Abstract
During replication, oral polio vaccine (OPV) can revert to neurovirulence and cause paralytic poliomyelitis. In individual vaccinees, it can acquire specific revertant point mutations, leading to vaccine-associated paralytic poliomyelitis (VAPP). With longer replication, OPV can mutate into vaccine-derived poliovirus (VDPV), which causes poliomyelitis outbreaks similar to those caused by wild poliovirus. After wild poliovirus eradication, safely phasing out vaccination will likely require global use of inactivated polio vaccine (IPV) until cessation of OPV circulation. Mexico, where children receive routine IPV but where OPV is given biannually during national immunization days (NIDs), provides a natural setting to study the duration of OPV circulation in a population primarily vaccinated with IPV. We developed a real-time PCR assay to detect and distinguish revertant and nonrevertant OPV serotype 1 (OPV-1), OPV-2, and OPV-3 from RNA extracted directly from stool and sewage. Stool samples from 124 children and 8 1-liter sewage samples from Orizaba, Veracruz, Mexico, collected 6 to 13 weeks after a NID were analyzed. Revertant OPV-1 was found in stool at 7 and 9 weeks, and nonrevertant OPV-2 and OPV-3 were found in stool from two children 10 weeks after the NID. Revertant OPV-1 and nonrevertant OPV-2 and -3 were detected in sewage at 6 and 13 weeks after the NID. Our real-time PCR assay was able to detect small amounts of OPV in both stool and sewage and to distinguish nonrevertant and revertant serotypes and demonstrated that OPV continues to circulate at least 13 weeks after a NID in a Mexican population routinely immunized with IPV.
INTRODUCTION
Since the World Health Organization (WHO) unveiled a plan to eradicate poliomyelitis in 1988, the number of reported annual global cases dropped from 350,000 to 1,606 in 2009 (1). The last case of wild poliovirus type 2 was reported in 1999, and all serotypes of indigenous wild poliovirus have been eliminated from all but four countries. This has been accomplished primarily with the use of the Sabin oral polio vaccine (OPV), used in most of the developing world because it is inexpensive and easy to administer and promotes herd immunity since it is shed in the stools of vaccinees and can spread to their close contacts. However, as a live, attenuated RNA virus, OPV can mutate into neurovirulent forms.
OPV can cause vaccine-associated paralytic poliomyelitis (VAPP) in vaccinees or their close contacts at a rate of about 1 case per 500,000 primary vaccinees (15). As OPV replicates in the gut, it rapidly acquires specific point mutations in stem-loop V of the 5′ untranslated region (OPV serotype 1 [OPV-1], 480 G to A; OPV-2, 481 A to G; OPV-3, 472 U to C) that reverts OPV back to the wild-type sequence at those positions. These mutations, though often encountered in healthy vaccinees, are known to be associated with VAPP and increased neurovirulence. More recently, it has been discovered that with prolonged replication, the OPV genome can mutate 1 to 15% to form vaccine-derived poliovirus (VDPV), capable of causing poliomyelitis outbreaks in undervaccinated communities (14). To date, 12 independent VDPV outbreaks have been identified. The attack rate and severity of disease associated with VDPV are similar to those seen with wild polioviruses (6).
There is debate about how to phase out the use of polio vaccine after wild-type polio eradication due to the risk of outbreaks from continued circulation of mutant forms of OPV. A safe but possibly expensive strategy would be to globally stop all OPV and administer inactivated polio vaccine (IPV) until all OPV circulation ceases. IPV is more expensive than OPV and is thought to provide inferior intestinal immunity, but it is effective at preventing poliomyelitis and does not have the potential to mutate into neurovirulent forms. However, there are few data regarding OPV circulation after IPV administration, especially in the era of PCR detection. Data from older, tissue culture-based studies may have underestimated the circulation of OPV and, thus, how long IPV would need to be given if there was a global switch to IPV. Children previously vaccinated with IPV are known to shed OPV for a longer period after an OPV challenge than children previously vaccinated with OPV, but population studies are lacking (2, 9). Mexico switched to a primary IPV regimen in 2007, but it still gives OPV during semiannual national immunization days (NIDs). With its unique vaccination practices, Mexico is an ideal setting to study the duration of OPV circulation in a population now vaccinated with IPV.
Traditionally, polioviruses are isolated from OPV vaccinees and VAPP patients by treating their stool specimens with chloroform and inoculating the supernatant into a variety of cell cultures (19). A second cell culture passage is generally required to identify the viral serotype. The virus grown in cell culture can then be further characterized using conventional or real-time PCR (7). The traditional method has the advantage of producing large concentrations of virus for characterization and sequencing and is well-suited for diagnostic purposes. However, for research purposes, this method has several disadvantages compared to performing reverse transcription and real-time PCR directly on RNA extracted from stool. First, growth in cell culture can lead to in vitro mutation of the virus, such that the virus identified after cell culture passage may not represent the genotype that was actually present in the patient's stool (8). Second, the cell culture step increases the time needed to obtain a result on the order of 1 to 3 weeks. Further, cell culture has been reported to be approximately 100-fold less sensitive than real-time PCR (3). Finally, including a cell culture step means that one cannot quantify the amount of virus present in the original sample, unlike with real-time PCR alone.
Our laboratory at Stanford previously reported a real-time PCR assay that could rapidly and efficiently detect and distinguish nonrevertant OPV-3 and revertant OPV-3 containing the characteristic point mutation in the 5′ untranslated region (4). We have since modified this assay to increase the sensitivity for revertant OPV-3 and have developed similar assays to detect and distinguish nonrevertant and revertant OPV-1 and -2. In this article, we describe this novel real-time PCR assay and report the results from our pilot study in which this assay was used to investigate the circulation of OPV and its revertant forms in stool and sewage in and around Orizaba, Veracruz, Mexico, 6 to 13 weeks after a NID.
MATERIALS AND METHODS
Study design and population.
This was a cross-sectional study conducted in four municipalities in or near Orizaba, Veracruz, Mexico (Rio Blanco, Orizaba, Rafael Delgado, and Tlilpanan). Enrollment occurred in August and September of 2009. The 124 children enrolled were evenly divided among the four municipalities and were between 0 and 2 years old. Inclusion criteria included having received or planning to receive IPV as their primary, scheduled vaccine regimen. Exclusion criteria included receipt of OPV as part of the primary vaccine regimen, international travel in the last 4 months, or unknown vaccination history. Receipt of OPV during the NID that occurred from 8 to 26 June 2009 was recorded but was neither an inclusion nor an exclusion criterion.
After informed consent was obtained from the parents or legal guardians of the study subjects, the following data were collected on each subject: age, vaccination history, travel history, breast-feeding and other nutritional history, general health, and household information. Anthropometric indices such as height, weight, and head circumference were taken. A stool sample was collected from each subject either at the initial visit or within a week of the initial visit. In addition, 1 liter of sewage was collected from each of four arroyos draining the four municipalities 6 and 13 weeks after the end of the NID (5 August and 23 September 2009). This study received approval from the Stanford Institutional Review Board, the Comisión de Ética and the Comisión de Bioseguridad of the Mexican National Institute of Public Health, and the Public Health Center of Orizaba, Veracruz, Mexico.
Sample processing.
The stool and sewage samples were aliquoted, labeled, and then frozen at −80°C until shipment on dry ice to Stanford. At Stanford, the sewage was thawed, separated into 100-ml aliquots, and concentrated with a vacuum filtration method using HAWG 0.45-μm-pore-size filter membranes (Millipore Inc., Billerica, MA) as previously described (17). The filtered sewage and filter membrane were added to cryovials containing 500 μl of guanidine isothiocyanate (GITC) buffer (GITC buffer contained 5 M guanidine thiocyanate, 100 mM EDTA, and 0.5% N-lauroyl sarcosine) and 350 μl of RLT buffer (Qiagen, Valencia, CA) containing β-mercaptoethanol, vortexed, and stored at −80°C.
After they were thawed, the stool and filtered sewage samples were disrupted or homogenized. The stool was disrupted by adding approximately 100 μl stool to 900 μl of the RLT buffer containing β-mercaptoethanol, aggressively vortexing the solution for 1 min, centrifuging the solution at 14,000 rpm for 3 min, and then collecting 700 μl of supernatant. The filtered sewage solution and filter membrane (ripped with sterile forceps) were homogenized by transferring them to a QIAshredder spin column, centrifuging for 2 min, and collecting the flow through (Qiagen, Valencia, CA).
RNA extraction.
The RNA extraction procedure was common to both the homogenized stool and sewage samples. An RNeasy minikit (Qiagen, Valencia, CA) was used according to the manufacturer's protocol. Positive- and negative-control reactions were included in each set of extractions. Extracted RNA preparations were stored at −80°C.
Reverse transcription.
After viral RNA extraction, reverse transcription was performed to create cDNA. Five microliters of extracted sample was combined with 1 μl random hexamer primer (3 μg/μl; Invitrogen, Carlsbad, CA), 1 μl 10 mM deoxynucleoside triphosphate (dNTP) mix (10 mM each dNTP), and 5 μl distilled water. This reaction mixture was heated to 65°C for 5 min and then quick chilled on ice. Four microliters 5× first-strand buffer (Invitrogen, Carlsbad, CA), 2 μl 0.1 M dithiothreitol, and 1 μl RNaseOut recombinant RNase inhibitor (40 units/μl; Invitrogen, Carlsbad, CA) were added, mixed with gentle pipetting, and incubated at 25°C for 2 min. Finally, 1 μl (200 units/μl) of SuperScript II reverse transcriptase (Invitrogen, Carlsbad, CA) was added and mixed with gentle pipetting. The mixture was incubated at 25°C for 10 min, 42°C for 50 min, and 70°C for 15 min. Forty-five microliters of distilled water was added to the reaction mixture, before proceeding to the PCR assay. Positive and negative controls were included with each set of reactions.
Real-time PCR method.
Each reverse transcription product then underwent real-time PCR analysis, with six different assays run in duplicate to detect both the revertant and nonrevertant OPV serotypes. The primers and probes used are presented in Table 1. Five microliters of the reverse transcription reaction mixture was combined with 10 μl TaqMan universal PCR master mix without uracil-N-glycosylase (Applied Biosystems, Foster City, CA), 0.05 μl TaqMan probe (Applied Biosystems, Foster City, CA), 0.9 μl each of forward and reverse primers, and 3.15 μl distilled water. Cycling conditions were as described previously (4). Samples which returned average cycle threshold (CT) values of >35 from duplicate assays were considered negative, due to an increased number of false-positive results above this CT.
Table 1.
Primers and probes for the real-time PCR assaysa
| Purpose and name of primer or probe | Sequence (5′ to 3′) | Location in genome (nt) |
|---|---|---|
| Assay for type 1 nonrevertants | ||
| P1 NM2 | CGGCTAATCCCAACCTCGTG | 461–480 |
| P1 REV | CACCATAAGCAGCCACAATAAAATAA | 566–591 |
| P1 probe | FAM-AAACACGGACACCCAAA-MGB | 545–561 |
| Assay for type 1 revertants | ||
| P1 MUT1 | CGGCTAATCCCAACCTCGTA | 461–480 |
| P1 REV | CACCATAAGCAGCCACAATAAAATAA | 566–591 |
| P1 probe | FAM-AAACACGGACACCCAAA-MGB | 545–561 |
| Assay for type 2 nonrevertants | ||
| P2 NM | CGGCTAATCCTAACCACGGCA | 461–481 |
| P2 REV | GGAAACACGGACACCCAAAG | 544–563 |
| P2 probe | FAM-TGACTGGCTTGTCGTAAC-MGB | 500–517 |
| Assay for type 2 revertants | ||
| P2 MUT | CGGCTAATCCTAACCACGGTG | 461–481 |
| P2 REV | GGAAACACGGACACCCAAAG | 544–563 |
| P2 probe | FAM-TGACTGGCTTGTCGTAAC-MGB | 500–517 |
| Assay for type 3 nonrevertants | ||
| P3 NM | CCCCTGAATGCGGCTAACT | 454–472 |
| P3 REV | CTGGCTGCTGGGTTGCA | 493–509 |
| P3 probe | FAM-TAACCATGGAGCAGGCA-MGB | 474–490 |
| Assay for type 3 revertants | ||
| P3 MUT1 | CCCCTGAATGCGGCTAAAC | 454–472 |
| P3 REV | CTGGCTGCTGGGTTGCA | 493–509 |
| P3 probe | FAM-TAACCATGGAGCAGGCA-MGB | 474–490 |
The nonrevertant and revertant assays for each serotype differ by only the last 2 nucleotides (nt) in the forward primer. FAM, 6-carboxyfluorescein; MGB, minor groove binder.
The following formula was used to calculate the revertant proportion (RP): 2−revCT/(2−revCT + 2−nonrevCT), where revCT and nonrevCT are revertant and nonrevertant CTs, respectively.
Development of PCR assay.
Using Primer Express software (version 3.0; Applied Biosystems, Foster City, CA), separate primer and probe sets were created for the three serotypes and their revertant forms. The forward primers were constructed using the principles of the mismatch amplification assay, such that the penultimate nucleotide overlaps with the revertant point mutation and the forward primer sequence for the revertant and nonrevertant assays for each serotype differed only by the last two nucleotides (10). We previously published the nonrevertant primer and probe combination for OPV-3 (4) and now describe the revertant OPV-3 forward primer and the primer and probe sets for nonrevertant and revertant OPV-1 and OPV-2.
Stocks of positive controls (Sabin type 1, GenBank accession no. AY184219; Sabin type 2, GenBank accession no. AY184220; Sabin type 3, GenBank accession no. AY184221; revertant type 1, GenBank accession no. V01149; revertant type 2, GenBank accession no. AY238473; revertant type 3, GenBank accession no. POL3L37) were obtained from the United States Food and Drug Administration (FDA) courtesy of Konstantin Chumakov. From these seed stocks, we grew stocks of positive controls in Hep2 cells using methods described in the WHO Polio Laboratory Manual (19). Isolates were sequenced (Sequetech, Mountain View, CA) to confirm that the region to be amplified in the 5′ untranslated region conformed to the known sequences for nonrevertant and revertant OPV-1, -2, and -3. These isolates were also tested with the real-time PCR assay to ensure that there was no cross-reaction with the other serotypes. The concentrations of our controls were calculated by inoculating plates of Hep2 cell cultures with serial dilutions of the control viruses in duplicate to calculate the 50% cell culture infectious dose (CCID50). Control stocks were used to compare different primer/probe sets until the most sensitive sets with minimal cross-reaction between serotypes were obtained (Table 1) and then to calculate the lower limit of detection of the assays.
Statistical analysis.
In analyzing characteristics between OPV shedders and nonshedders, Fisher's exact test was used for categorical outcomes, and the t test was used for continuous outcomes.
RESULTS
Sensitivity and discrimination of real-time PCR assays.
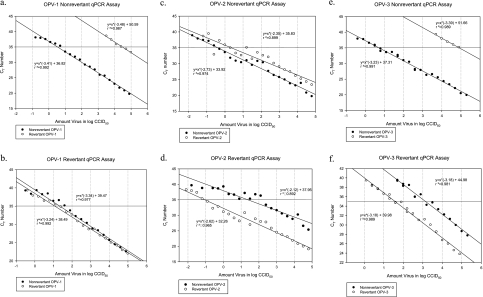
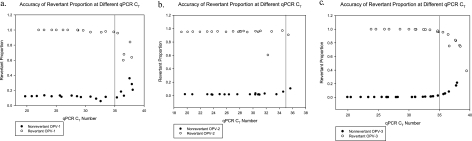
We spiked serial dilutions of control nonrevertant and revertant viral suspensions into control stool samples and performed a complete cycle of RNA extraction, reverse transcription, and real-time PCR to determine the sensitivity of our assays. We were able to detect as few as 3, 0.4, and 5 CCID50 virus in 100 μl of stool for nonrevertant OPV-1, -2, and -3, respectively, and 12, 0.09, and 37 CCID50 virus in 100 μl of stool for revertant OPV-1, -2, and -3, respectively (Fig. 1). Using our revertant proportion formula with the results from our nonrevertant and revertant real-time PCR assays, we were also able to distinguish nonrevertant and revertant strains up to a cycle threshold of 35, our cutoff for detection of OPV (Fig. 2). With the exception of one outlier among the revertant OPV-2, all of the serial dilutions of revertant OPV-1, -2, and -3 with cycle thresholds of ≤35 had a revertant proportion of >0.8, and all of the serial dilutions of nonrevertant OPV-1, -2, and -3 with cycle thresholds of ≤35 had a revertant proportion of <0.2. Further, in order to verify the discrimination of our assay, we shortened the forward primers so that they would not overlap with the revertant point mutation (which made the assay about 100-fold less sensitive) and sequenced the purified real-time PCR products using these shortened primers (Quintarabio, Albany, CA). While the concentrations of virus from the samples in the Mexican pilot study described here were too low for us to be able to sequence virus in this manner, we were able to sequence virus at higher concentrations from stool samples from another study (results not shown). In all cases, the sequencing results corresponded to our PCR assay results in terms of discrimination between the nonrevertant and revertant serotypes.
Fig. 1.
Lower limits of detection of OPV-1, -2, and -3 nonrevertant and revertant real-time PCR assays. qPCR, quantitative (real-time) PCR. (a, c, and e) Assays for OPV serotypes 1, 2, and 3, respectively, that are nonrevertant (without the point mutation associated with vaccine-associated paralytic poliomyelitis in the 5′ untranslated region); (b, d, and f) assays for revertant OPV serotypes 1, 2, and 3, respectively. Our revertant proportion equation, from which we determine if an isolate is primarily nonrevertant or revertant, takes into account the results of both the nonrevertant and revertant assays for each serotype. Consequently, although the revertant OPV-1 assay is not very specific for revertant OPV-1 (b), given the high specificity of the nonrevertant OPV-1 assay (a), revertant versus nonrevertant OPV-1 can reliably be distinguished using the revertant proportion equation. Each point represents a serial dilution of control nonrevertant or revertant OPV of a specific serotype that was spiked into stool and subsequently underwent RNA extraction, reverse transcription, and real-time PCR. The concentration where the plotted line crosses a CT of 35, our internal cutoff for a positive result, represents the lower limit of detection of that assay for that specific type of OPV.
Fig. 2.
Ability of the OPV-1, -2, and -3 real-time PCR assays to distinguish nonrevertant and revertant strains at different cycle thresholds. qPCR, quantitative (real-time) PCR. Revertant proportion is the proportion of virus that contains the revertant point mutation in the 5′ untranslated region that is associated with vaccine-associated paralytic poliomyelitis. A revertant proportion close to 1.0 indicates that most of the virus has the revertant point mutation, while a revertant point mutation close to 0.0 indicates that most of the virus has not developed this point mutation. Of note, a cycle threshold number above 35 is considered negative for the presence of OPV.
Of note, our laboratory has previously published a favorable side-by-side comparison of our real-time PCR assay to mutant analysis by PCR and enzyme cleavage, a method developed by the FDA, to detect and distinguish nonrevertant and revertant OPV-3 (4). We have since changed the sequence of the revertant forward primer, as we noted that this increased the sensitivity of our assay for revertant OPV-3 by approximately 60-fold.
Testing of samples from Mexican pilot study.
A total of 124 stool samples (1 from each enrolled child) and 5 1-liter samples of sewage were analyzed. Characteristics of the 124 enrolled children are described in Table 2. Study participants, on average, had received three doses of IPV, and only 27% of them had received OPV in the most recent NID. Four of the 124 children had detectable OPV in their stool (revertant OPV-1 at 7 weeks, revertant OPV-1 at 9 weeks, and both nonrevertant OPV-2 and OPV-3 in 2 children at 10 weeks after the NID; Table 3).
Table 2.
Characteristics of the 124 enrolled subjectsa
| Characteristic | Value |
|---|---|
| Gender | |
| Male | 57 (46) |
| Female | 67 (54) |
| Age | |
| Avg (mo) | 13 |
| 0-6 mo | 16 (13) |
| 7-12 mo | 50 (40) |
| 13-18 mo | 25 (20) |
| >18 mo | 33 (27) |
| Household | |
| Urban residence | 62 (50) |
| Rural residence | 62 (50) |
| Primary caretaker: mother | 118 (95) |
| Avg no. of other household children | 1.2 |
| Avg no. of other household children <5 yr old | 0.6 |
| In day care | 2 (2)b |
| Nutrition | |
| History of breast-feeding | 116 (94) |
| Currently breast-feeding | 76 (61) |
| Eating solid foods | 103 (83) |
| Water consumed | |
| Bottled | 71 (57) |
| Tap | 28 (23) |
| Boiled river, well, or tap | 23 (19) |
| Avg body mass index (kg/m2) | 17.4 |
| Vaccinations | |
| Up to date on vaccinations (per parent) | 120 (98)b |
| Avg no. of prior doses of IPV | 2.8 |
| Prior receipt of OPV in any NID | 55 (45)b |
| Receipt of OPV in most recent (June) NID | 33 (27) |
| Other household children with prior receipt of OPV | 62 (71)c |
| Health | |
| Immunodeficient person in household | 6 (5) |
| Child reported to generally be in good health | 124 (100) |
| Child reported to currently be ill | 3 (2) |
| Travel | |
| Any recent (prior 6 mo) international travel | 0 (0)b |
| Any recent international travel by household | |
| members | 0 (0) |
The only variable that showed any statistically significant difference between the 4 subjects with OPV isolated from their stools and the other 120 subjects, using Fisher's exact test for categorical variables and a t test for continuous variables, was average body mass index (20.0 versus 17.3 kg/m2, P = 0.0232). Unless indicated otherwise, data represent number (percent) of subjects.
n = 123, as the response was unknown for one enrollee.
n = 87, as the response was unknown for 37 enrollees.
Table 3.
Characteristics of study subjects shedding OPV and the OPV isolates detected in their stool
| Subject | Age (mo) | Gender | Municipality | No. of prior doses |
Receipt of OPV in June NID | Collection time (wk since NID) | Serotype isolated | Revertant proportion | |
|---|---|---|---|---|---|---|---|---|---|
| IPV | OPV | ||||||||
| 1 | 23 | Female | Orizaba (urban) | 3 | 1 | No | 7 | Revertant OPV-1 | 0.98 |
| 2 | 11 | Male | Orizaba (urban) | 3 | 0 | No | 9 | Revertant OPV-1 | 0.98 |
| 3 | 10 | Male | Orizaba (urban) | 3 | 2a | Yes | 10 | Nonrevertant OPV-2 | 0.070 |
| Nonrevertant OPV-3 | 0.23 | ||||||||
| 4 | 13 | Male | Rio Blanco (urban) | 3 | 0 | No | 10 | Nonrevertant OPV-2 | 0.044 |
| Nonrevertant OPV-3 | 0.037 | ||||||||
One of the prior doses of OPV for subject 3 was the one received in the June 2009 NID.
All three serotypes of OPV were detectable in 3/3 1-liter samples of sewage at 6 weeks and 1/2 1-liter samples of sewage at 13 weeks after the NID (Table 4). The liter of sewage that did not contain all three serotypes still contained two serotypes (OPV-2 and OPV-3). As in the stool samples, most of the OPV type 1 was revertant (10/17 positive aliquots had RPs of >0.5), while OPV types 2 and 3 were predominantly nonrevertant. The concentrations of OPV in both stool and sewage samples were relatively low, with CT numbers predominantly being in the 33 to 35 range.
Table 4.
Characteristics of sewage OPV isolatesa
| Arroyo where sewage was collected | Municipalities drained by this arroyo | Collection time (wk since NID) | OPV-1 |
OPV-2 |
OPV-3 |
|||
|---|---|---|---|---|---|---|---|---|
| % aliquots with detectable virus | Median revertant proportion | % aliquots with detectable virus | Median revertant proportion | % aliquots with detectable virus | Median revertant proportion | |||
| La Carbonera | Rio Blanco (urban) | 6 | 60 | 0.68 | 10 | 0.23 | 40 | 0.11 |
| La Carbonera | Rio Blanco (urban) | 13 | 0 | 60 | 0.11 | 30 | 0.059 | |
| Xalatl | Rafael Delgado and Tlilpanan (rural) | 6 | 60 | 0.50 | 70 | 0.13 | 70 | 0.11 |
| Escameda | Orizaba (urban) | 13 | 40 | 0.46 | 100 | 0.0090 | 100 | 0.17 |
| Totolitos | Orizaba (urban) | 6 | 10 | 0.36 | 20 | 0.092 | 40 | 0.14 |
Of note, each liter of sewage was divided into 10 100-ml aliquots that were analyzed separately, and the median revertant proportion of the 10 aliquots is listed. When it was detected, concentrations of OPV were low (cycle thresholds, between 32 and 35). A revertant proportion close to 1.0 indicates that most of the virus has the revertant point mutation, while a revertant point mutation close to 0.0 indicates that most of the virus has not developed this point mutation. Revertant proportions close to 0.5 indicate a mixture of revertant and nonrevertant isolates.
Although 8 1-liter samples of sewage were collected, we include analysis from only 5 of the 8 samples. We noted significant intraoperator variability in the sewage analysis, and we determined that including the filter membrane in the QIAshredder, as opposed to just the buffer in which the membrane was stored, markedly improved sensitivity. Consequently, we included only the 5 1-liter samples processed in this manner in our analysis. These 5 1-liter samples include 2 samples from arroyos draining urban municipalities at 6 weeks, the sample from the arroyo draining the rural municipalities at 6 weeks, and 2 samples from arroyos draining urban municipalities at 13 weeks after the NID (Table 4).
DISCUSSION
In this paper we describe our novel real-time PCR assay that can detect and accurately distinguish nonrevertant and revertant OPV serotypes 1, 2, and 3 at levels as low as or lower than 37 CCID50 virus in 100 μl of stool from RNA extracted directly from stool and sewage. Using this assay, we show that OPV continues to circulate at least 13 weeks after a NID in a Mexican community primarily vaccinated with IPV. IPV is felt to provide inferior intestinal immunity compared to OPV, and consequently, OPV circulation might continue for a longer period in this population than in a community primarily vaccinated with OPV. Children vaccinated with IPV have been found to have increased OPV shedding after an OPV challenge than children primarily vaccinated with OPV (37% IPV-vaccinated children with continued shedding at 3 weeks versus 5% of OPV-vaccinated children with continued shedding at 3 weeks) but less OPV shedding than previously unvaccinated children (of whom 81% were still shedding at 3 weeks) (2, 9).
The duration of shedding that we report is longer than that reported in a New Zealand study, which monitored stool and sewage for the presence of OPV after New Zealand changed to a national IPV vaccination regimen (5). In New Zealand, OPV was detected in stools at up to 4 weeks and in sewage at up to 12 weeks after the change. The longer duration of shedding that we report could be due to differences in climate and hygiene between the two countries or because the cell culture method to detect OPV used in the New Zealand study may have been less sensitive at detecting OPV than our PCR method. Our results appear to be similar to those from a study in Cuba, where IPV is not used and polio vaccination is done exclusively through semiannual NIDs with OPV (13). The Cuban study, using a cell culture technique to detect OPV, found that OPV was detectable in stool samples until between 7 and 15 weeks and in sewage until between 15 and 19 weeks after an NID. The proportions of children with detectable OPV in stool samples in the Cuban study were approximately 25% 3 to 6 weeks after vaccination, 13% by 7 weeks after vaccination, and 0% by 15 weeks after vaccination. However, since we did not continue the pilot study beyond 13 weeks, the duration of OPV circulation in the community that we studied in Mexico may extend beyond what we report. Indeed, in our study, sewage was collected during the rainy season and was visibly more dilute than at other times during the year, yet OPV was still detectable. This suggests that because OPV shedding was still prevalent at 13 weeks under such conditions, shedding may have continued for some weeks further. We are currently conducting a larger, longitudinal study in the same Mexican community to determine the precise duration of OPV circulation.
A surprising result of our study was that the OPV-2 and -3 detected in both stool and sewage 6 to 13 weeks after the NID were predominantly nonrevertant. This conflicts with historical data, from both Mexico and elsewhere, in which by 2 weeks after receipt of OPV, almost all of the OPV shed by vaccinees is revertant with the characteristic point mutations in the 5′ untranslated region (4, 12, 15). The etiology of this discrepancy is unclear. Mexico has recently changed from propagation of OPV seed strains in primary monkey kidney cells to Vero cells (16). However, there are no convincing data that propagation in Vero cells increases the stability of OPV, and indeed, one study showed no modification in the pattern of poliovirus excretion when children vaccinated with OPV propagated in Vero cells were compared with children vaccinated with OPV propagated in primary monkey kidney cells (11). We were unable to confirm the absence of the point mutations by sequencing our samples, due to the low concentrations of virus present. However, our real-time PCR assay detected primarily revertant OPV, as expected, from stool samples from a separate study in Africa looking at OPV shedding (data not shown). We were able to sequence some of these isolates from African stool samples that had high OPV concentrations, and in all cases the sequence results corresponded to our real-time PCR results in terms of the presence or absence of the revertant point mutations. This suggests that the etiology of the absence of revertant OPV-2 and OPV-3 is not due to a flaw in our assay but rather is inherent in the samples themselves.
One limitation of our study is that the conditions in the Mexican community approximate, but do not replicate, what might occur if wild poliovirus is eradicated and there is global OPV cessation and initiation of IPV. In the latter scenario, moving forward from the date of OPV cessation, any child too young to have been vaccinated would be vaccinated only with IPV. In the Mexican community that we studied, while all the children had received IPV as their primary regimen, 45% of them had also received OPV during a national immunization day. Consequently, they might have higher intestinal immunity than children vaccinated solely with IPV. In addition, while the environmental conditions in Mexico are similar to those in countries that most favor poliovirus spread, Mexico has very high vaccination rates. Consequently, Mexico is not 1 of the 16 countries, primarily located in Africa and southern Asia, that suffered an outbreak of VDPV in the last decade (18). These are countries where OPV circulation after global OPV cessation would be expected to continue the longest. However, until these countries implement a national change from OPV to IPV, the unique vaccination policy in Mexico best allows us to study OPV community circulation in a country that has switched to using IPV as its primary vaccination regimen.
In summary, our real-time PCR assay was able to detect very low concentrations of OPV in both stool and sewage and to distinguish nonrevertant and revertant serotypes. Using this assay, we were able to determine that OPV circulation continues for at least 13 weeks after a NID in a Mexican community primarily vaccinated with IPV. Further studies are ongoing to better define the duration of OPV circulation and mutation after a NID in Mexico.
ACKNOWLEDGMENTS
We thank the population, participants, and health care workers of the Orizaba Health Jurisdiction, Mexico, for their generous support and cooperation. We acknowledge Julia Janssen and Katie Nelson for their assistance with performing the laboratory assays, Ali Rowhani-Rahbar for assistance in performing the sample size calculations, and Amy Sturt for review of the manuscript.
This work was supported by a Thrasher Research Fund Young Investigator Award (to S. B. Troy), NIH T32 training grant 5 T32 AI 07502-13 (to S. B. Troy), and the Instituto Nacional de Salud Pública in Mexico.
Footnotes
Published ahead of print on 16 March 2011.
REFERENCES
- 1. Anonymous 2010. Progress toward interruption of wild poliovirus transmission—worldwide, 2009. MMWR Morb. Mortal. Wkly. Rep. 59:545-550 [PubMed] [Google Scholar]
- 2. Asturias E., et al. 2007. Randomized trial of inactivated and live polio vaccine schedules in Guatemalan infants. J. Infect. Dis. 196:692–698 [DOI] [PubMed] [Google Scholar]
- 3. Corless C., et al. 2002. Development and evaluation of a ‘real-time’ RT-PCR for the detection of enterovirus and parechovirus RNA in CSF and throat swab samples. J. Med. Virol. 67:555–562 [DOI] [PubMed] [Google Scholar]
- 4. Gnanashanmugam D., et al. 2007. Shedding and reversion of oral polio vaccine type 3 in Mexican vaccinees: comparison of mutant analysis by PCR and enzyme cleavage to a real-time PCR assay. J. Clin. Microbiol. 45:2419–2425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Huang Q., et al. 2005. Persistence of oral polio vaccine virus after its removal from the immunisation schedule in New Zealand. Lancet 366:394–396 [DOI] [PubMed] [Google Scholar]
- 6. Jenkins H., et al. 2010. Implications of a circulating vaccine-derived poliovirus in Nigeria. N. Engl. J. Med. 362:2360–2369 [DOI] [PubMed] [Google Scholar]
- 7. Kilpatrick D. R., et al. 2009. Rapid group-, serotype-, and vaccine strain-specific identification of poliovirus isolates by real-time reverse transcription-PCR using degenerate primers and probes containing deoxyinosine residues. J. Clin. Microbiol. 47:1939–1941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Laassri M., et al. 2005. Genomic analysis of vaccine-derived poliovirus strains in stool specimens by combination of full-length PCR and oligonucleotide microarray hybridization. J. Clin. Microbiol. 43:2886–2894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Laassri M., et al. 2005. Effect of different vaccination schedules on excretion of oral poliovirus vaccine strains. J. Infect. Dis. 192:2092–2098 [DOI] [PubMed] [Google Scholar]
- 10. Li B., Kadura I., Fu D., Watson D. 2004. Genotyping with TaqMAMA. Genomics 83:311–320 [DOI] [PubMed] [Google Scholar]
- 11. Mallet L., et al. 1997. Comparative study of poliovirus excretion after vaccination of infants with two oral polio vaccines prepared on Vero cells or on primary monkey kidney cells. J. Med. Virol. 52:50–60 [PubMed] [Google Scholar]
- 12. Martinez C., et al. 2004. Shedding of Sabin poliovirus type 3 containing the nucleotide 472 uracil-to-cytosine point mutation after administration of oral poliovirus vaccine. J. Infect. Dis. 190:409–416 [DOI] [PubMed] [Google Scholar]
- 13. Más Lago P., et al. 2003. Poliovirus detection in wastewater and stools following an immunization campaign in Havana, Cuba. Int. J. Epidemiol. 32:772–777 [DOI] [PubMed] [Google Scholar]
- 14. Minor P. 2009. Vaccine-derived poliovirus (VDPV): impact on poliomyelitis eradication. Vaccine 27:2649–2652 [DOI] [PubMed] [Google Scholar]
- 15. Minor P., Macadam A., Stone D., Almond J. 1993. Genetic basis of attenuation of the Sabin oral poliovirus vaccines. Biologicals 21:357–363 [DOI] [PubMed] [Google Scholar]
- 16. ProMexico 2010, posting date Negocios. Birmex: The Mexican Health Company, Mexico DF, Mexico: http://negocios.promexico.gob.mx/english/02-2010/art05.html [Google Scholar]
- 17. Walters S., Yamahara K., Boehm A. 2009. Persistence of nucleic acid markers of health-relevant organisms in seawater microcosms: implications for their use in assessing risk in recreational waters. Water Res. 43:4929–4939 [DOI] [PubMed] [Google Scholar]
- 18. WHO 22 March 2011, posting date Circulating vaccine-derived poliovirus (cVDPV) 2000–2011. World Health Organization, Geneva, Switzerland: www.globalpolioeradication.org/dataandmonitoring/poliothisweek/circulatingvaccinederivedpoliovirus.aspx [Google Scholar]
- 19. WHO 2004. Polio laboratory manual, 4th ed. Department of Immunization, Vaccines, and Biologicals, World Health Organization, Geneva, Switzerland [Google Scholar]