membrane protein), two adhesin-encoding genes (papC and fimH), two siderophore-related genes (iutA and fyuA), a serum survival-related gene (traT), and a pathogenicity-associated island (PAI) marker. Determination of the E. coli phylogenetic groups (A, B1, B2, and D) was carried out by a modified multiplex PCR (2, 12). These isolates underwent typing using the DiversiLab system (bioMérieux), following the manufacturer's instructions, and multilocus sequence typing (MLST) (16), according to the recommended procedure at the E. coli MLST website (http://mlst.ucc.ie/mlst/dbs/Ecoli).
From 33 ESBL-positive isolates received at CIDEIM, 18 harbored cluster 1 blaCTX-M, and DNA sequence analysis revealed the presence of blaCTX-M-15 in 14 isolates. Within the remaining four isolates, one harbored blaCTX-M-12 and the other three harbored blaCTX-M-12a. Table 1 summarizes the clinical, microbiological, and molecular data of patients and CTX-M-15 isolates.
Table 1.
Clinical data of patients and microbiological and molecular data of CTX-M-15-producing isolates
| Patient | Sexa | Age (yr) | Diagnosis | Underlying disease(s)b | ATB last 3 moc | MIC (μg/ml)f |
Virulence factor genes | Other bla genes detected | Presence ofe: |
Phylogenetic group | MLSTd |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CTX | CAZ | FEP | TZP | ETP | MEM | CIP | AMK | qnr | aac(6′)-Ib-cr | CC | ST | |||||||||
| 1 | F | 85 | Cystitis | R-UTI, UI | NIT, CIP | >32 | 8 | 8 | ≤8/4 | ≤0.5 | ≤0.12 | >8 | ≤8 | PAI, iutA, traT | None | − | + | A | 10 | 617 |
| 2 | F | 80 | Cystitis | R-UTI | No | >32 | 32 | 16 | ≤8/4 | ≤0.5 | ≤0.12 | >8 | ≤8 | PAI, iutA, traT | None | − | + | A | 10 | 617 |
| 3 | M | 30 | Peritonitis | Ulcerative colitis | No | >32 | 8 | 8 | ≤8/4 | ≤0.5 | ≤0.12 | >8 | ≤8 | fimH, iutA, traT | None | − | + | A | 10 | 44 |
| 4 | F | 80 | Cystitis | UI, HTN | CIP | 16 | ≤2 | 2 | ≤8/4 | ≤0.5 | ≤0.12 | 8 | 16 | fimH, iutA, traT | None | − | + | A | 10 | 44 |
| 5 | F | 69 | Cystitis | R-UTI | NIT | >32 | 16 | 32 | 16/4 | ≤0.5 | ≤0.12 | >8 | ≤8 | fimH, fyuA | blaTEM-1 | − | + | D | 405 | 405 |
| 6 | F | 75 | Cystitis | R-UTI, UI | NIT | >32 | 32 | 16 | ≤8/4 | ≤0.5 | ≤0.12 | >8 | ≤8 | fimH, fyuA | blaTEM-1 | − | + | D | 405 | 405 |
| 7 | F | 88 | Pyelonephritis | CRF, CHF, DM2 | No | 16 | 8 | 8 | ≤8/4 | ≤0.5 | ≤0.12 | >8 | ≤8 | PAI, fimH, fyuA, iutA, traT | None | − | + | D | 405 | 405 |
| 8 | F | 18 | Cystitis | None | No | >32 | 16 | 16 | ≤8/4 | ≤0.5 | ≤0.12 | >8 | ≤8 | ompT, PAI, fyuA, traT | blaTEM-1 | − | − | D | None | 2043 |
| 9 | F | 81 | Pyelonephritis | Alzheimer's disease | No | >32 | 32 | 32 | 16/4 | ≤0.5 | ≤0.12 | >8 | 16 | ompT, PAI, fyuA | blaTEM-1 | − | + | D | 648 | 648 |
| 10 | M | 80 | Cystitis | R-UTI, COPD | No | >32 | 32 | 32 | 32/4 | 2 | 0.25 | >8 | ≤8 | PAI, ompT, fimH, fyuA, traT | None | − | + | D | 648 | 648 |
| 11 | M | 2 | Cystitis | None | No | >32 | >32 | 64 | 32/4 | ≤0.5 | ≤0.12 | 8 | 32 | PAI, fimH, iutA, traT | None | − | + | D | None | 1722 |
| 12 | F | 2 | Cystitis | None | No | >32 | 8 | 16 | ≥128/4 | ≤0.5 | ≤0.12 | >8 | ≤8 | usp, ompT, PAI, fimH, fyuA, iutA, traT | blaTEM-1 | − | + | B2 | None | 131 |
| 13 | F | 69 | Cystitis | R-UTI, UI, HTN | CIP | >32 | >32 | 32 | 32/4 | ≤0.5 | ≤0.12 | >8 | ≤8 | usp, ompT, PAI, fimH, fyuA, iutA, traT | blaTEM-1 | − | + | B2 | None | 131 |
| 14 | F | 64 | Cystitis | UI, DM2, HTN | No | 32 | 32 | 2 | ≤8/4 | ≤0.5 | ≤0.12 | >8 | ≤8 | usp, ompT, PAI, papC, fimH, fyuA, iutA | blaTEM-1 | − | + | B2 | None | 2042 |
F, female; M, male.
R-UTI, repetitive urinary tract infection; UI, urinary incontinence; HTN, arterial hypertension; CRF, chronic renal failure, CHF, congestive heart failure; DM2, diabetes mellitus 2; COPD, chronic obstructive pulmonary disease.
ATB, antibiotic; NIT, nitrofurantoin; CIP, ciprofloxacine.
ST, sequence type; CC, clonal complex.
+, positive; −, negative.
CTX, cefotaxime; CAZ, ceftazidime; FEP, cefepime; TZP, piperacillin-tazobactam; ETP, ertapenem; MEM, meropenem; AMK, amikacin.
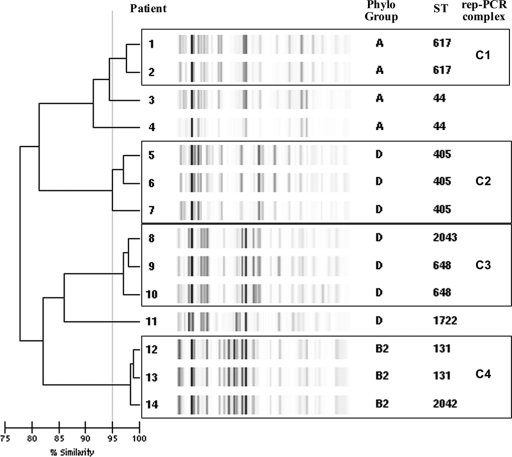
DiversiLab Rep-PCR, using 95% as a cutoff value, allowed the identification of four different repetitive PCR (rep-PCR) complexes of related isolates, while three strains yielded single rep-PCR types, as shown in Fig. 1. Within complex 4, two E. coli isolates (12 and 13) belong to ST131. The DiversiLab system was able to identify clone ST131, as previously reported by Pitout et al. (10); in addition, isolate 14 is a new single-locus variant (SLV) of ST131, designated ST2042, which might demonstrate a local variation of the ST131 clone. These three E. coli isolates had coresistance to quinolones (ciprofloxacin MIC, >8 μg/ml) harboring aac(6′)-Ib-cr without qnr determinants, as well as blaTEM-1. Like ST131 isolates recovered in other geographic areas, they were also found to carry the virulence factor genes fimH, fyuA, and usp (6, 14).
Fig. 1.
CTX-M-15-producing E. coli isolates typed by DiversiLab Rep-PCR using 95% as a cutoff value.
Isolates belonging to the virulent phylogenetic group D were grouped in two different rep-PCR complexes: complex 2 correspond to isolates belonging to the ST405, and complex 3 is formed by three isolates, two that belong to the ST648 and the other to the newly designated ST2043 (double-locus variant [DLV] of ST648). Isolate 11 (ST1722) represents a unique rep-PCR profile within phylogenetic group D. The remaining four isolates (isolates 1 to 4) belong to the phylogenetic group A and to the sequence type ST10 complex; two were ST617 and grouped within rep-PCR complex 1, and the other two were ST44. The CC10 has been also associated with other ESBLs such as CTX-M-14 or SHV-12 in isolates producing UTI in community patients (7). All previous isolates harbor several virulence factors and have coresistance to ciprofloxacin; all but one harbor aac(6′)-Ib-cr, and none harbored qnr determinants (Table 1).
Patients' ages ranged between 2 and 88 years, with most of them (10 of 14) over 60 years old; all but one presented with community-onset UTI. Recurrent UTI (6/11) and urinary incontinence (5/11) were the most common underlying conditions detected among the patients. Two patients had a history of previous intake of ciprofloxacin, two of nitrofurantoin, and one of both antibiotics during the last 3 months (Table 1).
We report for the first time in Colombia CTX-M-15-producing E. coli in several sequence types (STs), including the presence of ST131 and ST405, which are multidrug-resistant virulent clones involved in the intercontinental dissemination of blaCTX-M-15. As previously reported, ST131 has been associated with UTI (4, 11). The presence of CTX-M-15-producing E. coli causing COI and its potential spread in the community is an important public health concern, as Colombia may become part of the so-called “CTX-M pandemic” (1). There is a serious need to monitor the spread of these multidrug-resistant clones since, for instance, ST131 has been implicated in severe COI, including bacteremias (11). It is therefore mandatory to study the prevalence, clinical impact, and risk factors for the acquisition of CTX-M-producing E. coli in the community in Colombia and South America.
Acknowledgments
This study was supported by a research grant from Merck Sharp & Dohme. We also thank the Chicago Infectious Diseases Research Institute for money support.
We thank James Johnson for providing us with control strains for PCR procedures and Natalia Rosas, Elsa De la Cadena, and Alejandro Vargas of CIDEIM. We also thank the participating hospitals: Clínica Comfandi Tequendama (Rodrigo Lopez, Maribel Lopez, Geilli Sierra, Gloria Cuero, María Arboleda), Fundación Valle del Lili (Mónica Recalde Claudia Rocío Castañeda, Alejandra Toala, Luisa Fernanda Martínez, María Fernanda Peña, Alba Lucía Giraldo, Luz Stella Ortega), and Centro Médico Imbanaco (Beatriz Vanegas, Cristina Diaz, Armando Moreno, Lorna Castillo).
J. P. Quinn is an employee of Pfizer Global Research and Development (New London, CT); M. V. Villegas has received consulting fees and research grants from Merck Sharp & Dohme, Pfizer SA, Janssen-Cilag SA, Novartis, Merck Colombia, and AstraZeneca Colombia SA; E. Martinez has received consulting fees from Merck Sharp & Dohme, Pfizer SA, Janssen-Cilag SA, Bristol-Myers Squibb, Merck Colombia, AstraZeneca Colombia SA, and Glaxo Smith Kline; F. Rosso has received consulting fees from Merck Sharp & Dohme, Pfizer SA, Novartis, Roche, Merck Colombia, 3M, and Janssen-Cilag SA.
Footnotes
Published ahead of print on 16 February 2011.
REFERENCES
- 1. Canton R., Coque T. M. 2006. The CTX-M beta-lactamase pandemic. Curr. Opin. Microbiol. 9:466–475 [DOI] [PubMed] [Google Scholar]
- 2. Clermont O., Bonacorsi S., Bingen E. 2000. Rapid and simple determination of the Escherichia coli phylogenetic group. Appl. Environ. Microbiol. 66:4555–4558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Clinical and Laboratory Standards Institute 2010. Performance standards for antimicrobial susceptibility testing; twentieth informational supplement. CLSI document M100-S20U. Clinical and Laboratory Standards Institute, Wayne, PA [Google Scholar]
- 4. Coque T. M., et al. 2008. Dissemination of clonally related Escherichia coli strains expressing extended-spectrum beta-lactamase CTX-M-15. Emerg. Infect. Dis. 14:195–200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Johnson J. R., Delavari P., Kuskowski M., Stell A. L. 2001. Phylogenetic distribution of extraintestinal virulence-associated traits in Escherichia coli. J. Infect. Dis. 183:78–88 [DOI] [PubMed] [Google Scholar]
- 6. Nicolas-Chanoine M. H., et al. 2008. Intercontinental emergence of Escherichia coli clone O25:H4-ST131 producing CTX-M-15. J. Antimicrob. Chemother. 61:273–281 [DOI] [PubMed] [Google Scholar]
- 7. Oteo J., et al. 2009. Extended-spectrum beta-lactamase-producing Escherichia coli in Spain belong to a large variety of multilocus sequence typing types, including ST10 complex/A, ST23 complex/A and ST131/B2. Int. J. Antimicrob. Agents 34:173–176 [DOI] [PubMed] [Google Scholar]
- 8. Park C. H., Robicsek A., Jacoby G. A., Sahm D., Hooper D. C. 2006. Prevalence in the United States of aac(6′)-Ib-cr encoding a ciprofloxacin-modifying enzyme. Antimicrob. Agents Chemother. 50:3953–3955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Peirano G., Aseni M., Pitout J. D. D. 2010. Molecular characteristics of extended-spectrum β-lactamases (ESBL) Escherichia coli from Rio de Janeiro, Brazil, abstr. C2-680. Abstr. 50th Intersci. Conf. Antimicrob. Agents Chemother American Society for Microbiology, Washington, DC [Google Scholar]
- 10. Pitout J. D., et al. 2009. Using a commercial DiversiLab semiautomated repetitive sequence-based PCR typing technique for identification of Escherichia coli clone ST131 producing CTX-M-15. J. Clin. Microbiol. 47:1212–1215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Pitout J. D., Gregson D. B., Campbell L., Laupland K. B. 2009. Molecular characteristics of extended-spectrum-beta-lactamase-producing Escherichia coli isolates causing bacteremia in the Calgary Health Region from 2000 to 2007: emergence of clone ST131 as a cause of community-acquired infections. Antimicrob. Agents Chemother. 53:2846–2851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Pitout J. D., Laupland K. B., Church D. L., Menard M. L., Johnson J. R. 2005. Virulence factors of Escherichia coli isolates that produce CTX-M-type extended-spectrum beta-lactamases. Antimicrob. Agents Chemother. 49:4667–4670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Robicsek A., Strahilevitz J., Sahm D. F., Jacoby G. A., Hooper D. C. 2006. qnr prevalence in ceftazidime-resistant Enterobacteriaceae isolates from the United States. Antimicrob. Agents Chemother. 50:2872–2874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Rogers B. A., Sidjabat H. E., Paterson D. L. 2011. Escherichia coli O25b-ST131: a pandemic, multiresistant, community-associated strain. J. Antimicrob. Chemother. 66:1–14 [DOI] [PubMed] [Google Scholar]
- 15. Villegas M. V., Kattan J. N., Quinteros M. G., Casellas J. M. 2008. Prevalence of extended-spectrum beta-lactamases in South America. Clin. Microbiol. Infect. 14(suppl. 1):154–158 [DOI] [PubMed] [Google Scholar]
- 16. Wirth T., et al. 2006. Sex and virulence in Escherichia coli: an evolutionary perspective. Mol. Microbiol. 60:1136–1151 [DOI] [PMC free article] [PubMed] [Google Scholar]