Abstract
Patients with autoimmune polyendocrinopathy-candidiasis-ectodermal dystrophy (APECED) are prone to chronic mucocutaneous candidiasis, which is often treated with azoles. The purpose of this study was to characterize the oral Candida populations from 16 Irish APECED patients, who comprise approximately half the total number identified in Ireland, and to examine the effect of intermittent antifungal therapy on the azole susceptibility patterns of Candida isolates. Patients attended between one and four clinical evaluations over a 5-year period, providing oral rinses and/or oral swab samples each time. Candida was recovered from 14/16 patients, and Candida albicans was the only Candida species identified. Interestingly, clinical diagnosis of candidiasis did not correlate with microbiological evidence of Candida infection at 7/22 (32%) clinical assessments. Multilocus sequence typing analysis of C. albicans isolates recovered from the same patients on separate occasions identified the same sequence type each time. Fluconazole resistance was detected in isolates from one patient, and isolates exhibiting a progressive reduction in itraconazole and/or fluconazole susceptibility were identified in a further 3/16 patients, in each case correlating with the upregulation of CDR- and MDR-encoded efflux pumps. Mutations were also identified in the ERG11 and the TAC1 genes of isolates from these four patients; some of these mutations have previously been associated with azole resistance. The findings suggest that alternative Candida treatment options, other than azoles such as chlorhexidine, should be considered in APECED patients and that clinical diagnosis of oral candidiasis should be confirmed by culture prior to the commencement of anti-Candida therapy.
INTRODUCTION
Autoimmune polyendocrinopathy-candidiasis-ectodermal dystrophy (APECED) is a monogenic autosomal recessive genetic disease caused by mutations in the AIRE (AutoImmune REgulator) gene that plays a role in the induction of T cell tolerance in the thymus and is located on human chromosome 21q22.3 (21, 63). To date, more than 60 different mutations have been reported worldwide in this gene, particularly in Finnish, Sardinian, and Iranian Jewish populations (7).
The disease is characterized by autoimmunity to endocrine organs, ectodermal disorders such as hypoplasia of dental enamel, pitted nail dystrophy, and alopecia, and chronic mucocutaneous candidiasis (CMC). The latter of these features is one of the three major clinical signs that define the syndrome, along with hypoparathyroidism and Addison's disease. Typically, CMC is the first manifestation and most common feature of APECED, often occurring before the age of five years, and may affect the nails, mucous membranes, and skin. Mucocutaneous candidiasis can present clinically as pseudomembranous candidiasis, erythematous candidiasis, or chronic hyperplastic candidiasis. These presentations may be accompanied by angular cheilitis, which may also present on its own. Several cases of oral carcinoma have been associated with CMC of the oral cavity and esophagus in these patients (4, 24, 45, 47). To date, a comprehensive analysis of the prevalence of oral Candida species in patients with APECED is lacking; however, a study of Finnish APECED patients by Rautemaa et al. (46) recovered oral Candida from 42/56 (75%) of the patients investigated. Candida albicans, the most commonly identified Candida species in humans, was identified in 35/56 (63%) of the APECED patients. Non-C. albicans Candida species were recovered from 7/56 (13%) APECED patients examined (46).
Because of the high prevalence of CMC in APECED patients, lifelong management of candidiasis is frequently required, typically by intermittent treatment with prophylactic topical and/or systemic azole antifungal drugs (46). The application of a similar candidiasis prophylaxis management strategy in human immunodeficiency virus (HIV)-infected or AIDS patients during the 1990s frequently resulted in the development of resistance to the triazole antifungal fluconazole, as well as cross-resistance to other azoles. Such cases are well documented in both C. albicans and other Candida species (11, 32, 61). The extensive use of prophylactic azole therapy in HIV-infected patients and other patient cohorts also resulted in the selection of Candida species with inherent reduced susceptibility to azoles such as C. glabrata and C. krusei (3, 22, 36). Since many APECED patients are routinely treated with antifungal agents, it is not surprising that azole resistance has also been reported in isolates recovered from them. Rautemaa et al. (46) identified C. albicans isolates with decreased fluconazole susceptibility (fluconazole MIC, 4 to 32 μg/ml) in 11/56 (20%) Finnish APECED patients. That study also reported reduced fluconazole susceptibility in non-C. albicans isolates recovered from two APECED patients.
Due to the predominantly clonal mode of reproduction of pathogenic Candida species, the population structure of these species can provide useful epidemiological information regarding the provenance of isolates exhibiting antifungal drug resistance, as well as predicting common molecular mechanisms by which antifungal drug resistance is mediated. For example, multilocus sequence typing (MLST) has identified the presence of 17 main clades in the population structure of C. albicans, of which clade 1 has been associated with flucytosine resistance due to a common mutation in the FUR1 gene encoding uracil phosphoribosyltransferase that has spread throughout this clade (16, 35, 43). In Candida dubliniensis an MLST clade C3-specific mutation in the FCY1 gene encoding cytosine deaminase has also been shown to confer high-level flucytosine resistance (29, 30). The population structure of C. albicans is now well established according to the consensus MLST scheme, identifying diploid sequence types (DSTs) based on the nucleotide sequences of the AAT1, ACC1, ADP1, PMIb (formerly MPIb), ALA1 (formerly SYA1), VPS13, and ZWF1 genes (6, 35). The online database currently contains allelic and epidemiological information on more than 1,700 DSTs (http://calbicans.mlst.net/).
The molecular mechanisms of azole resistance in Candida species are well documented and are typically mediated (i) by failure to accumulate intracellular concentrations of the drug due to upregulation of the CDR- or MDR-encoded efflux pumps or (ii) by upregulation or conformational alteration of the lanosterol demethylase azole target encoded by the ERG11 gene (22, 23, 26, 32, 36, 38, 51). The most common mechanism of fluconazole resistance in clinical isolates of C. albicans is CaCDR1 upregulation, which also confers resistance to itraconazole and ketoconazole (12, 52–54). An association between the mating type of C. albicans isolates and azole resistance has previously been observed, due to the proximity of the MAT mating type gene to the TAC1 gene on chromosome 5 (14, 50). Gain-of-function (GOF) mutations occurring in the transcriptional activator TAC1 gene have been shown to be responsible for upregulation of CDR-encoded efflux pumps (15, 50, 56). Similarly, GOF mutations in the MRR1 transcriptional regulator gene have been associated with upregulation of the Mdr1 efflux pump in C. albicans and C. dubliniensis (18, 50).
The purpose of the present study was to correlate clinical signs of oral candidiasis with Candida species and cell densities and to investigate the long-term prevalence and persistence of Candida species in a well-characterized Irish cohort of 16 patients with APECED, representing the majority of such patients identified in Ireland. The oral health of this group of APECED patients has previously been reported (28). MLST was used to determine whether the same isolate persists or becomes replaced in APECED patients over prolonged periods of time, particularly following azole treatment, and to identify any cocolonization of patients with more than one type of strain. The effect of intermittent and repetitive antifungal therapy on the azole susceptibility patterns of these isolates was also investigated.
MATERIALS AND METHODS
Study group.
The study group consisted of 16 patients with APECED in Ireland who have been described previously (28) and who each had a confirmed mutation in the AIRE gene. This group represented approximately one-half of such patients identified in Ireland (17). These 16 patients had at least one oral examination at the Dublin Dental University Hospital between 2005 and 2010. Ethical approval was obtained from St. James's Hospital, and the Adelaide and Meath Hospital, incorporating the National Children's Hospital research ethics committee, and also from the research committee at Our Lady's Children's Hospital, Crumlin, Dublin. A calibrated pediatric dentist carried out the majority of the clinical examinations in 2005, with a follow-up examination by another calibrated dentist in 2010. Patients who attended more than one clinical assessment were examined at intervals that were at least 1 month apart; however, the majority were separated by 3-month intervals (Table 1). At each clinical assessment, patients underwent a clinical examination seated in a dental chair with a conventional dental light for illumination. The dentist undertook a standardized visual examination of the oral mucosa, periodontal tissues, and teeth. A comprehensive dental, medical, and drug history was recorded for each patient (Table 1).
Table 1.
Patients with APECED included in the study
| Patient | Sexa | Ageb | Date of clinical assessment (mo/yr) | Medical history-age (yr)c at diagnosis | Drug therapy |
|---|---|---|---|---|---|
| 1 | M | 7 | 3/05 | AI-6, AH-7 | Fludrocortisone, itraconazole, nystatin, miconazole |
| 6/05 | |||||
| 9/05 | |||||
| 2/10 | |||||
| 2 | F | 15 | 2/05 | NAD | None |
| 3 | M | 18 | 2/05 | HPT-13, AI-16, AH-3 | Hydrocortisone, fludrocortisones, nystatin |
| 3/10 | |||||
| 4 | F | 17 | 3/05 | HPT-3.5, AI-8.5 | Prednisolone, fludrocortisones, miconazole, fluconazole |
| 7/05 | |||||
| 10/05 | |||||
| 9/10 | |||||
| 5 | M | 14 | 4/05 | HPT-7, AI-7, Di-4, GI | Hydrocortisone, fludrocortisone, miconazole, fluconazole |
| 5/05 | |||||
| 7/05 | |||||
| 5/10 | |||||
| 6 | F | 14 | 2/05 | HPT-7, AI-11 | Fludrocortisone, hydrocortisone, nystatin, beclomethasone |
| 5/05 | |||||
| 8/05 | |||||
| 7/10 | |||||
| 7 | F | 6 | 2/05 | HPT-5, A, GI | Nystatin |
| 6/05 | |||||
| 12/05 | |||||
| 9/10 | |||||
| 8 | M | 2 | 2/05 | HPT-2, AI-2, GI | Fludrocortisone, nystatin, fluconazole |
| 6/05 | |||||
| 12/05 | |||||
| 9/10 | |||||
| 9 | M | 11 | 2/05 | HPT-5, AI-8, GI | Itraconazole, fluconazole, nystatin, miconazole |
| 10 | M | 15 | 3/05 | HPT-6, A | None |
| 5/05 | |||||
| 6/05 | |||||
| 7/10 | |||||
| 11 | M | 9 | 3/05 | AI-6 | Fludrocortisone, hydrocortisone, miconazole, fluconazole |
| 9/05 | |||||
| 10/05 | |||||
| 4/10 | |||||
| 12 | F | 39 | 3/05 | HPT-2, Di-28, A | Insulin, miconazole, nystatin, fluconazole, itraconazole |
| 9/05 | |||||
| 10/05 | |||||
| 13 | F | 13 | 3/05 | HPT-8, AI-12, A, GI | Fludrocortisone, prednisilone, nystatin, itraconazole |
| 5/05 | |||||
| 9/05 | |||||
| 14 | F | 7 | 8/05 | HPT-3.5 | Miconazole, fluconazole |
| 5/10 | |||||
| 15 | F | 21 | 4/05 | HPT-2.5, AI-8 | Fludrocortisone, hydrocortisone, prednisilone, miconazole, nystatin, fluconazole |
| 9/10 | |||||
| 16 | F | 15 | 5/05 | HPT-8, AI-9, A | Fluconazole, nystatin, itraconazole, fludrocortisone, hydrocortisone |
| 11/05 | |||||
| 12/05 |
F, female; M, male.
That is, the age of the patient in years when the study commenced.
Abbreviations: AI, adrenal insufficiency; AH, autoimmune hepatitis; NAD, no appreciable disease; HPT, hypoparathyroidism; Di, diabetes; GI, gastrointestinal disturbance; A, alopecia.
Assessment of oral candidiasis and identification of Candida isolates.
Lesions clinically diagnosed by the dentist as oral candidiasis were recorded according to both a descriptive classification of lesions by Sitheeque and Samaranayake (57) and lesion sites in the oral cavity. Oral candidiasis clinical presentations included pseudomembranous candidiasis, erythematous candidiasis, and hyperplastic candidiasis. An additional Candida-associated lesion, angular cheilitis, was also recorded if clinically present. During the initial clinical examinations in 2005, a quantitative determination of oral Candida cell density was made using oral rinse samples. Patients voluntarily performed a 30-s oral rinse with 10 ml of sterile water, which was then returned into a sterile container. Neat oral rinse samples and 10-fold dilutions prepared in sterile water were plated on CHROMagar Candida medium (CHROMagar, Paris, France) and incubated at 37°C for 48 h. After incubation, the plates were examined, and the number of colonies present on each plate and their colors and relative abundances were recorded. Nitrogen-gassed VI-PAK sterile swabs (Sarstedt, Wexford, Republic of Ireland) were also used to sample the oral mucosa and any lesions suggestive of oral candidiasis. These were inoculated onto CHROMagar Candida medium and incubated as for oral rinse samples. Only swab samples were taken from patients who attended a follow-up visit in 2010. Single-colony isolates were presumptively identified on the basis of colony color and morphology on this medium (34). Definitive identification was undertaken by determining their substrate assimilation profiles using the API ID 32C yeast identification system (bioMérieux, Marcy l'Etoile, France) as described previously (41). For the purposes of the present study, oral rinse samples from individuals that yielded ≥500 Candida CFU/ml were considered as microbiological evidence of Candida infection. Similar cutoff thresholds for differentiating between oral Candida carriage and infection have been used in previous studies (19, 39). In the present study, the threshold was defined based on clinical and microbiological analysis of oral candidiasis in a minimum of 20,000 patients attending the Dublin Dental University Hospital (DDUH) over the past 25 years. Control data obtained from the DDUH Oral Biosciences Clinical database showed that 100% of 91 oral-rinse samples taken from HIV-infected patients who were not receiving antiretroviral therapy and displaying clinical signs of oral candidiasis yielded ≥500 CFU/ml. In 96% of these samples, the Candida cell densities were >1,000 CFU/ml. In contrast, 100% of the 85 oral rinses obtained from normal healthy university students who were not receiving any medication and had no clinical signs of oral candidiasis yielded ≤500 CFU/ml, with 97% of these samples yielding ≤50 CFU/ml.
Routine Candida culture.
Candida isolates were routinely cultured on YPD (1% [wt/vol] yeast extract, 2% [wt/vol] Bacto neo-peptone [Sigma-Aldrich Ireland, Ltd., Dublin, Republic of Ireland], 2% [wt/vol] glucose; pH 5.5) agar or in YPD broth at 37°C. Liquid cultures were grown overnight at 37°C in an orbital incubator (Gallenkamp, Leicester, United Kingdom) with shaking at 200 rpm. After primary plating on CHROMagar Candida medium, swabs were incubated in YPD broth containing 20 μg of chloramphenicol (Sigma-Aldrich)/ml, followed by incubation overnight 37°C at 200 rpm to enrich for Candida. After incubation, cultures were diluted 1 in 1,000 in sterile water and plated out on CHROMagar Candida medium and then incubated as described above.
Chemicals, enzymes, and oligonucleotides.
Analytical-grade or molecular-biology-grade chemicals were purchased from Sigma-Aldrich Ireland, Ltd., or Fisher Scientific, Ltd. (Loughborough, United Kingdom). Enzymes were purchased from the Promega Corp. (Madison, WI), Applied Biosystems (Warrington, United Kingdom), or Invitrogen (Biosciences, Ltd., Dublin, Republic of Ireland). Fluconazole powder was a gift from Pfizer Central Research (Sandwich, United Kingdom), and itraconazole was a gift from Janssen Pharmaceuticals (Cork, Republic of Ireland).
Nucleic acid isolation.
In order to isolate DNA from C. albicans isolates, cells from 1.5 ml of YPD overnight culture were harvested by centrifugation at 14,000 × g. These cells were resuspended in 200 μl of cell breaking buffer (2% [vol/vol] Triton X-100, 1 mM EDTA, 1% [wt/vol] sodium dodecyl sulfate, 100 mM NaCl, 10 mM Tris; pH 8) and were then transferred to a 2-ml screw-cap tube (Sarstedt, Ltd., Wexford, Republic of Ireland) containing 0.3 g of acid-washed glass beads (Sigma-Aldrich). After the addition of 200 μl of a mixture of 24:24:1 phenol-chloroform-isoamyl alcohol, the cells were disrupted in a BIO101 FastPrep instrument (Qbiogene, Cambridge, United Kingdom) for 20 s at speed 4, followed by centrifugation at 14,000 × g for 10 min. The aqueous phase was removed and extracted twice with an equal volume of 24:1 chloroform-isoamyl alcohol. Finally, the nucleic acids were precipitated by the addition of 400 μl of 70% ethanol at −20°C. Purified DNA was collected by centrifugation at 14,000 × g for 10 min and resuspended in 50 μl of sterile water.
For quantitative gene expression analysis, RNA was extracted from isolates that were grown in YPD broth in the presence of a subinhibitory concentration of fluconazole (0.06 μg/ml). The RNAs were extracted by using an RNeasy minikit (Qiagen Science, West Sussex, United Kingdom) and treated with an Ambion Turbo DNA-free kit (Applied Biosystems) according to the manufacturer's instructions. The RNA samples were then reverse transcribed to cDNA by using a Superscript II reverse transcriptase kit (Invitrogen).
ABC genotyping.
Selected C. albicans isolates from each patient were assigned to genotypes A, B, or C based on the nucleotide sequence of the internal transcribed spacer region of the 25S rRNA gene as previously described (27).
Determination of mating types.
The mating type of C. albicans isolates recovered from each patient during the course of the present study was determined by multiplex PCR according to the method of Rustad et al. (48).
MLST.
At least one C. albicans isolate recovered from each patient at each clinical assessment was subjected to MLST analysis as previously described (5, 6, 59). Briefly, the seven gene loci AAT1a, ACC1, ADP1, PMIb, ALA1, VPS13, and ZWF1b were amplified by PCR for each isolate. These PCR products were purified by using a QIAquick 96-well PCR purification kit (Qiagen Science, MD) and sequenced using the same primers that were used for amplification. Sequencing reactions were performed commercially at Source BioScience LifeSciences (Dublin, Republic of Ireland) using an ABI 3730xl DNA analyzer. Sequence analysis was performed by examination of chromatogram files using the ABI prism Seqscape software (version 2.6; Applied Biosystems, Foster City, CA). Identification of genotypes, allelic profiles, and diploid sequence types (DSTs) was achieved using the consensus C. albicans MLST website (http://calbicans.mlst.net/).
Susceptibility testing.
All isolates that were recovered and stored from each APECED patient were streaked onto separate CHROMagar Candida plates containing fluconazole at 8 and 16 μg/ml and containing itraconazole at 1 μg/ml and 0.125 μg/ml in order to carry out large-scale, rapid screening of isolates for reduced susceptibility or resistance to azole drugs. These plates were incubated at 37°C for 48 h and then examined for the presence or absence of yeast colonies in comparison to drug-free control plates. Definitive azole MIC determinations for C. albicans isolates were performed in duplicate using broth microdilution assays according to the method of the Clinical and Laboratory Standards Institute (CLSI; formerly the National Committee for Clinical Laboratory Standards) document M27-A2 (9). Fluconazole was titrated from a concentration of 64 to 0.125 μg/ml, and itraconazole was titrated from a concentration of 16 to 0.03 μg/ml in RPMI 1640 medium (Sigma-Aldrich). MICs were determined as the lowest concentrations of the drug that reduced turbidity by 50% relative to the growth of the drug-free controls. Isolates exhibiting fluconazole MICs of ≥64 μg/ml were classified as resistant, fluconazole MICs between 16 and 32 μg/ml as dose-dependent susceptible, and fluconazole MICs of ≤8 μg/ml as susceptible. Isolates exhibiting itraconazole MICs of ≥1 μg/ml were classified as resistant, itraconazole MICs between 0.25 and 0.5 μg/ml as dose-dependent susceptible, and itraconazole MICs of ≤0.125 μg/ml as susceptible.
Gene expression analysis.
Quantitative real-time PCR (qRT-PCR) was carried out according to standard protocols in order to monitor the relative gene expression of the ERG11 gene and the CDR1-, CDR2-, and MDR1-encoded efflux pumps in C. albicans isolates exhibiting reduced azole susceptibilities (Table 2), specifically isolates recovered from patients 1, 11, 12, and 16. This was undertaken in order to determine whether significant upregulation of these genes was associated with reduced azole susceptibility. These data were compared to those of the two fluconazole-susceptible control isolates (fluconazole MIC = 0.125 μg/ml), one of which was recovered during the second clinical assessment of patient 4 (P4V2), and the C. albicans reference strain, SC5314 (8). The comparative amplification efficiencies of primer pairs were compared to the amplification efficiency of the ACT1 internal control primer pair (Table 2) prior to qRT-PCR using primer amplification efficiency plot analysis as previously described (40). RNAs were extracted from isolates that were grown in either the presence or absence of a subinhibitory concentration of fluconazole (0.06 μg/ml) in order to identify whether any gene upregulation was constitutive or facultative. Quantitative real-time PCRs were carried out with 0.3 μM concentrations of each primer (Table 2) and Fast SYBR green Master Mix in an ABI 7500 real-time PCR system (Applied Biosystems) according to the manufacturer's recommended protocols. Data analysis was carried out as described by Livak and Schmittgen (25), calculating 2−ΔCT values from the average CT values acquired from three replicates for both the test and ACT1 genes. Each qRT-PCR analysis was carried out in duplicate on two separate occasions.
Table 2.
Primers used in the present study
| Analysis and primer | Coordinatesa | Sequence (5′–3′) |
|---|---|---|
| qRT-PCR | ||
| RT ACT1F | +1429 → +1452 | AGCTCCAGAAGCTTTGTTCAGACC |
| RT ACT1R | +1579 → +1602 | CCCAGGTATTGCTGAACGTATGCA |
| RT CDR1F | +4204 → +4225 | TGTGCTGAACGTGAATATGTTT |
| RT CDR1R | +4329 → +4349 | CTGTCAAATGAGTTCCACCAA |
| RT CDR2F | +4197 → +4218 | TTGTGCACCTAGAGAATTGGTT |
| RT CDR2R | +4358 → +4380 | CCATCAATGCTTTGTTTAGTCAA |
| RT MDR1F | +683 → +705 | GTGGTGCTAGTGTTGCTGATGTG |
| RT MDR1R | +747 → +767 | TTTGGGTGCTGTTTGTGGTCC |
| RT ERG11F | +686 → +716 | CCCCTATTAATTTTGTTTTCCCTAATTTACC |
| RT ERG11R | +774 → +803 | GAAAGAAATTAAACTGAGAAGAGAACGTGG |
| Amplification | ||
| ERG11F | –527 → –511 | ATTGTACGTGGCGCGAGGTACTAGAAA |
| ERG11R | +1899 → +1924 | CATCTGCTAATATAGGACCAGGATTCGAC |
| TAC1F | –558 → –532 | GAAATTGTTAATGACGGTTCTACCTTC |
| TAC1R | +3192 → +3217 | TATTCATATACCCAACCGGAAATTGG |
| Sequencing | ||
| TAC1F2 | +256 → +274 | AATAAATCAACCGCCAATA |
| TAC1F3 | +1117 → +1133 | ATTACCACCCCTGCTTC |
| TAC1F4 | +1821 → +1842 | TTCCATATCCAATACTTTAGAA |
| TAC1R2 | +423 → +442 | TATGCATCATTCTCGACATT |
| ERG11F2 | +254 → +273 | TTATGTTATTAGGGAAAATT |
| ERG11F3 | +988 → +1009 | CAAGATGTTATTTATCAAGAAG |
| ERG11F4 | +1783 → +1802 | GAGGCGGAATCGGTTGATGG |
Primer coordinates are numbered based on the adenine residue of the ATG start codon at the 5′ end of the C. albicans gene being designated +1.
The expression data for each gene was normalized to the expression of the ACT1 gene for each isolate, which functioned as an internal control. The average 2−ΔCT values were obtained for isolates recovered from each patient in the presence or absence of fluconazole and were compared to the average 2−ΔCT value obtained from the fluconazole-susceptible control isolates, P4V2 and SC5314, also in the presence or absence of fluconazole. Two-tailed Student t tests were performed in order to assess the significance of the data.
Sequence analysis of the ERG11 and TAC1 genes.
The complete open reading frames of the CaERG11 and CaTAC1 genes (GenBank accession numbers DQ903898.1 and DQ837377, respectively) were amplified by using the pairs of oligonucleotides primers (Sigma Genosys Biotechnologies Europe, Ltd., Cambridgeshire, United Kingdom) listed in Table 2. These genes were amplified from the DNA of one isolate recovered from both the first and the last clinical assessment of patients 1, 11, 12, and 16. Sequence data were compared between each of two isolates recovered per patient in order to correlate any mutations with increased expression of CDR-encoded efflux pumps between clinical assessments. The nucleotide sequence of the ERG11 gene was examined in order to identify any polymorphisms that could be associated with azole resistance. All sequence data were also compared to those of the fluconazole-susceptible isolate (fluconazole MIC = 0.125 μg/ml) recovered during the second clinical assessment of APECED patient 4 (P4V2). Amplification reaction mixtures consisted of 0.2 μM concentrations of each deoxynucleotide triphosphate (Promega), 0.2 μM concentrations of each primer, 1 U of Phusion HotStart high-fidelity DNA polymerase (New England BioLabs, Ltd., Hertfordshire, United Kingdom), 1× Phusion HF buffer (New England Biolabs, Ltd.), and 100 ng of template DNA. These reaction mixtures underwent an initial denaturation step of 98°C for 1 min, followed by 35 cycles of 98°C for 30 s, 61°C for 30 s, and 72°C for 3 min 30s, followed by a final elongation step of 72°C for 10 min. For amplification of the CaERG11 gene, the repeated elongation step was performed at 72°C for 2 min 30 s instead of 3 min 30 s. The CaERG11 and CaTAC1 amplification products were 2.5 and 3.7 kb, respectively. These products were purified by using a GenElute PCR clean-up kit (Sigma-Aldrich) and were sequenced commercially by Source Bioscience Lifesciences (Dublin, Republic of Ireland) using the sequencing primers (Table 2) and a 3730xl DNA analyzer. Nucleotide and amino acid sequence alignments were carried out by using the CLUSTAL W2 sequence alignment computer program (60) available at the EMBL-EBI website (http://www.ebi.ac.uk/tools/msa/clustalw2/).
RESULTS
Clinical assessment of oral candidiasis.
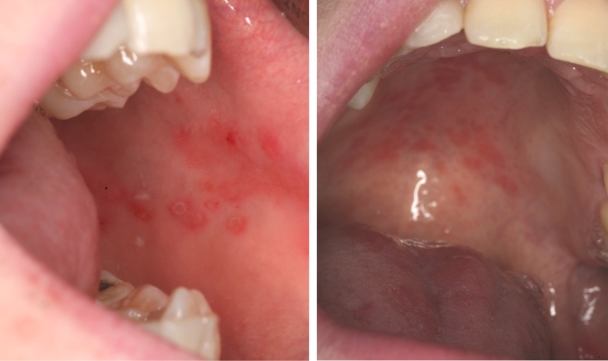
Patients were clinically assessed at the Dublin Dental University Hospital (DDUH) on at least one occasion, during which a full medical history was taken and a complete clinical assessment of the oral cavity was carried out. Several patients had multiple clinical presentations of candidiasis at separate clinical assessments. At the initial assessment in 2005, 7/16 (44%), 11/16 (69%), and 7/16 (44%) patients displayed clinical signs suggestive of pseudomembranous candidiasis (PC), erythematous candidiasis (EC), and angular cheilitis (AC), respectively (Table 3). In total, 11/16 (69%) patients showed clinical signs of oral Candida infection, and oral Candida isolates were recovered from 12/16 (75%) patients (Table 3). At the second assessment, 4/11 (36%), 4/11 (36%), and 2/11 (18%) patients displayed clinical signs suggestive of PC, EC, and AC, respectively (Table 3). In all, 5/11 (45%) showed clinical signs of oral Candida infection, and oral Candida isolates were recovered from 8/11 (73%) patients (Table 3). At the third assessment, 4/11 (36%), 4/11 (36%), and 4/11 (36%) patients displayed clinical signs indicative of PC, EC, and AC, respectively (Table 3). In total, 6/11 (54%) showed clinical signs of oral Candida infection, and oral Candida isolates were recovered from 7/11 (64%) patients (Table 3). Individuals from whom ≥500 Candida CFU/ml of oral rinse were recovered were considered as having microbiological evidence of Candida infection. Clinical signs suggestive of candidiasis did not always correlate with microbiological evidence of infection. Throughout the three assessments, clinical signs suggestive of candidiasis were noted at 22 individual assessments of the 16 patients studied and oral Candida cell densities indicative of Candida infection (range, 500 to >10,000 CFU/ml in oral rinse samples) were recovered from patients on 15/22 (68%) of these occasions (Table 3). A provisional clinical diagnosis of candidiasis was not supported by microbiological analysis of oral samples in the remaining 7/22 patient evaluations (32%), since Candida cell densities of between 0 and 380 CFU/ml were observed in oral rinse samples (Table 3). For example, at the first clinical assessment, patients 3, 6, 9, and 13 exhibited clinical signs suggestive of either pseudomembranous candidiasis, erythematous candidiasis, angular cheilitis, or a combination of more than one of these conditions, despite the recovery of only 80, 70, 380, and 80 CFU/ml, respectively, from these patients at this assessment (Table 3). At the second clinical assessment, 0 CFU/ml were recovered from patient 13, who exhibited clinical signs suggestive of erythematous candidiasis (Table 3). In contrast, at the third clinical assessment, patient 10 did not exhibit any clinical signs suggestive of candidiasis, and yet oral rinse samples yielded a Candida density of 500 CFU/ml (Table 3). The data recovered from direct swab sampling of lesions suggestive of oral candidiasis in several individual APECED patients reflected the findings of oral rinse sampling, but the yield of Candida recovered was on average five times lower. It should be noted that swab samples of the oral cavity yield significantly lower densities of Candida CFU than do oral rinse samples (10). Swabs used to directly sample oral lesions suggestive of erythematous candidiasis in patients 5, 6, and 10 during clinical assessments in 2010 all yielded fewer than 30 C. albicans CFU. For example, patient 5 exhibited clinical signs of erythematous candidiasis at the left buccal mucosa and palate; these were not supported by microbiological analysis of swab samples taken from these sites, which yielded 5 and 14 CFU/swab, respectively (Fig. 1). In contrast, a swab used to directly sample a lesion of the right buccal mucosa suggestive of erythematous candidiasis in patient 15 in 2010 yielded 200 CFU.
Table 3.
Clinical signs and C. albicans density in the oral cavities of patients with APECED
| Patient | OCa | Other CMCb | Data from 2005 (rinse)c |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Clinical assessment 1 |
Clinical assessment 2 |
Clinical assessment 3 |
|||||||||
| Clinical signs of OC | CFU/ml | No. stored | Clinical signs of OC | CFU/ml | No. stored | Clinical signs of OC | CFU/ml | No. stored | |||
| 1 | Y | Y | PC, EC, AC | >10,000 | 6 | PC, EC, AC | >10,000 | 3 | PC, EC, AC | 380 | 3 |
| 2 | N | Y | PC, EC, AC | 940 | 4 | ||||||
| 3 | Y | Y | PC, EC, AC | 80 | 3 | ||||||
| 4 | Y | Y | PC, EC | 500 | 2 | PC | 5,920 | 3 | PC | 1,700 | 1 |
| 5 | Y | Y | PC, EC, AC | >10,000 | 10 | N | 130 | 1 | EC, AC | 1,680 | 2 |
| 6 | Y | N | EC | 70 | 3 | PC, EC | 1,300 | 5 | EC, AC | 510 | 3 |
| 7 | Y | Y | N | 0 | 0 | N | 0 | 0 | N | 0 | 0 |
| 8 | Y | Y | N | 0 | 0 | N | 0 | 0 | N | 0 | 0 |
| 9 | Y | Y | EC, AC | 380 | 5 | ||||||
| 10 | N | Y | N | 0 | 0 | N | 2 | 2 | N | 500 | 1 |
| 11 | Y | Y | PC, EC, AC | 1280 | 9 | N | 120 | 1 | N | 0 | 0 |
| 12 | Y | Y | N | 140 | 10 | N | 50 | 3 | PC | >10,000 | 3 |
| 13 | Y | Y | EC | 80 | 2 | EC | 0 | 0 | N | 0 | 0 |
| 14 | Y | Y | EC | >10,000 | 2 | ||||||
| 15 | Y | Y | N | 0 | 0 | ||||||
| 16 | Y | Y | PC, EC, AC | 1840 | 9 | PC, EC, AC | 1,300 | 4 | PC, EC, AC | 360 | 2 |
Y, yes; N, no. Whether the patient had a history of oral candidiasis (OC) is indicated.
Y, yes; N, no. Whether the patient had a history of chronic mucocutaneous candidiasis (CMC) apart from OC is indicated.
Patients provided an oral rinse sample from at least one clinical assessment in 2005. “Clinical signs of OC” refers to clinical signs at the time of assessment. The CFU/ml data refer to the Candida density recovered from oral rinse samples. No. stored, the number of isolates stored from each visit for further analysis. Blank fields indicate clinical assessments that were not attended by patients. PC, pseudomembraneous candidiasis; EC, erythematous candidiasis; AC, angular cheilitis.
Fig. 1.
Appearance of oral lesions indicative of erythematous candidiasis in an APECED patient in the absence of microbiological culture evidence. View of left buccal mucosa (left) and palate (right) taken from patient 5 at a follow-up clinical assessment in 2010. Swab samples from these two sites yielded 5 (buccal mucosa) and 14 (palate) C. albicans CFU, respectively.
C. albicans was the only Candida species recovered from the APECED patients during the course of the study. Swabs used to sample the oral cavities of patients 5, 6, 10, and 15 during a follow-up assessment in 2010 from which only C. albicans was recovered on CHROMagar Candida medium underwent overnight broth enrichment in order to recover any other Candida species that may be present at lower densities. This was unsuccessful for all four patients, and C. albicans remained the only Candida species to be identified.
Analysis of C. albicans population in patients with APECED.
Isolates were recovered sequentially from 14/16 APECED patients during between one and three clinical assessments in 2005. A further follow-up assessment was carried out on 11 of these patients in 2010, and further isolates were recovered from the oral mucosa or from lesions suggestive of oral candidiasis during these assessments. A total of 54 of these C. albicans isolates recovered sequentially from 14/16 APECED patients underwent MLST analysis and ABC genotyping in order to determine whether the isolates persisted or were replaced in patients with APECED throughout the course of the study.
A total of 18 different DSTs were identified by MLST from the 14 patients from whom C. albicans isolates were recovered (see Fig. S1 in the supplemental material). Eight of these DSTs were previously identified, and the remaining 10 newly identified DSTs were added to the C. albicans MLST database. Isolates recovered at sequential clinical assessments were available for nine of the 14 C. albicans-positive patients, and MLST analysis yielded identical DSTs at each evaluation (Table 4), indicating long-term maintenance of the same DST over the 5-year study period in these APECED patients. The remaining three patients yielded isolates of more than one DST. Eight isolates identified as DST 1778 were recovered from patient 1 on the first three clinical assessment visits (in March, June, and September 2005), and two isolates identified as DST 1780 and DST 1224 were recovered from this patient at a follow-up assessment (in February 2010) (Table 4). Two isolates identified as DST 1784 were recovered from patient 11 on the first two assessment visits (in March and September 2005), and two isolates identified as DST 1785 were recovered from this patient on a follow-up assessment in 2010 (Table 4). Isolates were recovered from patient 13 only on the first of three clinical assessments; however, isolates recovered from the first visit were identified as DSTs 1786 and 1787. In relation to patients who yielded C. albicans isolates with different DSTs, the differences between DSTs occurred at only one or two loci and, in all cases, resulted from the loss of heterozygosity at between two and nine nucleotides, a finding indicative of microvariation or genotypic shuffling. A UPGMA (unweighted pair-group method with arithmetic averages) dendrogram was constructed comparing the allelic profiles and DST data of the C. albicans isolates recovered during the course of this study to those previously reported by Odds et al. (35). This was used to identify the MLST clade to which C. albicans isolates recovered from patients with APECED belonged. Isolates recovered from 5/14 patients belonged to the MLST clade 1 (Table 4), the most predominant MLST clade in the population structure of C. albicans. The second most predominant clade in the present study was clade 4, to which isolates recovered from 3/14 patients belonged (Table 4). Isolates recovered from two patients belonged to clade 15, and the isolates recovered from the remaining patients belonged to clades 2 and 8. Isolates recovered from patients 13 and 15 were identified as singletons (Table 4).
Table 4.
Population analysis of C. albicans isolates recovered from APECED patientsa
| Isolate (n) | Date of clinical assessment (mo/yr) | DST | ABC type | eBURST CC | UPGMA clade | MAT type (n) |
|---|---|---|---|---|---|---|
| P1V1 (3) | 3/05 | 1778 | A | 1 | 1 | α/α (1) |
| a/α (2) | ||||||
| P1V2 (3) | 6/05 | 1778 | A | 1 | 1 | a/α (3) |
| P1V3 (2) | 9/05 | 1778 | A | 1 | 1 | a/α (2) |
| P1V4 (2) | 2/10 | 1780 | A | 1 | 1 | a/α (2) |
| 1224 | A | 1 | 1 | |||
| P2V1 (2) | 2/05 | 1779 | A | S | 15 | a/α (2) |
| P3V1 (1) | 2/05 | 1781 | C | S | 15 | a/α (1) |
| P3V4 (1) | 3/10 | 1781 | C | S | 15 | a/α (1) |
| P4V1 (2) | 3/05 | 1782 | A | 1 | 1 | a/α (2) |
| P4V2 (3) | 7/05 | 1782 | A | 1 | 1 | a/α (3) |
| P4V3 (1) | 10/05 | 1782 | A | 1 | 1 | a/α (1) |
| P5V1 (1) | 4/05 | 666 | B | 24 | 8 | a/α (1) |
| P5V2 (1) | 5/05 | 666 | B | 24 | 8 | a/α (1) |
| P5V3 (1) | 7/05 | 666 | B | 24 | 8 | a/α (1) |
| P5V4 (1) | 5/10 | 666 | B | 24 | 8 | a/α (1) |
| P6V1 (1) | 2/05 | 228 | A | 24 | 4 | a/α (1) |
| P6V2 (1) | 5/05 | 228 | A | 24 | 4 | a/α (1) |
| P6V3 (1) | 8/05 | 228 | A | 24 | 4 | a/α (1) |
| P6V4 (1) | 7/10 | 228 | A | 24 | 4 | a/α (1) |
| P9V1 (2) | 2/05 | 1783 | A | S | 1 | a/α (2) |
| P10V2 (1) | 5/05 | 189 | C | 2 | 4 | a/α (1) |
| P10V3 (1) | 6/05 | 189 | C | 2 | 4 | a/α (1) |
| P10V4 (1) | 7/10 | 189 | C | 2 | 4 | a/α (1) |
| P11V1 (1) | 3/05 | 1784 | A | 1 | 1 | a/α (1) |
| P11V2 (1) | 9/05 | 1784 | A | 1 | 1 | a/a (1) |
| P11V4 (2) | 4/10 | 1785 | A | 1 | 1 | a/a (2) |
| P12V1 (3) | 3/05 | 315 | A | 1 | 2 | a/α (3) |
| P12V2 (2) | 9/05 | 315 | A | 1 | 2 | a/α (2) |
| P12V3 (2) | 10/05 | 315 | A | 1 | 2 | a/α (2) |
| P13V1 (2) | 3/05 | 1786 | A | 37 | S | a/α (2) |
| 1787 | A | 37 | S | |||
| P14V1 (2) | 8/05 | 363 | C | S | 4 | a/α (2) |
| P15V2 (3) | 9/10 | 1220 | C | 17 | S | a/α (3) |
| P16V1 (1) | 5/05 | 519 | A | 1 | 1 | a/α (1) |
| P16V2 (1) | 11/05 | 519 | A | 1 | 1 | a/a (1) |
| P16V3 (1) | 12/05 | 519 | A | 1 | 1 | a/a (1) |
Sequentially recovered isolates are defined according to patient number (P1 to P16) and clinical assessment number (V1 to V4). The numbers of isolates analyzed from each clinical assessment per patient are indicated in parentheses. Clonal clusters (CCs) were assigned to MLST-identified DSTs by eBURST analysis, and UPGMA clades were identified according to the method of Odds et al. (35).
Analysis of ABC genotypes of recovered isolates.
The predominant ABC type identified in isolates recovered from the 14 C. albicans-positive patients was genotype A, which was identified in isolates recovered from nine patients. Isolates recovered from one patient were identified as ABC genotype B, and isolates from the remaining four patients were identified as ABC genotype C (Table 4).
Azole susceptibility of isolates.
The fluconazole and itraconazole susceptibilities of 62 and 59 C. albicans isolates recovered sequentially from 14 patients were also determined, respectively, according to the CLSI protocol M27-A2. Fluconazole resistance was exhibited by eight isolates (fluconazole MIC = 64 μg/ml), recovered sequentially from patient 12 during three separate clinical assessments (in March, September, and October 2005). Seven of these isolates also exhibited dose-dependent susceptibility to itraconazole (itraconazole MIC = 0.5 μg/ml). Seven of these eight isolates were analyzed by MLST and identified as DST 315. Dose-dependent susceptibility (itraconazole MIC = 0.25 μg/ml) was also exhibited by isolates recovered from patient 16, despite the susceptibility (itraconazole MIC = 0.06 μg/ml) of an isolate recovered during an earlier assessment (in March 2005) of this patient (Table 5). All isolates recovered from this patient were identified as DST 519 (Table 4). The remaining isolates were all susceptible to both fluconazole and itraconazole (fluconazole MIC = 0.125 to 2 μg/ml and itraconazole MIC = 0.03 to 0.125 μg/ml); however, a gradual stepwise reduction in azole susceptibility was noted in isolates recovered from patients 1, 11, and 16 at sequential clinical assessments (Table 5). MLST analysis showed that the same DSTs persisted in each of these patients throughout these sequential clinical assessments (Table 4) undergoing microvariation rather than strain replacement.
Table 5.
Fluconazole and itraconazole MICs exhibited by isolates recovered sequentially from patients 1, 11, 12, and 16a
| Patient | Azole therapy | MAT type | Date of clinical assessment (mo/yr) | Total no. of isolates examined | FLC |
ITC |
Genes significantly upregulated (P ≤ 0.005) | Amino acid substitution(s) in: |
|||
|---|---|---|---|---|---|---|---|---|---|---|---|
| MIC (μg/ml) | No. of isolates examined | MIC (μg/ml) | No. of isolates examined | ERG11 | TAC1 | ||||||
| 1 | ITC, MC | a/α | 3/05 | 3 | 0.125 | 2 | 0.03 | 3 | A114V, D116E, K119N | L734L/F, N772N/K, D776D/N, N874N/G | |
| α/α | |||||||||||
| a/α | 6/05 | 2 | 0.5 | 2 | 0.03 | CDR1 (2.2), MDR1 (2.8) | |||||
| a/α | 9/05 | 2 | 0.5 | 2 | 0.03 | CDR1 (2.4), MDR1 (2.6) | |||||
| a/α | 2/10 | 5 | 1 | 5 | 0.125 | CDR1 (2.2), MDR1 (3.9) | |||||
| 11 | FLC, MC | a/α | 3/05 | 3 | 0.125 | 3 | 0.03 | 3 | D116E, K119K/T, K128T | A736V | |
| a/α | 9/05 | 1 | 0.125 | 1 | 0.03 | ||||||
| a/a | 4/10 | 2 | 2 | 2 | 0.03 | CDR1 (5.6), CDR2 (33.8) | |||||
| 12 | FLC, MC, ITC | a/α | 3/05 | 5 | 64 | 5 | 0.5 | 3 | CDR1 (3.4), CDR2 (30.2) | K128T, S405F, V437I | F189S, S199N, R206H, V207A, S236L, N398S, D776N, N894N/S |
| a/α | 9/05 | 1 | 64 | 1 | 0.5 | CDR1 (4.5), CDR2 (30.2) | |||||
| a/α | 10/05 | 2 | 64 | 1 | 0.5 | CDR1 (3.6), CDR2 (30.2) | |||||
| 16 | FLC, ITC | a/α | 5/05 | 1 | 0.125 | 1 | 0.06 | 3 | CDR1 (2.9), CDR2 (6.4) | N772K, D776D/N | |
| a/a | 11/05 | 1 | 1 | 1 | 0.25 | CDR1 (6.3), CDR2 (4.9) | |||||
| a/a | 12/05 | 1 | 1 | 1 | 0.25 | ||||||
Average fold increases in gene expression values were obtained by comparing the average 2−ΔCT values for isolates recovered from each patient in both the presence and absence of fluconazole to the average 2−ΔCT values obtained from the fluconazole-susceptible control isolates P4V2 and SC5314 also in both the presence and absence of fluconazole. Amino acid substitutions identified in the ERG11 and TAC1 genes of isolates exhibiting reduced azole susceptibility that were not present in P4V2 are indicated in the last column of the table. Mutations in the TAC1 and ERG11 genes that have previously been definitely associated with azole resistance in C. albicans are highlighted in boldface. ITC, itraconazole; FLC, fluconazole; MC, miconazole.
The mating types of all isolates subjected to MLST analysis were also determined. All isolates examined were a/α at the MAT locus with the exception of certain isolates recovered from patients 1, 11, and 16 (Table 4). One isolate recovered from patient 1 was identified as α/α at the MAT locus, despite being identified as the same DST as all other isolates recovered sequentially from this patient. Isolates recovered during the latter clinical assessments of patients 11 and 16 were identified as a/a at the MAT locus, in contrast to isolates recovered at earlier assessments (Tables 4 and 5). All patients from whom isolates exhibiting azole resistance or a gradual reduction in azole susceptibility were recovered were being treated with differing combinations of fluconazole, itraconazole, or miconazole (Tables 1 and 5).
CDR1 gene expression analysis.
The average CDR1 expression was significantly higher in isolates recovered from patient 1 (2.3-fold, P < 0.0005), patient 12 (3.8-fold, P < 0.00005), and patient 16 (4.5-fold, P < 0.05) compared to the fluconazole-susceptible control isolates (Table 5). A highly significant (P < 0.005) 7.9-fold increase in CDR1 expression was observed in an isolate recovered from the fourth clinical assessment of patient 11 compared to isolates recovered from the same patient at the first and second clinical assessments. This increase in CDR1 expression was 5.6-fold in comparison to the control isolates (Table 5). Patient 11 had recently completed a course of fluconazole therapy prior to the fourth clinical assessment (Table 5). No significant differences in CDR1 expression were observed in isolates upon supplementation of growth media with fluconazole (Table 5).
CDR2 gene expression analysis.
The average expression of the CDR2 gene was upregulated an average of 30-fold in isolates recovered from patient 12 compared to the fluconazole-susceptible control isolates (P < 0.005). Isolates recovered from patient 16 also exhibited an average 4.2-fold increase in CDR2 expression (P < 0.05) compared to the control isolates (Table 5). A highly significant (P = 0.0001) 35-fold increase in CDR2 expression was observed in an isolate recovered from the fourth clinical assessment of patient 11 compared to isolates recovered from the same patient at the first and second clinical assessments (Table 5). This increase in CDR2 expression was 33.8-fold compared to the control isolates (Table 5). This isolate had also exhibited a 7.9-fold increase in CDR1 expression. No significant difference in CDR2 expression was observed in isolates recovered from patient 1 compared to the fluconazole-susceptible control isolates (Table 5). No significant differences in CDR2 expression were observed in isolates upon supplementation of growth medium with fluconazole (Table 5).
MDR1 gene expression analysis.
Isolates recovered from patient 1 exhibited an average 3-fold increase in MDR1 expression compared to the fluconazole-susceptible control isolates (P < 0.0005). No other significant increases in MDR1 expression were observed in isolates recovered from patients 11, 12, or 16 compared to the fluconazole-susceptible control isolates (Table 5). Significant differences in MDR1 expression were also not observed in isolates upon supplementation of the growth medium with fluconazole (Table 5).
ERG11 gene expression analysis.
No significant differences in ERG11 expression were observed between isolates recovered from patients 1, 11, 12, or 16 in compared to fluconazole-susceptible control isolates. The presence of fluconazole reduced the expression (range, 1.7- to 2.1-fold) of ERG11 in all isolates recovered from patients 11 and 12 significantly (P < 0.01). No other significant differences were observed in isolates recovered from other patients.
Sequence analysis of the ERG11 and TAC1 genes.
The nucleotide sequence of the TAC1 gene of eight isolates exhibiting reduced azole susceptibility or azole resistance was analyzed and compared to that of the fluconazole-susceptible isolate recovered from patient 4, P4V2. These eight isolates consisted of the first and last isolate recovered from patients 1, 11, 12, and 16. Previous studies have shown that mutations often accumulate over time in genes associated with reduced azole susceptibility, such as TAC1 and ERG11 in C. albicans isolates recovered sequentially from individual patients receiving recurrent azole therapy (33, 50). Twelve amino acid substitutions were identified in the TAC1 genes of these eight isolates that were not present in the nucleotide sequence of the fluconazole-susceptible P4V2 isolate (Table 5). One of these amino acid substitutions, A736V, has previously been identified as a GOF mutation in the TAC1 gene of clinical C. albicans isolates, leading to upregulation of the CDR-encoded efflux pumps (15). This substitution occurred in the last isolate recovered from patient 11 during a follow-up clinical assessment in 2010 and correlated with significant constitutive upregulation of the CDR1 and CDR2 genes (Table 5).
Isolates recovered from patient 12 exhibited fluconazole resistance and dose-dependent susceptibility to itraconazole (Table 5). Although eight amino acid substitutions were identified in the TAC1 gene of these two isolates (Table 5), none of these have been definitively associated with CDR upregulation. Three amino acid substitutions were also identified in the ERG11 genes of these two isolates, and one of these substitutions (S405F) has previously been associated with azole resistance (33).
Amino acid substitutions were also identified in the ERG11 and TAC1 genes of isolates recovered from patient 1 and in the TAC1 gene of isolates recovered from patient 16 (Table 5). However, none of these mutations have been definitively associated with azole resistance.
In all, six amino acid substitutions were identified in the ERG11 gene of the eight isolates examined that were not present in the fluconazole-susceptible isolate P4V2. All six of these substitutions had previously been reported (33). However, only the S405F substitution (Table 5), which was observed in both isolates sequentially recovered from patient 12, has been directly associated with azole resistance (33).
DISCUSSION
Although oral candidiasis is well established as one of the primary features of APECED, complete and in-depth analyses of the Candida populations in these patients is mostly lacking. To date, a search of the literature revealed only two studies regarding the prevalence and population structures of Candida species recovered from patients with APECED. These two studies have both been published by the same group of researchers and are based on APECED patients in Finland (46, 56). The purpose of the present study was to carry out a thorough and in-depth analysis of the Candida species recovered from approximately half of the APECED patients definitively identified in Ireland (17). These patients were assessed at the DDUH over a period of 5 years, and the clinical signs of candidiasis as diagnosed by an experienced oral medicine physician were correlated with microbiological evidence of Candida infection. Two interesting findings resulted from this analysis; the first was that clinical signs suggestive of oral candidiasis were not supported by microbiological evidence of candidiasis on a total of 7/22 (32%) occasions in six individual patients. Similarly, direct sampling of lesions suggestive of erythematous candidiasis yielded very low Candida counts and did not support microbiological evidence of candidiasis in several patients during the follow-up clinical assessment in 2010. This suggests that the cause of these clinical signs on these occasions was not due to Candida infection but may have been due to infection with other microbial agents such as bacteria or viruses. Alternatively, the clinical lesions suggestive of oral candidiasis may have been due to abnormal T cell proliferation in these patients, leading to an exaggerated immune response to environmental stimuli such as the presence of yeast pathogens at low levels, as previously observed in AIRE-deficient mice (44). Further histological, immunological, and microbiological investigation of such non-Candida-associated oral mucosal lesions is therefore warranted.
APECED patients are often prescribed systemic or topical antifungal therapy prophylactically to control oral candidiasis, often based on the provisional clinical diagnosis of candidiasis and without prior microbiological confirmation. This is probably an unwise practice in light of past experience with the development of azole resistance in Candida species in HIV-infected and AIDS patients receiving azoles prophylactically (1, 49, 62).
The second interesting feature observed during the course of the study was the monospecies colonization of the oral cavities of these APECED patients with C. albicans. C. albicans is the most common cause of all types of candidiasis. However, during the 1990s other Candida species, such as C. glabrata, C. parapsilosis C. tropicalis, C. krusei, and C. dubliniensis, were associated with superficial and systemic infection more frequently, particularly in HIV-infected and AIDS patients who were receiving azole treatment (11, 13, 42, 58). Antiretroviral treatment has reduced the rates of opportunistic fungal infections in HIV-infected patients; however, non-C. albicans Candida species are still frequently associated with candidiasis in immunocompromised patients (20, 37, 55, 64). In particular, C. glabrata and C. krusei are often selected for in patients receiving azole therapy due to intrinsic or acquired reduced azole susceptibility (3, 22, 31, 36, 64). It is therefore very surprising that only C. albicans was recovered from the APECED patients, despite the fact that they received frequent antifungal therapy, not only with azoles (11/16 [69%] of patients) but also with the polyene antifungal drug nystatin (Table 1). In the present study, broth enrichment of oral swab samples from four patients harboring C. albicans was undertaken in an attempt to recover non-C. albicans isolates potentially present at low abundance in the original samples. However, no non-C. albicans isolates were recovered. An analysis of the Candida population in a group of Finnish patients with APECED reported the recovery of non-C. albicans Candida species in 7/56 (12.5%) patients investigated (46); however, the identity of these species was not specified. Given the relatively low level of non-C. albicans Candida species in the Finnish study, the most likely reason for the C. albicans monospecies colonization observed in our study group may be a result of the small size of the patient population available for study, although approximately half of the Irish population with APECED was represented (17).
The susceptibility of sequentially recovered isolates to fluconazole and itraconazole was examined using agar containing azoles at breakpoint concentrations, as well as using broth microdilution assays. In all, 11/16 (69%) of the APECED patients examined in the present study are currently known to be receiving treatment with fluconazole or itraconazole. Four of these eleven patients (36%) yielded isolates that exhibited either azole resistance or a gradual reduction in azole susceptibility. Broth microdilution assays identified the presence of isolates exhibiting fluconazole resistance and dose-dependent susceptibility to itraconazole in patient 12. This patient is currently 39 years old and has been treated with both topical and systemic azole therapy for many years. Two isolates recovered from this patient exhibited significant upregulation of the CDR1 and CDR2 genes. Eight amino acid substitutions were identified in the TAC1 gene of these two isolates (Table 5), although none of these have been definitively identified as GOF mutations. Three amino acid substitutions were also identified in the ERG11 gene of these two isolates also, one of which (S405F) has previously been associated with azole resistance (33). This suggests that the azole resistance exhibited by the isolates recovered from this patient is possibly mediated by multiple factors. Isolates recovered sequentially from patient 11 exhibited a gradual reduction in fluconazole susceptibility, and isolates recovered sequentially from both patients 1 and 16 exhibited a gradual reduction in susceptibility to both fluconazole and itraconazole (Table 5). These patients were also regularly treated with both topical and systemic azole drugs (Table 1). Although the MICs exhibited by these isolates were below the susceptible MIC breakpoints, gradual increases in CDR1, CDR2, and MDR1 expression correlated with the gradual reduction in azole susceptibility. A GOF mutation (A736V) in the TAC1 gene that has previously associated with CDR upregulation was identified in an isolate recovered from patient 11 in 2010 (Table 5). This mutation was not present in an isolate recovered from this patient in 2005, and the acquisition of this mutation correlated with significant upregulation of the CDR1 and CDR2 genes. Amino acid substitutions were also identified in the TAC1 and ERG11 genes of isolates recovered from patients 1 and 16, although none of these have been definitively identified as GOF mutations. It is possible that similar GOF mutations in the MRR1 gene are responsible for the MDR1-upregulation exhibited by isolates recovered from patient 1.
MLST analysis of isolates recovered sequentially from 9/16 of these patients showed that the same isolates persisted for prolonged periods of time, undergoing minor genetic variation rather than strain replacement, and azole susceptibility testing demonstrated that persistent isolates develop azole resistance gradually rather than undergoing replacement with resistant isolates. Similar findings were recently reported with C. albicans isolates from Finnish APECED patients (56).
The present study has demonstrated for the first time that clinical signs of candidiasis did not correlate with microbiological evidence of infection in 32% of clinical assessments carried out on Irish APECED patients. It should be noted that two calibrated dentists at the DDUH carried out these clinical assessments individually and that in both cases the reporting of clinical signs indicative of oral candidiasis did not always correlate with microbiological culture-based evidence of Candida infection. The present study highlights the necessity for both microbiological and clinical diagnosis of candidiasis prior to the commencement of antifungal therapy in APECED patients. Many of these patients are prescribed azoles prophylactically, a clinical practice that is probably best avoided. The use of regular topical treatments with biocides such as chlorhexidine may provide a more beneficial long-term prophylactic measure for controlling oral Candida in this patient cohort, as has been previously reported for HIV-infected children (2). Patients should only be prescribed antifungal therapies with azoles following microbiological culture analysis and confirmation of candidiasis to prevent the development of azole resistance in isolates persisting in these patients. This can be easily accomplished since microbiological evidence of candidiasis can be observed on CHROMagar Candida medium after incubation at 37°C for as little as 24 h. Furthermore, this diagnostic process enables presumptive species identification, which should also be taken into account when antifungal agents are prescribed due to the reduced susceptibility of some Candida species to azoles (3, 22, 36).
Supplementary Material
ACKNOWLEDGMENT
This study was supported by the Microbiology Research Unit, Dublin Dental University Hospital.
Footnotes
Supplemental material for this article may be found at http://jcm.asm.org/.
Published ahead of print on 2 March 2011.
REFERENCES
- 1. Baily G. G., Perry F. M., Denning D. W., Mandal B. K. 1994. Fluconazole-resistant candidosis in an HIV cohort. AIDS 8:787–792 [DOI] [PubMed] [Google Scholar]
- 2. Barasch A., Safford M. M., Dapkute-Marcus I., Fine D. H. 2004. Efficacy of chlorhexidine gluconate rinse for treatment and prevention of oral candidiasis in HIV-infected children: a pilot study. Oral Surg. Oral Med. Oral Pathol. Oral Radiol. Endod. 97:204–207 [DOI] [PubMed] [Google Scholar]
- 3. Bille J. 2000. Mechanisms and clinical significance of antifungal resistance. Int. J. Antimicrob. Agents 16:331–333 [DOI] [PubMed] [Google Scholar]
- 4. Bockle B. C., Wilhelm M., Muller H., Gotsch C., Sepp N. T. 2010. Oral mucous squamous cell carcinoma-an anticipated consequence of autoimmune polyendocrinopathy-candidiasis-ectodermal dystrophy (APECED). J. Am. Acad. Dermatol. 62:864–868 [DOI] [PubMed] [Google Scholar]
- 5. Bougnoux M. E., Morand S., d'Enfert C. 2002. Usefulness of multilocus sequence typing for characterization of clinical isolates of Candida albicans. J. Clin. Microbiol. 40:1290–1297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bougnoux M. E., et al. 2003. Collaborative consensus for optimized multilocus sequence typing of Candida albicans. J. Clin. Microbiol. 41:5265–5266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Buzi F., et al. 2003. Autoimmune polyendocrinopathy-candidiasis-ectodermal dystrophy syndrome: time to review diagnostic criteria? J. Clin. Endocrinol. Metab. 88:3146–3148 [DOI] [PubMed] [Google Scholar]
- 8. Cheng S., Clancy C. J., Nguyen K. T., Clapp W., Nguyen M. H. 2007. A Candida albicans petite mutant strain with uncoupled oxidative phosphorylation overexpresses MDR1 and has diminished susceptibility to fluconazole and voriconazole. Antimicrob. Agents Chemother. 51:1855–1858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Clinical and Laboratory Standards Institute 2002. Reference method for broth microdilution antifungal susceptibility testing of yeast; approved standard; M27-A2. Clinical and Laboratory Standards Institute, Wayne, PA [Google Scholar]
- 10. Coco B. J., et al. 2008. Mixed Candida albicans and Candida glabrata populations associated with the pathogenesis of denture stomatitis. Oral Microbiol. Immunol. 23:377–383 [DOI] [PubMed] [Google Scholar]
- 11. Coleman D. C., et al. 1993. Oral Candida in HIV infection and AIDS: new perspectives/new approaches. Crit. Rev. Microbiol. 19:61–82 [DOI] [PubMed] [Google Scholar]
- 12. Coleman D. C., Moran G. P., McManus B. A., Sullivan D. J. 2010. Mechanisms of antifungal drug resistance in Candida dubliniensis. Future Microbiol. 5:935–949 [DOI] [PubMed] [Google Scholar]
- 13. Coleman D. C., et al. 1997. Candidiasis: the emergence of a novel species, Candida dubliniensis. AIDS 11:557–567 [DOI] [PubMed] [Google Scholar]
- 14. Coste A., et al. 2006. A mutation in Tac1p, a transcription factor regulating CDR1 and CDR2, is coupled with loss of heterozygosity at chromosome 5 to mediate antifungal resistance in Candida albicans. Genetics 172:2139–2156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Coste A. T., Crittin J., Bauser C., Rohde B., Sanglard D. 2009. Functional analysis of cis- and trans-acting elements of the Candida albicans CDR2 promoter with a novel promoter reporter system. Eukaryot. Cell 8:1250–1267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Dodgson A. R., Dodgson K. J., Pujol C., Pfaller M. A., Soll D. R. 2004. Clade-specific flucytosine resistance is due to a single nucleotide change in the FUR1 gene of Candida albicans. Antimicrob. Agents Chemother. 48:2223–2227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Dominguez M., et al. 2006. Autoimmune polyendocrinopathy-candidiasis-ectodermal dystrophy (APECED) in the Irish population. J. Pediatr. Endocrinol. Metab. 19:1343–1352 [DOI] [PubMed] [Google Scholar]
- 18. Dunkel N., Blass J., Rogers P. D., Morschhauser J. 2008. Mutations in the multidrug resistance regulator MRR1, followed by loss of heterozygosity, are the main cause of MDR1 overexpression in fluconazole-resistant Candida albicans strains. Mol. Microbiol. 69:827–840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Epstein J. B., Pearsall N. N., Truelove E. L. 1980. Quantitative relationships between Candida albicans in saliva and the clinical status of human subjects. J. Clin. Microbiol. 12:475–476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Farah C. S., Lynch N., McCullough M. J. 2010. Oral fungal infections: an update for the general practitioner. Aust. Dent. J. 55(Suppl. 1):48–54 [DOI] [PubMed] [Google Scholar]
- 21. Fierabracci A. 2011. Recent insights into the role and molecular mechanisms of the autoimmune regulator (AIRE) gene in autoimmunity. Autoimmun. Rev. 10:137–143 [DOI] [PubMed] [Google Scholar]
- 22. Ghannoum M. A., Rice L. B. 1999. Antifungal agents: mode of action, mechanisms of resistance, and correlation of these mechanisms with bacterial resistance. Clin. Microbiol. Rev. 12:501–517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lamb D. C., et al. 1997. The mutation T315A in Candida albicans sterol 14α-demethylase causes reduced enzyme activity and fluconazole resistance through reduced affinity. J. Biol. Chem. 272:5682–5688 [DOI] [PubMed] [Google Scholar]
- 24. LeBoeuf N., Garg A., Worobec S. 2007. The autoimmune polyendocrinopathy-candidiasis-ectodermal dystrophy syndrome. Pediatr. Dermatol. 24:529–533 [DOI] [PubMed] [Google Scholar]
- 25. Livak K. J., Schmittgen T. D. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25:402–408 [DOI] [PubMed] [Google Scholar]
- 26. Marichal P., et al. 1997. Molecular biological characterization of an azole-resistant Candida glabrata isolate. Antimicrob. Agents Chemother. 41:2229–2237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. McCullough M. J., Clemons K. V., Stevens D. A. 1999. Molecular epidemiology of the global and temporal diversity of Candida albicans. Clin. Infect. Dis. 29:1220–1225 [DOI] [PubMed] [Google Scholar]
- 28. McGovern E., et al. 2008. Oral health in autoimmune polyendocrinopathy candidiasis ectodermal dystrophy (APECED). Eur. Arch. Paediatr. Dent. 9:236–244 [DOI] [PubMed] [Google Scholar]
- 29. McManus B. A., et al. 2008. Multilocus sequence typing reveals that the population structure of Candida dubliniensis is significantly less divergent than that of Candida albicans. J. Clin. Microbiol. 46:652–664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. McManus B. A., Moran G. P., Higgins J. A., Sullivan D. J., Coleman D. C. 2009. A Ser29Leu substitution in the cytosine deaminase Fca1p is responsible for clade-specific flucytosine resistance in Candida dubliniensis. Antimicrob. Agents Chemother. 53:4678–4685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Metwally L., et al. 2007. Trends in candidemia and antifungal susceptibility in a university hospital in Northern Ireland 2001–2006. J. Infect. 55:174–178 [DOI] [PubMed] [Google Scholar]
- 32. Moran G. P., et al. 1998. Identification and expression of multidrug transporters responsible for fluconazole resistance in Candida dubliniensis. Antimicrob. Agents Chemother. 42:1819–1830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Morio F., Loge C., Besse B., Hennequin C., Le Pape P. 2010. Screening for amino acid substitutions in the Candida albicans Erg11 protein of azole-susceptible and azole-resistant clinical isolates: new substitutions and a review of the literature. Diagn. Microbiol. Infect. Dis. 66:373–384 [DOI] [PubMed] [Google Scholar]
- 34. Odds F. C., Bernaerts R. 1994. CHROMagar Candida, a new differential isolation medium for presumptive identification of clinically important Candida species. J. Clin. Microbiol. 32:1923–1929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Odds F. C., et al. 2007. Molecular phylogenetics of Candida albicans. Eukaryot. Cell 6:1041–1052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Orozco A. S., et al. 1998. Mechanism of fluconazole resistance in Candida krusei. Antimicrob. Agents Chemother. 42:2645–2649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Peltroche-Llacsahuanga H., Dohmen H., Haase G. 2002. Recovery of Candida dubliniensis from sputum of cystic fibrosis patients. Mycoses 45:15–18 [PubMed] [Google Scholar]
- 38. Perea S., et al. 2002. Molecular mechanisms of fluconazole resistance in Candida dubliniensis isolates from human immunodeficiency virus-infected patients with oropharyngeal candidiasis. Antimicrob. Agents Chemother. 46:1695–1703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Pereira C. M., Pires F. R., Correa M. E., di Hipolito O., Junior, Almeida O. P. 2004. Candida in saliva of Brazilian hemophilic patients. J. Appl. Oral Sci. 12:301–306 [DOI] [PubMed] [Google Scholar]
- 40. Pfaffl M. W. 2001. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 29:e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Pincus D. H., et al. 1999. Rapid identification of Candida dubliniensis with commercial yeast identification systems. J. Clin. Microbiol. 37:3533–3539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Powderly W. G. 1992. Mucosal candidiasis caused by non-albicans species of Candida in HIV-positive patients. AIDS 6:604–605 [PubMed] [Google Scholar]
- 43. Pujol C., Pfaller M. A., Soll D. R. 2004. Flucytosine resistance is restricted to a single genetic clade of Candida albicans. Antimicrob. Agents Chemother. 48:262–266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Ramsey C., et al. 2002. Aire deficient mice develop multiple features of APECED phenotype and show altered immune response. Hum. Mol. Genet. 11:397–409 [DOI] [PubMed] [Google Scholar]
- 45. Rautemaa R., Hietanen J., Niissalo S., Pirinen S., Perheentupa J. 2007. Oral and oesophageal squamous cell carcinoma: a complication or component of autoimmune polyendocrinopathy-candidiasis-ectodermal dystrophy (APECED, APS-I). Oral Oncol. 43:607–613 [DOI] [PubMed] [Google Scholar]
- 46. Rautemaa R., et al. 2007. Decreased susceptibility of Candida albicans to azole antifungals: a complication of long-term treatment in autoimmune polyendocrinopathy-candidiasis-ectodermal dystrophy (APECED) patients. J. Antimicrob. Chemother. 60:889–892 [DOI] [PubMed] [Google Scholar]
- 47. Rautemaa R., Richardson M., Pfaller M., Perheentupa J., Saxen H. 2008. Reduction of fluconazole susceptibility of Candida albicans in APECED patients due to long-term use of ketoconazole and miconazole. Scand. J. Infect. Dis. 40:904–907 [DOI] [PubMed] [Google Scholar]
- 48. Rustad T. R., Stevens D. A., Pfaller M. A., White T. C. 2002. Homozygosity at the Candida albicans MTL locus associated with azole resistance. Microbiology 148:1061–1072 [DOI] [PubMed] [Google Scholar]
- 49. Sangeorzan J. A., et al. 1994. Epidemiology of oral candidiasis in HIV-infected patients: colonization, infection, treatment, and emergence of fluconazole resistance. Am. J. Med. 97:339–346 [DOI] [PubMed] [Google Scholar]
- 50. Sanglard D., Coste A., Ferrari S. 2009. Antifungal drug resistance mechanisms in fungal pathogens from the perspective of transcriptional gene regulation. FEMS Yeast Res. 9:1029–1050 [DOI] [PubMed] [Google Scholar]
- 51. Sanglard D., Ischer F., Calabrese D., Majcherczyk P. A., Bille J. 1999. The ATP binding cassette transporter gene CgCDR1 from Candida glabrata is involved in the resistance of clinical isolates to azole antifungal agents. Antimicrob. Agents Chemother. 43:2753–2765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Sanglard D., Ischer F., Monod M., Bille J. 1997. Cloning of Candida albicans genes conferring resistance to azole antifungal agents: characterization of CDR2, a new multidrug ABC transporter gene. Microbiology 143(Pt. 2):405–416 [DOI] [PubMed] [Google Scholar]
- 53. Sanglard D., Ischer F., Monod M., Bille J. 1996. Susceptibilities of Candida albicans multidrug transporter mutants to various antifungal agents and other metabolic inhibitors. Antimicrob. Agents Chemother. 40:2300–2305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Sanglard D., et al. 1995. Mechanisms of resistance to azole antifungal agents in Candida albicans isolates from AIDS patients involve specific multidrug transporters. Antimicrob. Agents Chemother. 39:2378–2386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Sebti A., et al. 2001. Candida dubliniensis at a cancer center. Clin. Infect. Dis. 32:1034–1038 [DOI] [PubMed] [Google Scholar]
- 56. Siikala E., et al. 2010. Persistent Candida albicans colonization and molecular mechanisms of azole resistance in autoimmune polyendocrinopathy-candidiasis-ectodermal dystrophy (APECED) patients. J. Antimicrob. Chemother. 65:2505–2513 [DOI] [PubMed] [Google Scholar]
- 57. Sitheeque M. A., Samaranayake L. P. 2003. Chronic hyperplastic candidosis/candidiasis (candidal leukoplakia). Crit. Rev. Oral Biol. Med. 14:253–267 [DOI] [PubMed] [Google Scholar]
- 58. Sullivan D. J., Westerneng T. J., Haynes K. A., Bennett D. E., Coleman D. C. 1995. Candida dubliniensis sp. nov.: phenotypic and molecular characterization of a novel species associated with oral candidosis in HIV-infected individuals. Microbiology 141(Pt. 7):1507–1521 [DOI] [PubMed] [Google Scholar]
- 59. Tavanti A., Gow N. A., Senesi S., Maiden M. C., Odds F. C. 2003. Optimization and validation of multilocus sequence typing for Candida albicans. J. Clin. Microbiol. 41:3765–3776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Thompson J. D., Higgins D. G., Gibson T. J. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673–4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Vanden Bossche H., Marichal P., Odds F. C., Le Jeune L., Coene M. C. 1992. Characterization of an azole-resistant Candida glabrata isolate. Antimicrob. Agents Chemother. 36:2602–2610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Vazquez J. A., et al. 2001. Evolution of antifungal susceptibility among Candida species isolates recovered from human immunodeficiency virus-infected women receiving fluconazole prophylaxis. Clin. Infect. Dis. 33:1069–1075 [DOI] [PubMed] [Google Scholar]
- 63. von Schnurbein J., Lahr G., Posovszky C., Debatin K. M., Wabitsch M. 2008. Novel homozygous AIRE mutation in a German patient with severe APECED. J. Pediatr. Endocrinol. Metab. 21:1003–1009 [DOI] [PubMed] [Google Scholar]
- 64. Warnock D. W. 2007. Trends in the epidemiology of invasive fungal infections. Nippon Ishinkin Gakkai Zasshi 48:1–12 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.