Abstract
We used several molecular typing methods to analyze 196 methicillin-resistant Staphylococcus aureus (MRSA) and 139 methicillin-susceptible S. aureus (MSSA) isolates collected between 1996 and 2005. The sequence type 72 MRSA has increased in frequency in the community in the Republic of Korea and in hospitals in recent years.
TEXT
Multilocus sequence typing (MLST) (4) and staphylococcal chromosomal cassette mec (SCCmec) typing (11) are widely used to identify molecular differences between Staphylococcus aureus isolates. Studies of the evolution of S. aureus over prolonged periods have shown that the predominant strains can change (1, 15). Methicillin-resistant S. aureus (MRSA) accounts for more than 60% of the nosocomial S. aureus isolates in the Republic of Korea (6), and a few MRSA clones (sequence type 5 [ST5]-MRSA-SCCmec II, ST239-MRSA-SCCmec III, and ST239-MRSA-SCCmec IIIA) predominate in Korean hospitals (8, 13). We performed the present analysis, based on MLST, SCCmec typing, and toxin gene profiling, to evaluate the molecular types of S. aureus in the Republic of Korea over a prolonged period.
(The present findings were presented at the 18th European Congress of Clinical Microbiology and Infectious Diseases, Barcelona, Spain, 19 to 22 April 2008 [abstr. P1437].)
We selected at random 13 to 14% of the MRSA isolates and 4 to 8% of the methicillin-susceptible S. aureus (MSSA) isolates stored in Korean investigations between 1996 and 2005, using the Microsoft Excel 2007 program. We also included 15 to 88% of the MRSA and MSSA isolates taken from the anterior nares of healthy individuals in the community in 1997-1998 and 2005, respectively. We did not select the same proportion of MRSA and MSSA isolates from each year. Details of these isolates are provided in the supplemental material.
Confirmatory tests were performed using the Vitek system (bioMérieux, Durham, NC). Antimicrobial susceptibility testing was carried out by the disk diffusion method according to Clinical and Laboratory Standards Institute guidelines (3). Ten antibiotics were tested, and S. aureus ATCC 25923 served as a control strain. We performed SCCmec typing (11); MLST (4); a multiplex PCR assay for genes encoding staphylococcal enterotoxins, staphylococcal exfoliative toxins, and toxic shock syndrome toxin (2); and a PCR assay to detect Panton-Valentine leucocidin (PVL) (10).
We evaluated 335 nonduplicated isolates selected from 2,901 S. aureus isolates, of which 196 were MRSA and 139 were MSSA. No MRSA isolates were found among 42 S. aureus isolates collected from the anterior nares of healthy people in 1997-1998, whereas 8% (18/224) of such isolates were MRSA in 2005. MLST clustered the 196 MRSA isolates into 24 STs and the 139 MSSA isolates into 39 STs. Twenty-nine (9%) of the 335 S. aureus isolates were determined to have novel polymorphisms in the seven allelic genes by MLST. Cluster analysis of the 335 S. aureus isolates was performed with the eBURST program, and clonal complexes (CCs) were defined using a criterion of six alleles out of seven. The S. aureus isolates examined were assigned to 8 CCs and 11 singletons. Seventy-two percent (242/335) of the S. aureus isolates belonged to one of three major CCs (CC1, CC5, or CC239). Among the MRSA isolates, 87% (171/196) clustered into these major CCs, as did 51% (71/139) of the MSSA isolates. Of the singletons, the ST72 clone (7%, 13/196) and the ST30 clone (9%, 13/139) were the most frequent among the MRSA and MSSA isolates, respectively. Most of the ST72 MRSA clones were isolated from the anterior nares of healthy individuals in 2005.
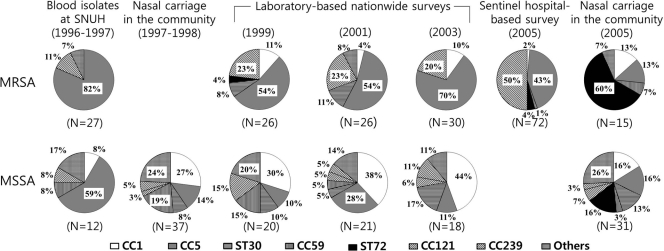
The proportions (percentages) of the major MRSA and MSSA clones in the various pools of isolates are shown in Fig. 1. Before 2003, we identified only one MRSA isolate of the ST72 clone, although this was the most common isolate among Korean community-associated MRSA (CA-MRSA) isolates (ST72-MRSA-SCCmec IV/IVA) (5). The ST72 clone was not found among the MSSA isolates from the anterior nares of healthy people during 1997-1998, but in 2005, it was common among the MSSA (5/31) and MRSA (9/15) isolates from the anterior nares of healthy people.
Fig. 1.
Distribution of the CCs and STs of singletons among MRSA and MSSA isolates in the Republic of Korea from 1996 through 2005. SNUH, Seoul National University Hospital.
Details of the major S. aureus clones in the Republic of Korea are shown in Table 1. Susceptibility to antibiotics other than β-lactams differed, depending on oxacillin resistance and the ST. The MRSA isolates displayed different antimicrobial susceptibilities, depending on the ST; thus, while the ST5 and ST239 strains were resistant to most of the antibiotics tested except rifampin and sulfamethoxazole-trimethoprim, the ST72 strains were more susceptible to clindamycin, ciprofloxacin, and gentamicin.
Table 1.
Molecular characteristics, antimicrobial susceptibilities, and toxin gene profiles of major S. aureus clones isolated in the Republic of Korea from 1996 through 2005a
| MLST (allelic profile) | CC | Oxacillin susceptibility | SCCmec types (n) | No. of isolates | No. (%) of isolates susceptible to: |
Major toxin gene profile (n) | PVL (n) | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| EM | CM | CFLX | GM | RFP | SXT | |||||||
| 1 (1-1-1-1-1-1-1) | 1 | MSSA | 30 | 13 (43) | 27 (90) | 29 (97) | 21 (70) | 30 (100) | 30 (100) | sea-seh (18), sea-seh-pvl (4) | 6 | |
| MRSA | IV (8), IVA (1) | 9 | 3 (33) | 7 (78) | 6 (67) | 1 (11) | 9 (100) | 9 (100) | seh (7) | 0 | ||
| 5 (1-4-1-4-12-1-10) | 5 | MSSA | 6 | 3 (50) | 5 (83) | 6 (100) | 6 (100) | 6 (100) | 6 (100) | sec-seg-sei-tst (2) | 0 | |
| MRSA | I (1), II (54), IIA (34), IV (1), IVA (1), NT (3), NA (1) | 95 | 1 (1) | 11 (12) | 1 (1) | 7 (7) | 83 (87) | 91 (96) | sec-seg-sei-tst (53), seg-sei-tst (28) | 0 | ||
| 6 (12-4-1-4-12-1-3) | 5 | MSSA | 16 | 10 (63) | 12 (75) | 16 (100) | 16 (100) | 16 (100) | 15 (94) | sea (11) | 0 | |
| 15 (13-13-1-1-12-11-13) | 15 | MSSA | 5 | 5 (100) | 5 (100) | 5 (100) | 5 (100) | 5 (100) | 5 (100) | None (3) | 0 | |
| 30 (2-2-2-2-6-3-2) | Singleton | MSSA | 13 | 5 (38) | 10 (77) | 13 (100) | 13 (100) | 13 (100) | 12 (92) | sea-seg-sei-tst (6), sec-seg-sei-tst (2) | 2 | |
| MRSA | IV (2), IVA (2), NT (1) | 5 | 0 (0) | 2 (40) | 4 (80) | 1 (20) | 5 (100) | 4 (80) | sea-seg-sei-pvl (3) | 3 | ||
| 59 (19-23-15-2-19-20-15) | 59 | MSSA | 10 | 4 (40) | 4 (40) | 9 (90) | 10 (100) | 10 (100) | 9 (90) | None (8) | 1 | |
| 72 (1-4-1-8-4-4-3) | Singleton | MSSA | 5 | 1 (20) | 3 (60) | 5 (100) | 5 (100) | 5 (100) | 5 (100) | None (3) | 0 | |
| MRSA | IV (6), IVA (4), NA (3) | 13 | 2 (20) | 8 (62) | 13 (100) | 11 (85) | 13 (100) | 13 (100) | seg-sei (10) | 0 | ||
| 239 (2-3-1-1-4-4-3) | 239 | MRSA | IIA (1), III (23), IIIA (18), NT (2), NA (2) | 46 | 0 (0) | 5 (11) | 1 (2) | 0 (0) | 44 (96) | 10 (22) | sea (34), none (8) | 0 |
| 254 (3-32-1-1-4-4-3) | 239 | MRSA | IIIA (1), IV (4), IVA (1) | 6 | 0 (0) | 2 (33) | 1 (17) | 1 (17) | 1 (17) | 2 (33) | sea-seb (4) | 0 |
| 580 (3-35-48-19-20-26-39) | NA | MSSA | 6 | 1 (17) | 3 (50) | 5 (83) | 4 (67) | 6 (100) | 5 (83) | None (3), seg-sei (2) | 0 | |
NT, nontypeable; NA, not available; EM, erythromycin; CM, clindamycin; CFLX, ciprofloxacin; GM, gentamicin; RFP, rifampin; SXT, sulfamethoxazole-trimethoprim.
Toxin gene analysis was performed on 335 S. aureus isolates. While 93% (183/196) of the MRSA isolates harbored at least one toxin gene, this was true of only 78% (108/139) of the MSSA isolates. PCR amplification of the PVL toxin gene was positive in only 14 (4%) of the S. aureus isolates, of which 3 and 11 were MRSA and MSSA isolates, respectively. Each of the three PVL-positive MRSA isolates was an ST30-MRSA-SCCmec IV/IVA strain. The PVL-positive MSSA isolates were assigned to ST1 (n = 6), ST30 (n = 2), ST59 (n = 1), ST89 (n = 1), and ST121 (n = 1) by MLST. Toxin gene profiles also differed, depending on oxacillin resistance and the ST (Table 1).
The ST72-MRSA-SCCmec IV/IVA clone was the most common CA-MRSA clone in the Republic of Korea (5). CA-MRSA is generally thought to have evolved from CA-MSSA clones that acquire the genes for SCCmec IV. The ST72-MRSA-SCCmec IV/IVA clone circulating in the community in the Republic of Korea might also develop from CA-MSSA. However, a boundary between CA-MRSA and hospital-associated MRSA (HA-MRSA) is sometimes unclear in the Republic of Korea in that the most common nosocomial MRSA strains such as ST5 and ST239 were spreading in the community and CA-MRSA strains such as ST72 were isolated from patients in hospital settings (5, 12). ST72 MSSA isolates also formed a group genetically distinct from ST72 MRSA isolates in a recent study (9). Further work is needed to establish the origin of the ST72-MRSA-SCCmec IV/IVA clone because of the lack of a well-designed study of CA-MRSA in the Republic of Korea.
The MRSA isolates carried more toxin genes than the MSSA isolates (93% versus 78%), and the toxin gene profiles depended on the molecular types of the strains (Table 1), as noted in another study (7). We also found that PVL was infrequent among both the MRSA (2%, 3/196) and MSSA (8%, 11/139) isolates. All of our PVL-positive MRSA isolates were clustered in the form of ST30-MRSA-SCCmec IV/IVA clones. This clone was originally known as the Southwest Pacific clone and was also found in Japan (14). Evidently, a potentially pandemic PVL-positive ST30 CA-MRSA strain also exists in the Republic of Korea.
This study has some limitations. First, the S. aureus isolates were selected from diverse pools obtained in community and hospital settings. Moreover, they were not selected evenly. Therefore, there may be some selection bias and the results may not reflect the nationwide trend. Second, we only gathered clinical information for the isolates from nasal swabs and for those from a previous study (5). Third, we did not perform pulsed-field gel electrophoresis, spa typing, or the new method for differentiating nontypeable SCCmec strains.
In conclusion, the ST72 MRSA strain has become more frequent in the community in the Republic of Korea and in hospitals in recent years.
Supplementary Material
Acknowledgments
The isolates harboring the specific toxin genes were obtained through the Network on Antimicrobial Resistance in S. aureus (NARSA) program supported under NIAID/NIH contract N01-AI-95359. This work was supported by a grant from the Korea Centers for Disease Control and Prevention.
We are grateful to J. O. Cha and J. S. Kim for the gift of control strains.
Footnotes
Supplemental material for this article may be found at http://jcm.asm.org/.
Published ahead of print on 16 March 2011.
REFERENCES
- 1. Blanc D. S., et al. 2007. Changing molecular epidemiology of methicillin-resistant Staphylococcus aureus in a small geographic area over an eight-year period. J. Clin. Microbiol. 45:3729–3736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Cha J. O., et al. 2006. Molecular analysis of Staphylococcus aureus isolates associated with staphylococcal food poisoning in South Korea. J. Appl. Microbiol. 101:864–871 [DOI] [PubMed] [Google Scholar]
- 3. Clinical and Laboratory Standards Institute 2006. Performance standards for antimicrobial disk susceptibility tests, approved standard, 9th ed. Clinical and Laboratory Standards Institute, Wayne, PA [Google Scholar]
- 4. Enright M. C., Day N. P., Davies C. E., Peacock S. J., Spratt B. G. 2000. Multilocus sequence typing for characterization of methicillin-resistant and methicillin-susceptible clones of Staphylococcus aureus. J. Clin. Microbiol. 38:1008–1015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Kim E. S., et al. 2007. A survey of community-associated methicillin-resistant Staphylococcus aureus in Korea. J. Antimicrob. Chemother. 60:1108–1114 [DOI] [PubMed] [Google Scholar]
- 6. Kim H. B., et al. 2004. In vitro activities of 28 antimicrobial agents against Staphylococcus aureus isolates from tertiary-care hospitals in Korea: a nationwide survey. Antimicrob. Agents Chemother. 48:1124–1127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kim J. S., et al. 2006. Association between the methicillin resistance of clinical isolates of Staphylococcus aureus, their staphylococcal cassette chromosome mec (SCCmec) subtype classification, and their toxin gene profiles. Diagn. Microbiol. Infect. Dis. 56:289–295 [DOI] [PubMed] [Google Scholar]
- 8. Ko K. S., et al. 2005. Distribution of major genotypes among methicillin-resistant Staphylococcus aureus clones in Asian countries. J. Clin. Microbiol. 43:421–426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ko K. S., et al. 2011. Sequence type 72 meticillin-resistant Staphylococcus aureus isolates from humans, raw meat, and soil in South Korea. J. Med. Microbiol. 60(Pt. 4):442–445 [DOI] [PubMed] [Google Scholar]
- 10. Lina G., et al. 1999. Involvement of Panton-Valentine leukocidin-producing Staphylococcus aureus in primary skin infections and pneumonia. Clin. Infect. Dis. 29:1128–1132 [DOI] [PubMed] [Google Scholar]
- 11. Oliveira D. C., de Lencastre H. 2002. Multiplex PCR strategy for rapid identification of structural types and variants of the mec element in methicillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 46:2155–2161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Song J.-H., et al. 20 February 2011, posting date Spread of methicillin-resistant Staphylococcus aureus between the community and the hospitals in Asian countries: an ANSORP study. J. Antimicrob. Chemother. doi:10.1093/jac/dkr024 [DOI] [PubMed] [Google Scholar]
- 13. Soo Ko K., et al. 2005. Genotypic diversity of methicillin-resistant Staphylococcus aureus isolates in Korean hospitals. Antimicrob. Agents Chemother. 49:3583–3585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Takizawa Y., et al. 2005. A Panton-Valentine leucocidin (PVL)-positive community-acquired methicillin-resistant Staphylococcus aureus (MRSA) strain, another such strain carrying a multiple-drug resistance plasmid, and other more-typical PVL-negative MRSA strains found in Japan. J. Clin. Microbiol. 43:3356–3363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Wisplinghoff H., et al. 2005. Molecular evolution of methicillin-resistant Staphylococcus aureus in the metropolitan area of Cologne, Germany, from 1984 to 1998. J. Clin. Microbiol. 43:5445–5451 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.