Abstract
A modified multiple-locus variable-number tandem-repeat analysis (MMLVA) method was validated on Clostridium difficile-infected stool specimens from institutional outbreaks. The method allows simultaneous detection of toxin genes, deletions, and tandem repeats from cultured isolates or stool specimens. Results were used to aid institutional outbreak investigation by identifying clusters of NAP1/027.
TEXT
Clostridium difficile infection (CDI) is a major cause of gastrointestinal disease in patients treated with antibiotics in hospitals (10, 15). Effective control of CDI outbreaks requires identification of true clusters of infected patients. The typing method must have sufficient discriminatory power to differentiate outbreak isolates from sporadic isolates, especially because the epidemic strain North American Pulsotype (NAP) 1/ribotype 027 is extremely common in North America and responsible for the vast majority of institutional outbreaks (13, 16). A previous comparison of genotyping methods revealed that multiple-locus variable-number tandem-repeat analysis (MLVA) was the most discriminatory and had the potential to identify true clusters of CDI (6). However, MLVA is technically challenging, requires considerable expertise beyond the medical laboratory technologist level, and relies on prior culture of the organism, resulting in significant delays in reporting during institutional outbreak investigation (5, 7, 12, 19). In our initial attempts to validate MLVA, we noted that there was no variability observed in the F3Cd and H9Cd loci for CDI outbreak isolates in our region, and, thus, these two loci added no value in discriminating among the isolates (our unpublished observations). Additionally, MLVA does not indicate whether the isolate is the epidemic strain NAP1/027, which contains the tcdC deletion and elaborates binary toxin (cdtB) in addition to toxins A (tcdA) and B (tcdB), all constituents of the pathogenicity locus (PaLoc) of toxigenic strains. Finally, MLVA does not identify NAP7, a hypervirulent emerging strain in North America, due to DNA sequence variation (14). Thus, MLVA still requires complementary pulsed-field gel electrophoresis (PFGE) or ribotyping in order to confirm that it is NAP1/027. In this study, we have developed a modified MLVA (MMLVA) method which enables simultaneous detection of CDI and identification of hypervirulent epidemic strains, as well as providing typing information which enables rapid identification of clusters of outbreak isolates in institutions.
CDI specimens associated with institutional outbreaks (defined as ≥6 cases per ward per month) were received by our laboratory for typing by PFGE between December 2008 and December 2009. A case was defined as the presence of toxin A/B by enzyme immunoassay (EIA) or molecular test in a patient with clinical diarrhea. Outbreak-associated specimens (n = 155) were subjected to culture using C. difficile moxalactam-norfloxacin (CDMN) agar as described previously (2, 16). For PFGE, extraction of DNA from colonies grown on CDMN agar was performed using a commercial extraction method (InstaGene Matrix, Bio-Rad Laboratories, Hercules, CA). For MMLVA, DNA was extracted from colonies with the InstaGene Matrix (Bio-Rad, Hercules, CA). A single colony was resuspended in 100 μl of InstaGene, heated at 100°C for 10 min, vortexed for 10 s, and centrifuged at 10,000 rpm for 2 min. The supernatant was used for PCR. For MMLVA performed directly from stool, DNA was extracted using the QIAamp stool minikit (Qiagen, Valencia, CA) adapted to the QIAcube platform. In order to amplify five tandem repeat loci (denoted A6Cd, B7Cd, C6Cd, E7Cd, and G8Cd according to the method of van den Berg [19]), the tcdC deletion, toxin genes (tcdA, tcdB, cdtB), and the housekeeping gene tpi, three multiplex PCRs were conducted under identical cycling conditions. C. difficile genome sequences from GenBank (630: taxonomy ID [TID], 272563; M120: TID, 699035; NAP07: TID, 525258; NAP08: TID, 525259; CD196: TID, 645462; R20291: TID, 645463) were used to modify primer sequences to accommodate a broader set of strain types (Table 1). Each primer stock solution was prepared at a concentration of 100 μM and mixed with other primers to make the primer mixes (PM) indicated in Table 1. Each PM (1 μl) was added to 7 μl of the Type-it Microsatellite master mix (Qiagen, Valencia, CA) for a total reaction volume of 8 μl. The cycling conditions were as follows: hot start at 90°C for 5 min, then 36 cycles of 30 s at 90°C, 60 s at 50°C, and 30 s at 72°C, with a final extension step of 30 min at 60°C. Amplicons were diluted 1:20 with distilled water, and 1 μl was transferred to 11 μl of formamide mixed with LIZ500 DNA ladder (0.1 μl per well), heated for 3 min at 95°C, and snap-cooled to 4°C. The Applied Biosystems (Carlsbad, CA) 3130xl genetic analyzer was used to separate the fragments, and GeneScan software was used to identify their fragment length. BioNumerics (Applied Maths, Austin, TX) software was used to perform cluster analysis and generate dendrograms of MMLVA types compared to results of PFGE. Cluster analysis was performed using the Manhattan distance measure (numerical coefficient which sums differences in repeat units and deletions) and the Ward clustering algorithm to generate the dendrogram (6). To facilitate the interpretation, a cluster was defined as having <5% difference based on summed tandem repeat differences (STRD) at all five loci.
Table 1.
Primers used in modified multiple-locus variable-number tandem-repeat analysis (MMLVA)a
| Marker (PM) | Primer | Primer sequence (5′ to 3′) | Reference |
|---|---|---|---|
| A6Cd (1) | A6-F | FAM-TTAATTGAGGGAGAATGTTAAA | 19 |
| A6-R | AAATACTTTTCCCACTTTCATAA | 19 | |
| B7Cd (1) | B7-F | VIC-TTAATACTAAACTAACTCTAACCAGTAA | Modified from reference 19 |
| B7-NAP7/8-F | VIC-TTAATATTAAACTAACTCTAACCAGTAA | Modified from reference 19 | |
| B7-universal-R | TTATATTTTATGGGYATGTTAAA | Modified from reference 19 | |
| C6Cd (2) | C6-630, 91, and 96-F | VIC-GTTTAGAATCTACARCATTATTTGA | Modified from reference 19 |
| C6-NAP7/8-F | ATTTAGAATCTATACTATTATTTGA | Modified from reference 19 | |
| C6-R | ATTGGAATTGAATGTAACAAAA | 19 | |
| C6-NAP7/8-R | AGCGGAATTGAATGTAACAAAA | Modified from reference 19 | |
| E7Cd (2) | E7-F | FAM-TGGAGCTATGGAAATTGATAA | 19 |
| E7-R | CAAATACATCTTGCATTAATTCTT | 19 | |
| E7-NAP7/8-R | CAAATACATCTTGCACTAGTTCTT | Modified from reference 19 | |
| G8Cd (1) | G8-F | NED-TGTATGAAGCAAGCTTTTTATT | 19 |
| G8-R | ACCAAAAATTTCTAACCCAAC | 3 | |
| G8-NAP7/8-R | ACCAAAATTTTCTAACCCAAC | Modified from reference 3 | |
| tpi (3) | tpi-F | FAM-AAGAAGCTACTAAGGGTACAAA | 9 |
| tpi-R | CATAATATTGGGTCTATTCCTAC | 9 | |
| tcdA (3) | tcdA-F | FAM-AGATTCCTATATTTACATGACAATAT | 9 |
| tcdA-R | GTATCAGGCATAAAGTAATATACTTT | 9 | |
| tcdB (3) | tcdB-F | FAM-GGAAAAGAGAATGGTTTTATTAA | 9 |
| tcdB-R | ATCTTTAGTTATAACTTTGACATCTTT | 9 | |
| cdtB (3) | cdtB-F | FAM-CTTAATGCAAGTAAATACTGAG | 18 |
| cdtB-R | AACGGATCTCTTGCTTCAGTC | 18 | |
| tcdC deletion (2) | tcdC-F | FAM-AAGCTATTGAAGCTGAAAATC | 1 |
| tcdC-R | GCTAATTGGTCATAAGTAATACC | 1 |
Modifications to published primers are indicated as underlined nucleotides. Three multiplex reactions were used simultaneously under identical cycling conditions. Primer mix (PM) combinations and forward (F) and reverse (R) primers are indicated. Fluorophores are named next to each relevant primer sequence.
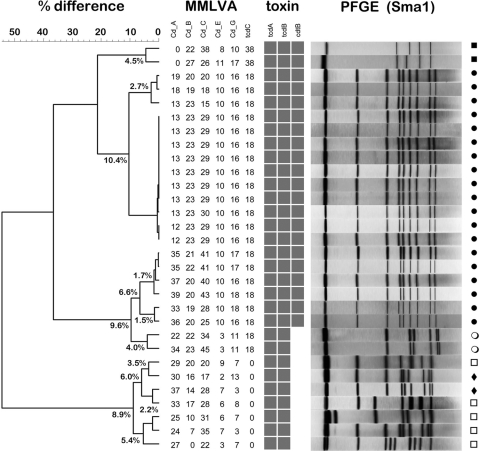
MMLVA types were compared with traditional PFGE for all outbreak-associated CDI. A representative dendrogram from a group of hospital outbreak specimens (n = 30) is depicted in Fig. 1. Confirmation of the presence of toxin genes is demonstrated using a matrix array format. By including the 18-bp NAP1 tcdC deletion in the MMLVA analysis, the NAP1/027 strain responsible for all outbreaks in our region is immediately separated from non-NAP1/027 strains. This includes NAP1/027 variants with single-nucleotide polymorphisms (SNPs) upstream of the deletion (11). Good correlation between MMLVA and PFGE was observed, with MMLVA being able to further segregate four clusters within 19 NAP1/027 isolates and two NAP7 isolates forming a cluster, as well as four other non-NAP1 pairs. A pair of NAP11 isolates was also noted but did not form a cluster by MMLVA, highlighting the discriminatory power of MMLVA for non-NAP1 isolates as well. The absence of tandem repeats at locus A6Cd and the 38-bp deletion at tcdC is typical of the NAP7 strain. A numerical measure (Manhattan) rather than categorical coefficient was used in our cluster analysis because categorical metrics tend to inflate variability when subtle changes occur at multiple loci. A more liberal cutoff value of <5% difference by Manhattan coefficient was used because the previously reported cutoff value of 2 STRD to define a “clone” is likely too stringent in the context of an outbreak (4). For example, we and others have observed that a single stool specimen with a polyclonal infection can contain greater variation than 2 STRD (17). As shown in this representative outbreak, variability in tandem repeats can be observed for clusters at loci A6Cd, B7Cd, and C6Cd, but little or no variability is seen at loci E7Cd and G8Cd within outbreak clusters. The fact that not all loci demonstrate equal variability hampers facile interpretation of true clusters and reinforces the fact that sound epidemiological investigation is required to confirm or refute the laboratory typing. Similar results were obtained whether using cultured isolates or those directly from stool, with the exception that certain stool specimens may produce more than one MMLVA type, but this does not interfere with the cluster analysis when using a <5% cutoff value (our unpublished observations). Ten stool specimens can be run concurrently when using a liquid handler for DNA extraction and capillary electrophoresis to separate fragments. CDI identification and typing results are obtained within 5 h and reported back to the institution (Table 2).
Fig. 1.
Dendrogram depicting MMLVA types compared to pulsotypes (n = 30) for an institutional outbreak. Clusters were defined as <5% difference based on the Manhattan distance measure. Within the NAP1 pulsotype (closed circles), four clusters are identified. A pair of NAP7 (closed squares) isolates formed a cluster, whereas a pair of NAP11 (closed diamonds) isolates did not form a cluster. Other unclassified pulsotypes (open circles, open squares) are noted. A matrix array format is used to identify the presence or absence of toxin genes (the tpi control gene is used as an amplification control and is not shown). Note the unusual pair of CDI cases (open circles) which has the tcdC deletion but not the binary toxin. PFGE (SmaI digest) results correlated closely with those of MMLVA.
Table 2.
Summary of the performance characteristics and costs of commercially available molecular detection and typing methods for C. difficile compared to those of MMLVA
| Assay | Manufacturer | Function | Test method | Batch size | Instrumentation for: |
Interpretation | Concern(s) | Advantage(s) | Avg time | Costa | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| DNA extraction | Detection | ||||||||||
| BD GeneOhm C. difficile | Becton Dickinson | Detects C. difficile toxin B gene | PCR | 8 | Heat lysis | SmartCycler | Cepheid SmartCycler software | Cost, throughput | Good specificity | 2-3 h | $25-$30 |
| ProGastro CD | GenProbe | Detects C. difficile toxin B gene | PCR | 8 | Automated extraction MagNa Pure | SmartCycler | Cepheid SmartCycler software | Cost, throughput | Good specificity | 2-3 h | $25-$30 |
| Illumigene C. difficile | Meridian Bioscience | Detects PaLoc gene segment; codes for both toxin A and B genes | Loop-mediated amplification technology (LAMP) | 10 | Illumigene sample prepn apparatus, heat lysis | Illumipro-10 | Illumipro-10 ratio of initial to final signal (turbidity) | Newer methodology | Minimal equipment required; rapid | 40 min | $25-$30 |
| Xpert C. difficile | Cepheid | Detects C. difficile toxin B gene | PCR | 10 cartridges, 120 cartridges | GeneXpert System | GeneXpert System | Cepheid SmartCycler software | Cost, throughput | Good specificity | 2 h | $35-$40 |
| MMLVA | This study—in-house test | Detects C. difficile, toxin A, toxin B, binary toxin, tcdC gene plus VNTRsb | PCR sequencing and fragment analysis | 1 to 48 | QIAcube (Qiagen)/InstaGene Matrix | 3130xl genetic analyzer (Life Technologies) | GeneMapper software | High setup cost, technical expertise | Highly discriminatory detection and typing | 5 h | $5-$10 |
| GenoType CDiff | Hain Lifescience | Detects C. difficile, toxin A, toxin B, binary toxin, tcdC gene, resistance to moxifloxacin | DNA strip technology | 12 | GenoType CDiff | GenoType CDiff | Visual assessment | Typing lacks discriminatory power | Detection and typing performed simultaneously | 5 h (stool direct) | $30-$35 |
Costs are in Canadian dollars and meant only to be approximate for reagnets alone.
VNTRs, variable-number tandem repeats.
Due to an ongoing epidemic of NAP1/027 resulting in CDI institutional outbreaks in our region, we have been conducting PFGE to support outbreak investigation and infection control (16). However, PFGE lacks discriminatory power to distinguish outbreak clusters from sporadic NAP1/027 and requires laborious culture (8). More discriminatory methods such as MLVA are highly specialized, result in amplification of uninformative loci (from the point of view of outbreak investigation), and do not identify the critical NAP1/027 strains per se. MMLVA couples the discriminatory power of tandem-repeat loci with the capacity to identify toxin genes and deletions as well as determine whether an isolate is related to the NAP1/027 epidemic strain in a single run. Identification of MMLVA clusters early in the course of an outbreak enables infection prevention and control workers to pinpoint possible breakdowns in strict barrier precautions, reinforcement of hand hygiene practices with soap and water, enhanced environmental cleaning, and equipment disinfection on the affected units that may have led to the increase in case load. The method is not restricted to NAP1 clusters, as the tandem-repeat units will also identify clusters of other pulsotypes. Importantly, the method works from cultured isolates or from stool, eliminating the need for culture, which can result in delays for infection prevention and control measures. We believe this method provides rapid identification of current outbreak clusters to aid investigation which may curb the spread of the NAP1/027 epidemic strain as well as other emerging strains.
Acknowledgments
We thank the staff of the Molecular Surveillance Department of the Public Health Laboratory for expert technical assistance.
Footnotes
Published ahead of print on 16 March 2011.
REFERENCES
- 1. Antikainen J., et al. 2009. Detection of virulence genes of Clostridium difficile by multiplex PCR. APMIS 117:607–613 [DOI] [PubMed] [Google Scholar]
- 2. Arroyo L. G., et al. 2005. Use of a selective enrichment broth to recover Clostridium difficile from stool swabs stored under different conditions. J. Clin. Microbiol. 43:5341–5343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Fawley W. N., et al. 2008. Use of highly discriminatory fingerprinting to analyze clusters of Clostridium difficile infection cases due to epidemic ribotype 027 strains. J. Clin. Microbiol. 46:954–960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Freeman J., et al. 2010. The changing epidemiology of Clostridium difficile infections. Clin. Microbiol. Rev. 23:529–549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Goorhuis A., et al. 2009. Application of multiple-locus variable-number tandem-repeat analysis to determine clonal spread of toxin A-negative Clostridium difficile in a general hospital in Buenos Aires, Argentina. Clin. Microbiol. Infect. 15:1080–1086 [DOI] [PubMed] [Google Scholar]
- 6. Hill T., Lewicki P. 2007. Statistics. Methods and applications. StatSoft, Tulsa, OK [Google Scholar]
- 7. Janezic S., Rupnik M. 2010. Molecular typing methods for Clostridium difficile: pulsed-field gel electrophoresis and PCR ribotyping. Methods Mol. Biol. 646:55–65 [DOI] [PubMed] [Google Scholar]
- 8. Killgore G., et al. 2008. Comparison of seven techniques for typing international epidemic strains of Clostridium difficile: restriction endonuclease analysis, pulsed-field gel electrophoresis, PCR-ribotyping, multilocus sequence typing, multilocus variable-number tandem-repeat analysis, amplified fragment length polymorphism, and surface layer protein A gene sequence typing. J. Clin. Microbiol. 46:431–437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Lemee L., et al. 2004. Multiplex PCR targeting tpi (triose phosphate isomerase), tcdA (toxin A), and tcdB (toxin B) genes for toxigenic culture of Clostridium difficile. J. Clin. Microbiol. 42:5710–5714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Loo V. G., et al. 2005. A predominantly clonal multi-institutional outbreak of Clostridium difficile-associated diarrhea with high morbidity and mortality. N. Engl. J. Med. 353:2442–2449 [DOI] [PubMed] [Google Scholar]
- 11. MacCannell D. R., et al. 2006. Molecular analysis of Clostridium difficile PCR ribotype 027 isolates from Eastern and Western Canada. J. Clin. Microbiol. 44:2147–2152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Marsh J. W., et al. 2006. Multilocus variable-number tandem-repeat analysis for investigation of Clostridium difficile transmission in hospitals. J. Clin. Microbiol. 44:2558–2566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. McDonald L. C., et al. 2005. An epidemic, toxin gene-variant strain of Clostridium difficile. N. Engl. J. Med. 353:2433–2441 [DOI] [PubMed] [Google Scholar]
- 14. Mulvey M. R., et al. 2010. Hypervirulent Clostridium difficile strains in hospitalized patients, Canada. Emerg. Infect. Dis. 16:678–681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Pepin J., et al. 2004. Clostridium difficile-associated diarrhea in a region of Quebec from 1991 to 2003: a changing pattern of disease severity. CMAJ 171:466–472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Pillai D. R., Longtin J., Low D. E. 2010. Surveillance data on outbreaks of Clostridium difficile infection in Ontario, Canada, in 2008-2009. Clin. Infect. Dis. 50:1685–1686 [DOI] [PubMed] [Google Scholar]
- 17. Tanner H. E., Hardy K. J., Hawkey P. M. 2010. Coexistence of multiple multilocus variable-number tandem-repeat analysis subtypes of Clostridium difficile PCR ribotype 027 strains within fecal specimens. J. Clin. Microbiol. 48:985–987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Terhes G., Urban E., Soki J., Hamid K. A., Nagy E. 2004. Community-acquired Clostridium difficile diarrhea caused by binary toxin, toxin A, and toxin B gene-positive isolates in Hungary. J. Clin. Microbiol. 42:4316–4318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. van den Berg R. J., Schaap I., Templeton K. E., Klaassen C. H., Kuijper E. J. 2007. Typing and subtyping of Clostridium difficile isolates by using multiple-locus variable-number tandem-repeat analysis. J. Clin. Microbiol. 45:1024–1028 [DOI] [PMC free article] [PubMed] [Google Scholar]