Abstract
In this study, the prevalence of Enterocytozoon bieneusi in China was investigated. Twelve genotypes of E. bieneusi were identified, of which 10 were novel genotypes. Further, 41.6% of the genotypes were found in both humans and animals. This is the first report of E. bieneusi in China.
TEXT
Microsporidia are a diverse group of obligate intracellular pathogens consisting of approximately 1,300 formally described species in 160 genera that infect a wide range of invertebrate and vertebrate hosts, including humans (8, 16). Enterocytozoon bieneusi, the most frequently diagnosed microsporidial species in humans, was first reported in an AIDS patient in 1985 (4). Over the last 2 decades, E. bieneusi has been detected in humans, other mammals, and birds in more than 30 countries (1, 6, 10, 12). However, the prevalence of this parasite in China has been unclear.
Currently, sequencing of the internal transcribed spacer (ITS) region of the rRNA gene is regarded as the standard method for species identification and genotyping of E. bieneusi isolates (17). More than 90 genotypes or variants of E. bieneusi that infect humans and animals have been identified (16). Recent molecular epidemiological studies indicated that some genotypes are host specific (17). Further, 8 genotypes (WL15, D, Peru6, EbpC, Peru10, Peru16, WL11, and Type IV) have been found in both humans and animals, indicating the potential for zoonotic transmission and the importance for surveillance of the epidemiology of E. bieneusi (17).
We investigated the prevalence of E. bieneusi infection in humans and animals in China. A total of 220 fecal samples were collected. Among them, 40 fecal samples were from diarrheal children in the First Hospital of Jilin University in Changchun City, northeast China, 61 were from pigs in a livestock production facility, 26 were from dogs in a pet market, and 93 were from cows. Both human and animal samples were collected in the same area around Changchun City; however, the human samples were not from the same farm as the animal samples. The collection of human and animal stool samples was approved by the Ethics Committee of the Institute of Zoonosis, Jilin University. DNA was extracted from each fecal sample with a modified protocol as described previously (7).
A nested PCR with primers based on the specific ITS sequences of E. bieneusi was applied for pathogen identification as previously reported (2, 5). E. bieneusi ITS sequences were determined, and a multiple alignment was performed using the ClustalX program. To assess the extent of genetic diversity and evolutionary relationships among the previously known genotypes of Enterocytozoon spp. and the novel genotypes, a neighbor-joining tree based on the evolutionary distances calculated by the Kimura two-parameter model (15) was constructed using the MEGA 4.0 program (19).
E. bieneusi-specific sequences were amplified from 56 of the 220 fecal samples (9 from humans, 2 from dogs, 35 from cows, and 10 from pigs) (Table 1). The 56 sequences were classified into 12 genotypes, which included the previously reported genotypes I and J (9, 13, 18) and 10 novel types (named CHN1 to CHN10) (Table 1). Surprisingly, genotypes I and J, originally detected in cattle (18), were identified in both cow and human samples in this study. Genotype I was found in samples from 3 children and 8 cows, and genotype J was found in samples from 3 children and 9 cows. Interestingly, the two genotypes coinfected with other genotypes in all positive samples. Of the novel genotypes, CHN1, which was detected in 5 children, 9 cows, and 4 pigs, was the most common genotype found in this study. Genotype CHN2 was found only in human samples. Genotypes CHN3 and CHN4 were also found in both human and cow samples.
Table 1.
Prevalence of E. bieneusi in 220 samples
| Origin of sample | No. of samples examined | No. of samples positive by PCR (%) | Genotypes (no. of positive hosts) | Genotypes from coinfected hosts (no. of positive hosts) |
|---|---|---|---|---|
| Humans | 40 | 9 (22.5) | I (3), J (3), CHN1 (5), CHN2 (2), CHN3 (4), CHN4 (3) | I, CHN2, CHN3 (1); I, CHN3 (1); J, CHN1 (1); J, CHN1, CHN2, CHN4 (1); CHN1, CHN4 (1); I, CHN1, CHN4 (1) |
| Dogs | 26 | 2 (7.8) | CHN5 (1), CHN6 (1) | |
| Cattle | 93 | 35 (37.6) | I (8), J (9), CHN1 (10), CHN3 (14), CHN4 (2) | I, J (5); CHN1, CHN3 (1); I, J, CHN1 (1); CHN1, CHN4 (1); J, CHN1 (1); CHN3, CHN4 (1) |
| Pigs | 61 | 10 (16.4) | CHN1 (4), CHN7 (4), CHN8 (1), CHN9 (1), CHN10 (2) | CHN1, CHN10 (1); CHN1, CHN9 (1) |
| Total | 220 | 56 (25.5) |
Genotypes CHN5 and CHN6 were found only in dogs in this study. Similarly, genotypes CHN7 to CHN10 were likely pig specific. CHN7 and CHN8 were found in monoinfections, while CHN9 and CHN10 coinfected with other genotypes.
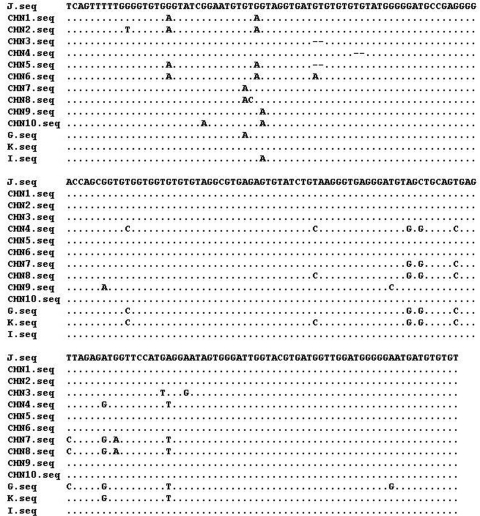
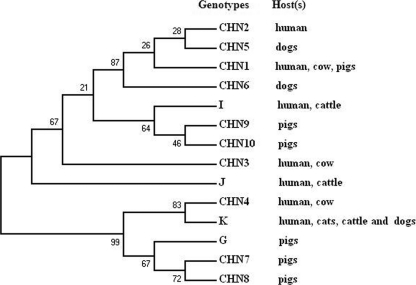
The ITS sequences of the 10 new genotypes were highly homologous to those of genotypes I, J, K, and G published earlier (3, 9, 11, 14, 18) (Fig. 1). Types CHN1, CHN2, CHN3, CHN5, and CHN6 differed from J by one to four positions, while type CHN4 is 2 bp shorter than type K. Types CHN7 and CHN8 differed from G by one and two nucleotides, respectively. Similarly, types CHN9 and CHN10 differed from type I by one and two nucleotides. A neighbor-joining tree was constructed by aligning the ITS sequences of 10 new genotypes and that of genotypes I, J, K, and G (Fig. 2). Due to the high level of similarity of the sequences, no obvious clusters related to host preference were observed. Thus, E. bieneusi is a mammalian parasite with the ability to infect multiple species.
Fig. 1.
Sequence variation in the ITS region of the rRNA gene of E. bieneusi isolates. The ITS sequences of the 10 distinct new genotypes (CHN1 to CHN10) identified in this study were aligned with that of the 4 known genotypes (I, J, K, and G). Dots indicate sequence identity to J, while dashes indicate deletions.
Fig. 2.
Phylogenetic relationships of the 10 distinct new genotypes (CHN1 to CHN10) identified in this study with 4 known genotypes (I, J, K, and G) of E. bieneusi inferred by a neighbor-joining analysis of internal transcribed spacer sequences based on genetic distances calculated by the Kimura two-parameter model. Numbers on the branches are percent bootstrapping values from 1,000 replicates.
In this study, we conducted the first investigation on the prevalence of E. bieneusi in China. Though E. bieneusi was first identified in a patient with clinical HIV infection (4), there is still no direct correlation between E. bieneusi infection and clinical disease. Of the 40 samples from the diarrheal children, only 22.5% were E. bieneusi positive, and all of the children with positive samples were infected with two or more genotypes of the parasite, which indicates that the disease may not be directly caused by E. bieneusi. Further studies in this area are obviously necessary.
In an earlier study, it was reported that an unusual E. bieneusi genotype was found in seven guinea pigs and a 2-year-old child in the same household in the United States in 2007, which strongly suggests the possibility of zoonotic transmission (3). In this study, among the 12 genotypes identified, 6 genotypes were found in children, and 5 of them were also detected in cows and pigs. Further, we also found human infections with genotypes I and J, which were found only in cattle in other countries. Due to the fact that all samples were collected from the same location, the data provide direct evidence that E. bieneusi isolates of animal origin are infective in humans. Interestingly, only two genotypes (CHN5 and CHN6) were found in the samples collected from dogs, and no human infections by these two genotypes were found. Taken together, these results provide further evidence that E. bieneusi isolates of animal origin may be infective in humans. The data argue for the importance of epidemiological control and prevention from both the agricultural and medical sectors.
Nucleotide sequence accession numbers.
The GenBank accession numbers for CHN1 to CHN6 are HM992509 to HM992514, and for CHN7 to CHN10, HM992516 to HM992519.
Acknowledgments
We are very grateful to the medical doctors who helped to collect and keep the samples from the children and to the farmers who provided the animal samples. The study was reviewed and approved by the Ethics Committee of Jilin University, China.
Q.C. was supported by a Distinguished Young Scientists Award from the NSFC (30625029) and by a National Science and Technology Project (China) grant (2008ZX-10004-011).
Footnotes
Published ahead of print on 9 March 2011.
The authors have paid a fee to allow immediate free access to this article.
REFERENCES
- 1. Abe N., Kimata I., Iseki M. 2009. Molecular evidence of Enterocytozoon bieneusi in Japan. J. Vet. Med. Sci. 71:217–219 [DOI] [PubMed] [Google Scholar]
- 2. Buckholt M. A., Lee J. H., Tzipori S. 2002. Prevalence of Enterocytozoon bieneusi in swine: an 18-month survey at a slaughterhouse in Massachusetts. Appl. Environ. Microbiol. 68:2595–2599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Cama V. A., et al. 2007. Transmission of Enterocytozoon bieneusi between a child and guinea pigs. J. Clin. Microbiol. 45:2708–2710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Desportes I., et al. 1985. Occurrence of a new microsporidian: Enterocytozoon bieneusi n.g., n. sp., in the enterocytes of a human patient with AIDS. J. Protozool. 32:250–254 [DOI] [PubMed] [Google Scholar]
- 5. Drosten C., Laabs J., Kuhn E. M., Schottelius J. 2005. Interspecies transmission of Enterozytozoon bieneusi supported by observations in laboratory animals and phylogeny. Med. Microbiol. Immunol. 194:207–209 [DOI] [PubMed] [Google Scholar]
- 6. Jeong D. K., et al. 2007. Occurrence and genotypic characteristics of Enterocytozoon bieneusi in pigs with diarrhea. Parasitol. Res. 102:123–128 [DOI] [PubMed] [Google Scholar]
- 7. Kahler A. M., Thurston-Enriquez J. A. 2007. Human pathogenic microsporidia detection in agricultural samples: method development and assessment. Parasitol. Res. 100:529–538 [DOI] [PubMed] [Google Scholar]
- 8. Keeling P. 2009. Five questions about microsporidia. PLoS Pathog. 5:e1000489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Lee J. H. 2007. Prevalence and molecular characteristics of Enterocytozoon bieneusi in cattle in Korea. Parasitol. Res. 101:391–396 [DOI] [PubMed] [Google Scholar]
- 10. Leelayoova S., et al. 2009. Genotypic characterization of Enterocytozoon bieneusi in specimens from pigs and humans in a pig farm community in Central Thailand. J. Clin. Microbiol. 47:1572–1574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Liguory O., David F., Sarfati C., Derouin F., Molina J. M. 1998. Determination of types of Enterocytozoon bieneusi strains isolated from patients with intestinal microsporidiosis. J. Clin. Microbiol. 36:1882–1885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Mathis A., Weber R., Deplazes P. 2005. Zoonotic potential of the microsporidia. Clin. Microbiol. Rev. 18:423–445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Rinder H., et al. 2000. Close genotypic relationship between Enterocytozoon bieneusi from humans and pigs and first detection in cattle. J. Parasitol. 86:185–188 [DOI] [PubMed] [Google Scholar]
- 14. Rinder H., Katzwinkel-Wladarsch S., Thomschke A., Loscher T. 1998. Strain differentiation in microsporidia. Tokai J. Exp. Clin. Med. 23:433–437 [PubMed] [Google Scholar]
- 15. Saitou N., Nei M. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4:406–425 [DOI] [PubMed] [Google Scholar]
- 16. Santin M., Fayer R. 9 August 2010, posting date Microsporidiosis: Enterocytozoon bieneusi in domesticated and wild animals. Res. Vet. Sci. [Epub ahead of print.] doi:10.1016/j.rvsc.2010.07.014 [DOI] [PubMed] [Google Scholar]
- 17. Santín M., Fayer R. 2009. Enterocytozoon bieneusi genotype nomenclature based on the internal transcribed spacer sequence: a consensus. J. Eukaryot. Microbiol. 56:34–38 [DOI] [PubMed] [Google Scholar]
- 18. Sulaiman I. M., et al. 2004. Molecular characterization of Enterocytozoon bieneusi in cattle indicates that only some isolates have zoonotic potential. Parasitol. Res. 92:328–334 [DOI] [PubMed] [Google Scholar]
- 19. Tamura K., Dudley J., Nei M., Kumar S. 2007. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24:1596–1599 [DOI] [PubMed] [Google Scholar]