Abstract
We evaluated two commercial F1 antigen capture-based immunochromatographic dipsticks, Yersinia Pestis (F1) Smart II and Plague BioThreat Alert test strips, in detecting plague bacilli by using whole-blood samples from mice experimentally infected with Yersinia pestis CO92. To assess the specificities of these dipsticks, an in-frame F1-deficient mutant of CO92 (Δcaf) was generated by homologous recombination and used as a negative control. Based on genetic, antigenic/immunologic, and electron microscopic analyses, the Δcaf mutant was devoid of a capsule. The growth rate of the Δcaf mutant generally was similar to that of the wild-type (WT) bacterium at both 26 and 37°C, although the mutant's growth dropped slightly during the late phase at 37°C. The Δcaf mutant was as virulent as WT CO92 in the pneumonic plague mouse model; however, it was attenuated in developing bubonic plague. Both dipsticks had similar sensitivities, requiring a minimum of 0.5 μg/ml of purified F1 antigen or 1 × 105 to 5 × 105 CFU/ml of WT CO92 for positive results, while the blood samples were negative for up to 1 × 108 CFU/ml of the Δcaf mutant. Our studies demonstrated the diagnostic potential of two plague dipsticks in detecting capsular-positive strains of Y. pestis in bubonic and pneumonic plague.
INTRODUCTION
Yersinia pestis is the causative agent of plague, occurring primarily in rodents and their fleas. The organism is transmitted to humans mainly through infected flea bites and close contact with infected animals. The three major plague pandemics resulted in more than 200 million deaths worldwide (13). Plague still is prevalent in many countries of the world today and is categorized as a reemerging human disease by the World Health Organization (10, 48).
There are three clinical forms of plague: bubonic, primary septicemic, and pneumonic. In bubonic plague, the lymph nodes in the region of the flea bite become swollen (the bubo) because of bacterial invasion and multiplication. If untreated, the disease has a mortality rate of 50% (37). In primary septicemic plague, the infected flea bites lead directly to the entry of the organism into the bloodstream without the development of a bubo, which is virtually always fatal due to poor diagnosis and the rapid progression of the disease. Pneumonic plague typically develops as a complication of bubonic plague, with the spread of Y. pestis to the lungs (37). The production of purulent sputum in the infected patient (or animal) can lead to highly contagious aerosol droplet transmission, where the disease can be spread from person to person. This form of the disease, termed primary pneumonic plague, is almost always fatal if antibiotic treatment is not initiated within 24 h of the onset of symptoms (9, 35).
The high fatality rate associated with pneumonic plague, the worldwide distribution of the organism, the ease of aerosol dissemination, and the potential use of the organism as a biological weapon have raised concerns and necessitated the development of rapid plague diagnostic tools (21). The prompt diagnosis of this fulminant disease is of key importance in reducing the mortality rate and effective control of its spread during plague epidemics and/or under a bioterrorist attack (9).
Traditionally, the clinical diagnosis of plague relies on the isolation of the organism from biological specimens and seroconversion, i.e., the development of F1-specific antibodies to Y. pestis (51). However, the slow growth of the plague bacilli and the time necessary for F1 antibodies to appear in serum at significant titers (usually 1 week) limit the early detection of the disease, because most human fatalities occur within a week after infection (51).
To circumvent such time constraints, techniques such as immunofluorescence (IF), enzyme-linked immunosorbent assay (ELISA), and immunochromatographic dipsticks have been developed recently to rapidly detect the F1 antigen in biological samples to expedite the diagnosis of plague in patients (8–10, 32, 44, 46, 51). The F1 antigen (Caf or fraction 1) is a fimbrial protein that accumulates on the bacterial surface to form an amorphous capsule and is unique to Y. pestis (38). Thus, the capsule represents a high-molecular-weight polymer consisting of linear fibers of a single protein subunit, Caf1, assembled on the surface of the bacterial cell (54). The assembly of such fibrillar organelles is accomplished by a highly conserved periplasmic chaperone/usher pathway in which caf1-encoded F1 fimbrial subunits are translocated from bacterial cytoplasm to periplasm, where they interact with Caf1M chaperone and dimerize prior to export onto the bacterial surface by the outer-membrane usher protein Caf1A (28, 55). The further addition of Caf1 dimers results in capsule formation, and caf1 is one of the most highly expressed genes during infection in mammals (38). The organism is surrounded by the F1 capsule in vivo, and free F1 antigen can be detected in tissues after being shed from the surface of bacteria (8, 9, 12, 36, 38, 39). Indeed, by using the above-mentioned technologies, the presence of the F1 antigen in patient specimens (i.e., blood, bubo, sputum, and urine) has been reported in several studies (3, 4, 8–10), and the methods were deemed sensitive and specific (8–10). However, information is lacking or inadequate regarding the application of such technologies among rodents, the natural reservoir of plague. Further, the sensitivity of such techniques in detecting Y. pestis strains, which either have low levels of F1 antigen or are devoid of it, has not been evaluated. Indeed, such strains exist in nature and have virulence similar to that of the F1-positive Y. pestis strains (18, 38, 49, 50, 53).
In this study, two F1 antigen detection kits, the Yersinia Pestis (F1) Smart II (New Horizons Diagnostics, Columbia, MD) and Plague BioThreat Alert test strips (Tetracore, Inc., Rockville, MD), were investigated. These kits are commercially available in the immunochromatographic dipstick format for easy and rapid application. As an appropriate negative control, we first generated and characterized an F1 antigen-deficient mutant (Δcaf) of wild-type (WT) Y. pestis CO92. Subsequently, mice were intranasally (i.n.) or subcutaneously (s.c.) challenged with either the WT or its Δcaf mutant to evoke pneumonic and bubonic plague, respectively. The mouse whole-blood samples were analyzed after different infection time points with the dipsticks for the presence of F1 antigen, and the results were compared with the bacterial loads in the corresponding blood samples. We also used purified F1 antigen, a commercially available F1-LcrV fusion protein (Biodefense and Emerging Infections Research Resources Repository [BEI Resources], Manassas, VA), as well as pure Y. pestis cultures to determine the detection limit of the dipsticks in in vitro assays. Our goal was to provide proof of concept with regard to the possible use of such dipsticks in monitoring plague bacilli among field rodents during epidemiological studies and/or under plague surveillance programs.
MATERIALS AND METHODS
Bacterial strains and reagents.
The WT Y. pestis CO92 strain is highly virulent and was first isolated in 1992 from a deceased pneumonic plague patient (14, 26, 40). It was acquired from the Centers for Disease Control and Prevention (CDC), Atlanta, GA. Y. pseudotuberculosis YPIII was purchased from the American Type Culture Collection (ATCC), Manassas, VA, and also was used as a negative control in this study in addition to the Δcaf mutant. Heart infusion broth (HIB) medium and plates (Difco, Voigt Global Distribution Inc., Lawrence, KS) were used to grow Y. pestis and Y. pseudotuberculosis YPIII either at 26 to 28°C or at 37°C with 2.5 mM calcium chloride. We used Luria-Bertani (LB) medium for the growth of other bacterial cultures (e.g., recombinant Escherichia coli) at 37°C with shaking (180 rpm). Molecular reagents, such as restriction endonucleases and T4 DNA ligase, were obtained from Promega (Madison, WI). The Advantage cDNA PCR kit was purchased from Clontech (Palo Alto, CA). Digested plasmid DNA or DNA fragments from agarose gels were purified by using QIAquick kits (Qiagen, Inc., Valencia, CA). All experiments involving live Y. pestis were conducted in approved, restricted-entry biosafety level 2 (BSL-2) or BSL-3 laboratory or in our animal biosafety level 3 (ABSL-3) facility located in the Galveston National Laboratory.
Generation and characterization of the caf1 mutant (Δcaf) of Y. pestis CO92.
The Δcaf mutant of CO92 was generated with CDC approval by using the gene deletion protocol adapted from the Lambda Red recombinase mutagenesis system, in which the target genes first were replaced with an antibiotic resistance marker located between the FRTs (flippase recombination sites) and subsequently removed with the FLP recombinase (11). The resulting deletion of 1,176 bp included a part of the caf1A and most of the caf1 gene encoding the usher protein and F1 subunit of the capsule, respectively. The mutagenesis was performed by using primers containing a prolonged 85-bp homology region to target the gene to facilitate recombination. The following primers were used: caf1A_lng_F, GCTAGGTAATAACTCAATAAATTCAAATTACCAAATGACATCAGATTCTCATGGTAACACTACCCATGAGGTAGGTGTGTACGGTGTGTAGGCTGGAGCTGCTTCG; and caf1_lng_R, CGCCTTTGGAACCAATTGAGCGAACAAAGAAATCCTGGCTGCCCGTAGCCAAGACGACGTCATCCCCCACAAGGTTCTCACCGTTATGGGAATTAGCCATGGTCCAT. The size of the deletion was confirmed by PCR by using primers flanking the removed region (caf1A_F, TCCTCTCAGTCGCTGGCTAGGT; caf1_R, CTGCTGCAAGTTTACCGCCTTT), followed by the sequencing of the corresponding fragments. All helper plasmids providing Lambda Red and FLP recombinases were cured from the constructed mutants as recommended elsewhere (11).
In addition, we analyzed the generated Δcaf mutant for the correct phenotype by performing genetic, antigenic/immunological, and electron microscopic testing.
(i) PCR analysis.
A PCR assay was performed to verify the deletion within the coding region of the caf1 gene. The genomic DNA from both WT CO92 and its Δcaf mutant was isolated and used as a template for PCR. We used specific primers (cafF, 5′ GCTAGCAAAAAAATCAGTTCCGTTATCGCC 3′; cafR, 5′ AGATCTGGATTATTGGTTAGATACGGTTACGG 3′) with the following amplification program: 94°C for 2 min (denaturation), followed by 30 cycles of 94°C for 1 min and 68°C for 3 min. The final extension was performed at 68°C for 7 min. The PCR-amplified products were resolved on a 0.7% agarose gel, visualized by ethidium bromide staining, and confirmed by DNA sequence analysis.
(ii) Antigenic/immunological analyses.
Western blot analysis, flow cytometry (FC), IF staining, and transmission electron microscopic (TEM) studies were performed to examine the production of the F1 antigen by the Y. pestis strains. The WT and Δcaf mutant of CO92 were grown in 3 ml of HIB medium at 37°C overnight with constant shaking (180 rpm). The bacilli were harvested by centrifugation (8,500 × g) and washed with phosphate-buffered saline (PBS). For Western blot analysis, the bacterial cells were lysed in SDS-PAGE sample buffer and probed with the primary anti-F1 antigen antibodies (generated in the laboratory) at a dilution of 1:2,500, followed by horseradish peroxidase (HRP)-labeled, goat anti-mouse secondary antibodies (Southern Biotech, Birmingham, Alabama) at a dilution of 1:20,000. The blots were developed by using a SuperSignal West Pico chemiluminescent substrate (Pierce, Rockford, IL) (40). Similarly, the production of V antigen (LcrV), a component of the type III secretion system (T3SS) (19), in the WT and Δcaf mutant of CO92 was analyzed by using antibodies to LcrV (generated in the laboratory) at a dilution of 1:2,500.
For the fluorescent analysis of F1 production by IF and FC techniques, the harvested bacteria were fixed with 4% formaldehyde (Polysciences Inc., Warrington, PA). Following fixation, bacteria (106 CFU/sample) were washed with PBS and incubated with a primary antibody to F1 antigen (Abcam, Cambridge, MA) or an isotype-matched irrelevant antibody (Sigma-Aldrich, St. Louis, MO) as a negative control. Cells were washed with PBS and further labeled with a secondary antibody (goat anti-mouse IgG1) conjugated to Alexa Fluor 488. Lastly, cells were washed and resuspended in 400 μl of 2% ultrapure formaldehyde (Polysciences Inc.) in PBS before analysis. To perform IF, a 10-μl aliquot of labeled cells was adsorbed onto a poly-l-lysine-coated slide. The slides were dried and mounted with a coverslip and viewed under a fluorescent microscope.
For FC, we acquired a total of 30,000 cells on a FACS Canto (BD Biosciences, San Diego, CA), and compensation was performed using FACS DIVA software (BD Biosciences). Analysis was conducted using FCS Express version 3 (De Novo Software).
(iii) TEM.
For TEM, the cultures were fixed in a mixture of 2.5% formaldehyde and 0.1% glutaraldehyde in 0.05 M cacodylate buffer, pH 7.3, to which 0.03% trinitrophenol and 0.03% CaCl2 were added, postfixed in 1% OsO4 in 0.1 M cacodylate buffer, stained en bloc with 2% aqueous uranyl acetate, dehydrated in ethanol, and embedded in Poly/Bed 812 epoxy resin (Polysciences). Ultrathin sections were cut on a Reichert-Leica Ultracut S ultramicrotome, stained with lead citrate, and examined in a Philips 201 electron microscope at 60 kV.
(iv) Growth curve studies.
An aliquot (10 to 20 μl) of each of the Y. pestis cultures (i.e., WT and Δcaf strains or Y. pseudotuberculosis) from −70°C stocks were streaked onto 5% sheep blood agar (SBA) plates (Teknova, Hollister, CA) and incubated at 28°C for 48 h. A single isolated colony from the blood agar plate of each culture then was inoculated into 5 to 10 ml HIB medium in a 50-ml HEPA filter TOP conical tube and grown at 28°C with constant shaking (180 rpm) overnight (12 to 16 h). Subsequently, the cultures grown overnight were diluted at a ratio of 1:40 with 100 ml of fresh HIB medium into a 500-ml HEPA filter TOP polycarbonate Erlenmeyer culture flask (Triforest Labware, Irvine, CA). The optical density at 600 nm (OD600) of the culture dilutions then was measured with the spectrophotometer (Jenway, Staffordshire, United Kingdom). The dilution ratios were adjusted to ensure that the ODs were around 0.05 to 0.1 at this initial time point. The diluted Y. pestis or Y. pseudotuberculosis cultures in 500-ml flasks then were grown at either 26 or 37°C with constant shaking. Samples from each flask were taken at 1-h intervals until the cultures reached their plateau phase. The OD600s at different time points were recorded and the CFU determined by plating, as we described previously (40).
Animal studies.
Female 5- to 6-week-old BALB/c mice were purchased from Jackson Laboratories (Bar Harbor, ME) and infected in the ABSL-3 facility either by the i.n. or s.c. route under an approved UTMB IACUC protocol to mimic pneumonic and bubonic plague, respectively (40). The animals were infected with either WT CO92 or its Δcaf mutant and monitored for up to 30 to 35 days for mortality. We used 30 mice in each group, and 10 animals from each group were assessed for morbidity or mortality during the duration of each experiment. Blood samples were collected from mice at 24, 48, 72, and 96 h postinfection, and 5 mice were used at each indicated time point.
Detection of the F1 antigen.
Two commercially available plague detection kits, the Yersinia Pestis (F1) Smart II (New Horizons Diagnostics) and Plague BioThreat Alert test strips (Tetracore, Inc.), were used to detect the F1 antigen from various sources. We spiked blood samples from naïve animals with a known number of bacteria or concentration of purified F1 or F1-LcrV fusion protein antigen (obtained from BEI Resources). Once the samples were spiked with bacteria, we used WT CO92 and its Δcaf mutant or Y. pseudotuberculosis (also as a negative control) grown at 37°C at log incremental doses of 1 × 103 to 1 × 108 CFU/ml. The purified F1 antigen or the F1-LcrV fusion protein, either spiked at doses of 0.0625 to 21 μg/ml in blood samples or diluted in PBS, was used to determine the detection limits of these kits.
Alternatively, we obtained infected blood samples at various time points (24, 48, 72, and 96 h) from mice that were challenged via the s.c. or the i.n. route with the indicated doses of WT CO92 or its Δcaf mutant. Subsequently, an aliquot (100 μl) of the blood sample was applied to the F1 antigen detection dipsticks. These detection kit assays are lateral-flow based, and the results were interpreted according to the manufacturer's instructions. Each blood sample was analyzed with both dipsticks at least in duplicate, and, where appropriate, the corresponding blood samples were serially diluted in PBS and cultured on SBA plates to determine the bacterial load (40).
Statistical analysis.
The animal mortality data were evaluated by using the Kaplan-Meier survival estimate between WT- and Δcaf mutant-infected mouse groups, and P ≤ 0.05 was considered significant.
RESULTS AND DISCUSSION
The Δcaf mutant of Y. pestis CO92 was devoid of F1 antigen production.
As shown in Fig. 1, the 513-bp caf1 gene and its encoded protein F1 antigen (20 kDa) could be detected only in the WT bacterium (Fig. 1A to C, lane 2), and it was missing from the Δcaf mutant (Fig. 1A to C, lane 1) based on PCR (Fig. 1A) and Western blot (Fig. 1B) analysis data, respectively. Figure 1C is a Western blot showing that the Δcaf mutant was not defective in the synthesis of the LcrV antigen, as it produced amounts of it similar to that of the WT bacterium (Fig. 1A to C, lanes 1 and 2; also see below). The surface-localized F1 antigen was visualized by IF on the WT bacterium but not on the Δcaf mutant (Fig. 1D). These data matched those derived from the fluorescent-activated cell sorter (FACS) analysis in which only the WT bacterium producing the F1 antigen exhibited a strong fluorescent signal following labeling with antibody to F1 and detection with a fluorescence-labeled secondary antibody (Fig. 1E). In contrast, the signal observed following the labeling of the Δcaf mutant with antibody to F1 and fluorescence-labeled secondary antibody was not greater than the background fluorescence observed following the labeling of both strains with an isotype-matched control antibody.
Fig. 1.
Genetic and antigenic/immunologic analyses of the Δcaf mutant of Y. pestis CO92. (A) Genomic DNA from both the WT and its Δcaf mutant was isolated, and the specific primers were used to PCR amplify the coding region of the caf1 gene. (B and C) Western blot analysis with specific antibodies to the F1 antigen (B) or the LcrV antigen (C) were used for examining the production of corresponding proteins in both the WT and its Δcaf mutant. (D) Both the WT (left) and its Δcaf mutant (right) were subjected to staining with specific anti-F1 antibody, followed by detection with Alexa-488-labeled (green) secondary antibody using a fluorescent microscope. (E) The production of F1 by the WT (green line) and the Δcaf mutant (black line) was further evaluated by flow cytometry using antibody specific to F1 or an isotype-matched control (red line), followed by detection with fluorescence-labeled (Alexa-488) secondary antibody.
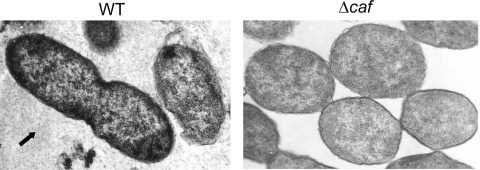
These results indicated that the generated Δcaf mutant was devoid of F1, which we further confirmed by performing TEM. As can be noted from Fig. 2, the Δcaf mutant lacked the capsule, as there was no capsular material surrounding the bacteria, while a significant amount of the capsular material was noted around and between the WT bacteria when grown at 37°C (Fig. 2, arrow).
Fig. 2.
Electron microscopic analysis showing the presence of capsular material in Y. pestis cultures. Both WT CO92 and its Δcaf mutant were grown on SBA plates at 37°C for 48 h. The bacilli then were fixed and examined with a Philips 201 electron microscope at 60 kV. The arrow indicates the shedding of the capsular material for the WT CO92, but no such capsular material was seen surrounding the Δcaf mutant.
Production of LcrV antigen by WT Y. pestis CO92 and its Δcaf mutant.
The type 3 secretion system (T3SS) plays an important role in the pathogenesis of yersiniae infections (24). Therefore, we examined the expression of the gene encoding the LcrV antigen in WT CO92 and its Δcaf mutant with the goal of evaluating whether the T3SS was intact in the mutant strain. Our Western blot analysis data with LcrV-specific antibodies (Fig. 1C) indeed revealed similar amounts of V antigen produced in both the WT (lane 2) and its Δcaf mutant (lane 1). These data indicated that the T3SS was not affected during the mutagenesis process used to delete the caf gene.
Growth curves for WT Y. pestis CO92 and its Δcaf mutant.
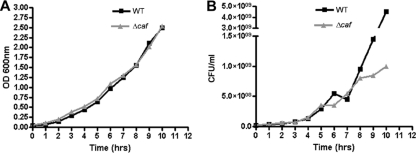
We then examined the growth of WT CO92 and its Δcaf mutant at both 37 and 26°C. No significant differences in growth patterns were observed between WT CO92 and its Δcaf mutant at 37°C based on optical density measurements (Fig. 3A); however, the viability of the Δcaf mutant was reduced at later growth stages (Fig. 3B). The Δcaf mutant displayed growth and viability characteristics similar to those of the WT bacterium at 26°C (data not shown). As the F1 antigen was produced only at 37°C (38), it is plausible that the F1 capsular antigen protected the WT strain from accumulated metabolic wastes in the culture medium. The F1 antigen would not have been produced at 26°C, and consequently no difference in viability would have been observed, as was indeed found to be the case (data not shown).
Fig. 3.
Growth curves of Y. pestis CO92 WT and its Δcaf mutant. The bacilli were grown at 37°C in HIB medium with 2.5 mM CaCl2. Samples were taken at 1-h intervals until the cultures reached their saturation phases. The OD600s of the samples at different time points were recorded (A), and their actual counts (in CFU) (B) were determined by plating (40).
Virulence of the Δcaf mutant of Y. pestis CO92 in bubonic and pneumonic plague mouse models.
Several studies have shown that the capsule acts in concert with the T3SS in making Y. pestis highly resistant to phagocytosis (6, 12, 17, 52). However, contradictory results have been reported pertaining to the overall attenuated virulence of naturally and/or experimentally derived caf mutants compared to that of their corresponding parental strains in animal models (5, 12, 15, 16, 18, 47, 50, 53). Therefore, to address this discrepancy, we tested WT CO92 and its Δcaf mutant for virulence in bubonic and pneumonic plague mouse models.
As shown in Fig. 4, all of the animals infected with either the WT or its Δcaf mutant died after subcutaneous (Fig. 4A) and intranasal (Fig. 4B) challenge. The mice were infected at a dose of 7.5 × 104 to 9.0 × 104 50% lethal doses (LD50) (10 bacteria represent 1 LD50 by the s.c. route) or 17.5 LD50 (100 CFU represents 1 LD50 by the i.n. route), respectively. We initially chose high doses of bacteria for infection studies so that the animals would die by day 4 due to bacteremia to better gauge the sensitivity of the dipstick test. Interestingly, an increase in mean time to death with the Δcaf mutant-infected mice (12 days) was observed compared to that seen in animals infected with the WT bacterium (5 days) when challenged by the s.c. route (Fig. 4A). When the challenge dose of WT CO92 and its Δcaf mutant was reduced to 100 to 1,000 LD50, a protective effect of the capsule (both in terms of mean time to death and survival) was clearly seen in the bubonic plague mouse model (Fig. 4C). However, virtually identical death curves were observed when animals were infected via the i.n. route, with all of the mice dying within 4 to 5 days (Fig. 4B). Even lowering the infection dose to 5 LD50 showed no statistically significant differences in mortality between the WT and its Δcaf mutant by the i.n. route (data not shown). These data suggested that the attenuation of the Δcaf mutant occurred only in the bubonic plague model.
Fig. 4.
Virulence potential of the Δcaf mutant of Y. pestis CO92 in both bubonic and pneumonic plague mouse models. The BALB/c mice were challenged subcutaneously (A and C) or intranasally (B) with the WT or Δcaf mutant of CO92 at the indicated doses. Mice were assessed for mortality during the duration of each experiment, and their percentages of survival were plotted. The Kaplan-Meier survival estimate was used to statistically analyze survival rates between WT- and Δcaf-infected mouse groups, and the corresponding P values are presented.
Earlier studies showed that some genetically undefined F1-negative strains were less virulent (5, 15, 18, 47, 50, 53), while several reports using genetically defined, caf-negative mutant strains uniformly demonstrated that the lack of F1 antigen had no effect on virulence in mouse and guinea pig models of bubonic plague or in the African Green monkey model of pneumonic plague (12, 16, 18). These contradictory data could be due to some other virulence factors, in addition to the F1 antigen, that were affected in the genetically undefined F1-negative strains.
In a recent study, Sebbane et al. reported that the caf-negative Y. pestis 195/P mutant was not impaired in either flea colonization or virulence in mice after intradermal inoculation (38). However, the absence of the caf operon decreased bubonic plague incidence after a flea bite, which eventually led to a reduction in the transmissibility and the potential of plague for epidemic spread (38). They suggested that transmission by flea bite represents a more sensitive way to detect small differences in the LD50, as the 50% lethal doses of Y. pestis for subcutaneous and intradermal challenges are extremely low to only a few bacilli, which is technically difficult to measure with confidence during an experimental challenge (38).
Although the exact role of F1 in Y. pestis infections is not fully understood, it has been reported to have an antiphagocytosis function (5–7, 17, 18, 33, 52). Further, the F1 antigen may have immunomodulatory effects, as interactions between F1 dimers bound to its chaperone Caf1M and interleukin-1 (IL-1) receptors on epithelial cells and macrophages have been demonstrated (1, 2, 56). Further, proinflammatory cytokine induction due to macrophage activation by F1 also has been reported (41–43), although these results are contradictory to other published reports (23, 25). The possibility exists that the F1 capsule is shielding other Y. pestis surface antigens from the immune system, and its role in depleting circulating F1 antibodies also may contribute to the virulence of Y. pestis (27, 38).
Nonetheless, it is clear that the ability of F1-negative strains to cause disease in both laboratory mice and humans is compatible with that of the F1-positive strains, and therefore there is a need to develop diagnostic tools capable of detecting both F1-positive and -negative strains of Y. pestis in biological samples in epidemiological settings. Currently, assay tools are available only for detecting F1 antigen, and this is the basis of the study described below.
Evaluation of F1 antigen capture-based dipsticks.
Two commercially available plague detection kits (Smart II and BioThreat Alert test strips) were assessed for their abilities to detect F1 antigen in various samples.
As shown in Table 1, the minimum amount of purified F1 and LcrV-F1 fusion protein required to demonstrate a positive reaction in either PBS or naïve blood following the use of either of the dipsticks was 0.5 μg/ml. It is worth mentioning that although the detection limit for both F1 and F1-LcrV fusion protein was the same, the actual amount of F1 antigen present in the fusion protein was lower. The increased sensitivity in detecting F1 antigen in the fusion protein could have been related to the better presentation of the F1 epitopes and interaction with the antibodies on the dipsticks.
Table 1.
Minimum detection limit of dipsticks in various samples tested
| Dipstick name | Minimum detection limit for: |
|||
|---|---|---|---|---|
| Infected mouse whole blood (CFU/ml) | PBS spiked with purified F1 antigen (μg/ml) | PBS spiked with LcrV-F1 fusion protein (μg/ml) | PBS or normal mouse blood spiked with WT Y. pestis CO92 grown in vitro at 37°C (CFU/ml) | |
| Plague BioThreat Alert test strips | 1 × 105–5 × 105 | 0.5 | 0.5 | 1 × 105–5 × 105 |
| Yersinia Pestis (F1) Smart II | 1 × 105–5 × 105 | 0.5 | 0.5 | 1 × 105–5 × 105 |
When PBS or blood samples were spiked with various doses of WT CO92, the limit of detection for both of the kits was 1 × 105 to 5 × 105 CFU/ml (Table 1). Even at the dose of 1 × 108 CFU/ml, the Δcaf mutant of Y. pestis or WT Y. pseudotuberculosis (nonencapsulated) did not show any reaction, indicating the specificity of the kits for the F1 antigen. Although the limits of detection were similar for both kits, we observed reactions to be quicker (resulting in stronger bands) with the Smart II strip than with the BioThreat Alert test strip (Fig. 5). These data indicated that the mouse blood did not interfere with the dipstick reactions.
Fig. 5.
Immunochromatographic reactions of dipsticks. The plague detection abilities of two dipsticks, Yersinia Pestis (F1) Smart II and Plague BioThreat Alert test strips, were evaluated in various biological samples. The diluted (1:10) whole-blood samples from naïve mice spiked with 1 ×107 CFU/ml of in vitro-grown (37°C) WT Y. pestis CO92 (A) and the whole-blood samples from the WT Y. pestis CO92-infected mice at 72 h postinfection and containing 1.05 × 109 CFU/ml of the bacilli (B) were employed. Each testing had the same reaction time of 2 min. The solid and open arrows indicated the control and sample positive reactions, respectively.
We initially chose the s.c. infection model (7.5 × 105 CFU [75,000 LD50] of WT CO92) (Fig. 4A) to demonstrate the sensitivity of the kits in detecting the minimal number of bacteria in the blood. Between 48 and 72 h (more often at 72 h), we could detect more than 1 × 107 CFU/ml of plague bacilli in the blood, and both kits exhibited a positive reaction. Although all of the animals died by day 4, 60% of the mice were bacteremic between 48 and 72 h. These data indicate that even without bacteremia, the animals could succumb to infection due to serious damage to the regional lymph nodes where the organisms first enter during bubonic plague. Alternatively, it is plausible that the organisms enter different body organs via the lymphatics, bypassing the blood. With the Δcaf mutant, only 25% of the animals were bacteremic between 48 and 72 h, with 1 × 104 to 5.5 × 104 CFU/ml detected in the blood, and as expected, both kits were negative.
We then examined the number of bacteria in blood after the i.n. infection of mice with WT CO92 and its Δcaf mutant (1,750 CFU [17.5 LD50]) (Fig. 4B). By 72 h, 70% of the animals were bacteremic, with a bacterial load as high as 1 × 109 CFU/ml. Likewise by 72 h, 75% of mice infected with the Δcaf mutant were bacteremic, with the highest bacterial load of 2.2 × 108 CFU/ml exhibiting a negative dipstick test. The minimal number of WT CO92 that led to a positive dipstick reaction was in the range of 1 × 105 to 5 × 105 CFU/ml. Thus, the sensitivity of the dipstick tests were very similar when bacteria were grown in vitro and spiked with PBS or naïve mouse blood or when directly isolated from blood samples of infected mice (Table 1). These data indicate that a similar amount of F1 antigen was displayed on the bacterial surface when grown in vitro and in vivo, and the mouse blood did not interfere with the dipstick's reactions.
We tested these dipsticks with up to 190 μg/ml of purified F1 antigen and 1 × 108 CFU/ml of WT CO92 spiked with PBS or naïve mouse whole blood; they all showed positive results without a prozone effect. The absence of prozone phenomenon is essential to avoid false-negative results (9). However, we did notice that mouse blood samples obtained late in the course of infection became more viscous, and some of them showed false-negative results. This might be due to the blockage of either the capillary flow or the antigen-antibody interactions on the dipsticks. Consequently, we diluted all of the blood samples 1:5 or 1:10 for optimal results.
Studies from other investigators reported much better sensitivity with methods similar to those we used here for the detection of the F1 antigen. For example, the F1 antigen could be detected at a concentration of 0.5 ng/ml, while 3 × 103 CFU/ml of plague bacilli gave a positive reaction (9, 10, 13, 46). Specifically, a recent study with the same Plague BioThreat Alert test strips from Tetracore reported a detection limit of 37 ng/ml for the purified F1 antigen and 6 × 103 CFU/ml for the bacteria (46). These numbers are much lower than what we have reported in Table 1. These sensitivity differences could be related to the purity of the F1 antigen used and the different strains of Y. pestis used, as the amount of the F1 antigen on the bacterial surface may vary among various Y. pestis strains. On the other hand, the different affinities of anti-F1 antibodies coated on the strips tested may contribute to the difference as well. Our studies are the first to use these kits with a highly virulent CO92 Y. pestis strain and its authentic F1 mutant.
The presence of the F1 antigen in plague patient sera has been reported in several studies and ranged from as low as 4 ng/ml to as high as 50,000 ng/ml (8, 46, 51). However, antigenemia was not present in all patients with acute plague, and only 20% of their sera were positive for the Fl antigen (51). In our study, regardless of the possibility of shedding F1 antigen from the bacterial surface, the positive results with both of the dipsticks always corresponded to the development of bacteremia in both bubonic and pneumonic plague mouse models. The lack of uniformity in the presence of antigenemia or bacteremia during infection may be the limiting factor for using F1 antigen detection-based methods in the diagnosis of plague. However, this issue can be circumvented by using other biological specimens, such as bubo aspirates, sputum, and urine, instead of blood (9, 10, 44, 46). We chose blood specimens in our study because blood is the most accessible and frequently assayed body fluid, and the most dangerous clinical presentations of plague are primarily from pneumonic and septicemic cases in which no bubos are present (44).
The data generated with the Δcaf mutant indicated that these dipsticks were very specific for the F1 antigen, and there are no cross-reactivities between the F1 antigen and other Y. pestis components. However, with infected blood samples, one of the dipsticks gave, on occasion, false-positive reactions after 10 min of incubation. Although the instructions for these kits indicated that the reactions can be recorded between 15 and 30 min, we believe chances of error increase after 10 min of incubation. Out of a total of 58 infected blood samples tested, we noted that 5/58 (8.6%) showed false-positive results by using the Smart II strips (New Horizons Diagnostics), while no false-positive results were obtained by employing the Plague BioThreat Alert test strips (Tetracore, Inc.).
Another caveat of using these dipsticks in the diagnosis of plague is that they cannot detect F1-negative Y. pestis strains. Such isolates do exist in nature and have virulence similar to that of the F1-positive strains (18, 38, 49, 50, 53), which we also demonstrated in this study. Studies have implicated the emergence of F1-negative strains during climatic conditions that favor a high flea burden, and such capsular-negative strains were isolated from fatal plague patients, thus emphasizing the importance of developing diagnostic tools for Y. pestis F1-negative strains (38, 53).
In addition to immunochromatographic dipsticks, ELISA, IF, and PCR techniques have been used for the detection of the F1 antigen or its gene (9, 10, 20, 22, 29–32, 34, 44–46, 51). Although ELISA is specific, it lacks sensitivity in all countries in which plague is endemic (9), and PCR generally is less sensitive than ELISA (34). The use of IF is somewhat limited because it detects only the bacterial surface-bound antigen, hence the quantification of the F1 antigen is difficult (51). More importantly, all of these techniques require sophisticated equipment and trained personnel. Therefore, most of these methods are not applicable in the remote rural areas where most of the world's 3,000 to 4,000 annually reported plague cases occur (13, 48). In this regard, the dipstick method could be the most viable option.
The rapid diagnosis of a fulminating disease with a strong epidemic potential, such as plague, improves the chances for the recovery of the patient through the early use of appropriate therapy and for the prevention of new cases by the timely application of effective control measures (9, 51). This is particularly important in cases of bioterrorist attacks with plague as a biological weapon (9, 21). Unfortunately, the current plague diagnostic tools do not quite meet these requirements, and therefore the development of simpler, quicker, and more specific methods with the capability of detecting both F1-negative and -positive Y. pestis strains is crucial in dealing with the challenges we face today.
ACKNOWLEDGMENTS
This work was supported by NIH/NIAID contract N01 AI40097 and grant AI064389. Our studies were conducted in the Galveston National Laboratory, and we acknowledge the UC7 grant awarded by NIH/NIAID.
We thank M. Susman for editing the manuscript.
Footnotes
Published ahead of print on 2 March 2011.
REFERENCES
- 1. Abramov V. M., et al. 2002. Structural and functional properties of Yersinia pestis Caf1 capsular antigen and their possible role in fulminant development of primary pneumonic plague. J. Proteome Res. 1:307–315 [DOI] [PubMed] [Google Scholar]
- 2. Abramov V. M., et al. 2001. Structural and functional similarity between Yersinia pestis capsular protein Caf1 and human interleukin-1 beta. Biochemistry 40:6076–6084 [DOI] [PubMed] [Google Scholar]
- 3. Baker E. E., Sommer H., Foster L. E., Meyer E., Meyer K. F. 1952. Studies on immunization against plague. I. The isolation and characterization of the soluble antigen of Pasteurella pestis. J. Immunol. 68:131–145 [PubMed] [Google Scholar]
- 4. Brubaker R. R. 1970. Interconversion of purine mononucleotides in Pasteurella pestis. Infect. Immun. 1:446–454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Burrows T. W. 1957. Virulence of Pasteurella pestis. Nature 179:1246–1247 [DOI] [PubMed] [Google Scholar]
- 6. Burrows T. W., Bacon G. A. 1956. The basis of virulence in Pasteurella pestis: the development of resistance to phagocytosis in vitro. Br. J. Exp. Pathol. 37:286–299 [PMC free article] [PubMed] [Google Scholar]
- 7. Cavanaugh D. C., Randall R. 1959. The role of multiplication of Pasteurella pestis in mononuclear phagocytes in the pathogenesis of flea-borne plague. J. Immunol. 83:348–363 [PubMed] [Google Scholar]
- 8. Chanteau S., et al. 1998. F1 antigenaemia in bubonic plague patients, a marker of gravity and efficacy of therapy. Trans. R. Soc. Trop. Med. Hyg. 92:572–573 [DOI] [PubMed] [Google Scholar]
- 9. Chanteau S., et al. 2003. Development and testing of a rapid diagnostic test for bubonic and pneumonic plague. Lancet 361:211–216 [DOI] [PubMed] [Google Scholar]
- 10. Chanteau S., et al. 2000. Early diagnosis of bubonic plague using F1 antigen capture ELISA assay and rapid immunogold dipstick. Int. J. Med. Microbiol. 290:279–283 [DOI] [PubMed] [Google Scholar]
- 11. Datsenko K. A., Wanner B. L. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U. S. A. 97:6640–6645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Davis K. J., et al. 1996. Pathology of experimental pneumonic plague produced by fraction 1-positive and fraction 1-negative Yersinia pestis in African green monkeys (Cercopithecus aethiops). Arch. Pathol. Lab. Med. 120:156–163 [PubMed] [Google Scholar]
- 13. Dennis D. T., Chu M. C. 2003. A major new test for plague. Lancet 361:191–192 [DOI] [PubMed] [Google Scholar]
- 14. Doll J. M., et al. 1994. Cat-transmitted fatal pneumonic plague in a person who traveled from Colorado to Arizona. Am. J. Trop. Med. Hyg. 51:109–114 [DOI] [PubMed] [Google Scholar]
- 15. Donavan J. E., Ham D., Fukui G. M., Surgalla M. J. 1961. Role of the capsule of Pasteurella pestis in bubonic plague in the guinea pig. J. Infect. Dis. 109:154–157 [DOI] [PubMed] [Google Scholar]
- 16. Drozdov I. G., et al. 1995. Virulent non-capsulate Yersinia pestis variants constructed by insertion mutagenesis. J. Med. Microbiol. 42:264–268 [DOI] [PubMed] [Google Scholar]
- 17. Du Y., Rosqvist R., Forsberg A. 2002. Role of fraction 1 antigen of Yersinia pestis in inhibition of phagocytosis. Infect. Immun. 70:1453–1460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Friedlander A. M., et al. 1995. Relationship between virulence and immunity as revealed in recent studies of the F1 capsule of Yersinia pestis. Clin. Infect. Dis. 21(Suppl. 2):S178–S181 [DOI] [PubMed] [Google Scholar]
- 19. Hamad M. A., Nilles M. L. 2007. Roles of YopN, LcrG and LcrV in controlling Yops secretion by Yersinia pestis. Adv. Exp. Med. Biol. 603:225–234 [DOI] [PubMed] [Google Scholar]
- 20. Hinnebusch J., Schwan T. G. 1993. New method for plague surveillance using polymerase chain reaction to detect Yersinia pestis in fleas. J. Clin. Microbiol. 31:1511–1514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Inglesby T. V., et al. 2000. Plague as a biological weapon: medical and public health management. Working Group on Civilian Biodefense. JAMA 283:2281–2290 [DOI] [PubMed] [Google Scholar]
- 22. Jonas D., et al. 2000. Comparative evaluation of three different genotyping methods for investigation of nosocomial outbreaks of Legionnaires' disease in hospitals. J. Clin. Microbiol. 38:2284–2291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kingston R., et al. 2007. The fraction 1 and V protein antigens of Yersinia pestis activate dendritic cells to induce primary T cell responses. Clin. Exp. Immunol. 149:561–569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Koornhof H. J., Smego R. A., Jr., Nicol M. 1999. Yersiniosis. II. The pathogenesis of Yersinia infections. Eur. J. Clin. Microbiol. Infect. Dis. 18:87–112 [DOI] [PubMed] [Google Scholar]
- 25. Krakauer T., Heath D. 1998. Lack of IL-1 receptor antagonistic activity of the capsular F1 antigen of Yersinia pestis. Immunol. Lett. 60:137–142 [DOI] [PubMed] [Google Scholar]
- 26. Lathem W. W., Price P. A., Miller V. L., Goldman W. E. 2007. A plasminogen-activating protease specifically controls the development of primary pneumonic plague. Science 315:509–513 [DOI] [PubMed] [Google Scholar]
- 27. MacIntyre S., Knight S. D., Fooks L. J. 2004. Structure, assembly and applications of the polymeric F1 antigen of Yersinia pestis, p. 363–408 In Hinnebusch E. C. B. J. (ed.), Yersinia molecular and cellular biology. Horizon Sciences, Norfolk, United Kingdom [Google Scholar]
- 28. MacIntyre S., et al. 2001. An extended hydrophobic interactive surface of Yersinia pestis Caf1M chaperone is essential for subunit binding and F1 capsule assembly. Mol. Microbiol. 39:12–25 [DOI] [PubMed] [Google Scholar]
- 29. Neubauer H., et al. 2000. A combination of different polymerase chain reaction (PCR) assays for the presumptive identification of Yersinia pestis. J. Vet. Med. B Infect. Dis. Vet. Public Health 47:573–580 [DOI] [PubMed] [Google Scholar]
- 30. Neubauer H., et al. 2000. Serodiagnosis of human plague by an anti-F1 capsular antigen specific IgG/IgM ELISA and immunoblot. Epidemiol. Infect. 125:593–597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Norkina O. V., et al. 1994. Development of a diagnostic test for Yersinia pestis by the polymerase chain reaction. J. Appl. Bacteriol. 76:240–245 [DOI] [PubMed] [Google Scholar]
- 32. Phillips A. P., Morris B. C., Hall D., Glenister M., Williams J. E. 1988. Identification of encapsulated and non-encapsulated Yersinia pestis by immunofluorescence tests using polyclonal and monoclonal antibodies. Epidemiol. Infect. 101:59–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Pujol C., Bliska J. B. 2005. Turning Yersinia pathogenesis outside in: subversion of macrophage function by intracellular yersiniae. Clin. Immunol. 114:216–226 [DOI] [PubMed] [Google Scholar]
- 34. Rahalison L., Vololonirina E., Ratsitorahina M., Chanteau S. 2000. Diagnosis of bubonic plague by PCR in Madagascar under field conditions. J. Clin. Microbiol. 38:260–263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Ratsitorahina M., Chanteau S., Rahalison L., Ratsifasoamanana L., Boisier P. 2000. Epidemiological and diagnostic aspects of the outbreak of pneumonic plague in Madagascar. Lancet 355:111–113 [DOI] [PubMed] [Google Scholar]
- 36. Runco L. M., Myrczek S., Bliska J. B., Thanassi D. G. 2008. Biogenesis of the fraction 1 capsule and analysis of the ultrastructure of Yersinia pestis. J. Bacteriol. 190:3381–3385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Russell P., et al. 1997. Laboratory diagnosis of plague. Br. J. Biomed. Sci. 54:231–236 [PubMed] [Google Scholar]
- 38. Sebbane F., Jarrett C., Gardner D., Long D., Hinnebusch B. J. 2009. The Yersinia pestis caf1M1A1 fimbrial capsule operon promotes transmission by flea bite in a mouse model of bubonic plague. Infect. Immun. 77:1222–1229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Sebbane F., et al. 2006. Adaptive response of Yersinia pestis to extracellular effectors of innate immunity during bubonic plague. Proc. Natl. Acad. Sci. U. S. A. 103:11766–11771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Sha J., et al. 2008. Braun lipoprotein (Lpp) contributes to virulence of yersiniae: potential role of Lpp in inducing bubonic and pneumonic plague. Infect. Immun. 76:1390–1409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Sharma R. K., Sodhi A., Batra H. V. 2005. Involvement of c-Jun N-terminal kinase in rF1 mediated activation of murine peritoneal macrophages in vitro. J. Clin. Immunol. 25:215–223 [DOI] [PubMed] [Google Scholar]
- 42. Sharma R. K., Sodhi A., Batra H. V., Tuteja U. 2005. Phosphorylation of p42/44 MAP kinase is required for rF1-induced activation of murine peritoneal macrophages. Mol. Immunol. 42:1385–1392 [DOI] [PubMed] [Google Scholar]
- 43. Sodhi A., Sharma R. K., Batra H. V., Tuteja U. 2004. Recombinant fraction 1 protein of Yersinia pestis activates murine peritoneal macrophages in vitro. Cell Immunol. 229:52–61 [DOI] [PubMed] [Google Scholar]
- 44. Splettstoesser W. D., Rahalison L., Grunow R., Neubauer H., Chanteau S. 2004. Evaluation of a standardized F1 capsular antigen capture ELISA test kit for the rapid diagnosis of plague. FEMS Immunol. Med. Microbiol. 41:149–155 [DOI] [PubMed] [Google Scholar]
- 45. Thullier P., Guglielmo V., Rajerison M., Chanteau S. 2003. Short report: serodiagnosis of plague in humans and rats using a rapid test. Am. J. Trop. Med. Hyg. 69:450–451 [PubMed] [Google Scholar]
- 46. Tomaso H., et al. 2007. Comparison of hand-held test kits, immunofluorescence microscopy, enzyme-linked immunosorbent assay, and flow cytometric analysis for rapid presumptive identification of Yersinia pestis. J. Clin. Microbiol. 45:3404–3407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Welkos S. L., Andrews G. P., Lindler L. E., Snellings N. J., Strachan S. D. 2004. Mu dI1(Ap lac) mutagenesis of Yersinia pestis plasmid pFra and identification of temperature-regulated loci associated with virulence. Plasmid 51:1–11 [DOI] [PubMed] [Google Scholar]
- 48. WHO 2000. Human plague in 1998 and 1999. Wkly. Epidemiol. Rec. 75:338–339 [PubMed] [Google Scholar]
- 49. Williams J. E., Cavanaugh D. C. 1983. Chronic infections in laboratory rodents from inoculation of nonencapsulated plague bacilli (Yersinia pestis). Experientia 39:408–409 [DOI] [PubMed] [Google Scholar]
- 50. Williams J. E., Cavanaugh D. C. 1984. Potential for rat plague from nonencapsulated variants of the plague bacillus (Yersinia pestis). Experientia 40:739–740 [DOI] [PubMed] [Google Scholar]
- 51. Williams J. E., Gentry M. K., Braden C. A., Leister F., Yolken R. H. 1984. Use of an enzyme-linked immunosorbent assay to measure antigenaemia during acute plague. Bull. World Health Organ. 62:463–466 [PMC free article] [PubMed] [Google Scholar]
- 52. Williams R. C., Jr., Gewurz H., Quie P. G. 1972. Effects of fraction I from Yersinia pestis on phagocytosis in vitro. J. Infect. Dis. 126:235–241 [DOI] [PubMed] [Google Scholar]
- 53. Winter C. C., Cherry W. B., Moody M. D. 1960. An unusual strain of Pasteurella pestis isolated from a fatal human case of plague. Bull. World Health Organ. 23:408–409 [PMC free article] [PubMed] [Google Scholar]
- 54. Zavialov A. V., et al. 2003. Structure and biogenesis of the capsular F1 antigen from Yersinia pestis: preserved folding energy drives fiber formation. Cell 113:587–596 [DOI] [PubMed] [Google Scholar]
- 55. Zavialov A. V., et al. 2002. Donor strand complementation mechanism in the biogenesis of non-pilus systems. Mol. Microbiol. 45:983–995 [DOI] [PubMed] [Google Scholar]
- 56. Zav'yalov V. P., et al. 1995. Specific high affinity binding of human interleukin 1 beta by Caf1A usher protein of Yersinia pestis. FEBS Lett. 371:65–68 [DOI] [PubMed] [Google Scholar]