Abstract
Pneumocystis jirovecii pneumonia (PCP) is a common opportunistic infection. Microscopic diagnosis, including diagnosis using the Merifluor-Pneumocystis direct fluorescent antigen (MP-DFA) test, has limitations. Real-time PCR may assist in diagnosis, but no commercially validated real-time PCR assay has been available to date. MycAssay Pneumocystis is a commercial assay that targets the P. jirovecii mitochondrial large subunit (analytical detection limit, ≤3.5 copies/μl of sample). A multicenter trial recruited 110 subjects: 54 with transplants (40 with lung transplants), 32 with nonmalignant conditions, 13 with leukemia, and 11 with solid tumors; 9 were HIV positive. A total of 110 respiratory samples (92% of which were bronchoalveolar lavage [BAL] specimens) were analyzed by PCR. Performance was characterized relative to investigator-determined clinical diagnosis of PCP (including local diagnostic tests), and PCR results were compared with MP-DFA test results for 83 subjects. Thirteen of 14 subjects with PCP and 9/96 without PCP (including 5 undergoing BAL surveillance after lung transplantation) had positive PCR results; sensitivity, specificity, and positive and negative predictive values (PPV and NPV, respectively) were 93%, 91%, 59%, and 99%, respectively. Fourteen of 83 subjects for whom PCR and MP-DFA test results were available had PCP; PCR sensitivity, specificity, PPV, and NPV were 93%, 90%, 65%, and 98%, respectively, and MP-DFA test sensitivity, specificity, PPV, and NPV were 93%, 100%, 100%, and 98%. Of the 9 PCR-positive subjects without PCP, 1 later developed PCP. The PCR diagnostic assay compares well with clinical diagnosis using nonmolecular methods. Additional positive results compared with the MP-DFA test may reflect low-level infection or colonization.
INTRODUCTION
Pneumocystis pneumonia (PCP) remains a formidable infection for both AIDS and non-AIDS patients, with mortality rates in excess of 20% (5, 9). The clinical presentations of the two groups differ, as do the principal differential diagnoses (9, 13, 30). While oxygen desaturation on exercise followed by hypoxemia are universal in PCP, these are not specific features, just as an elevated serum lactate dehydrogenase (LDH) level and pulmonary infiltrates are not (30). Therapy with high-dose trimethoprim plus sulfamethoxazole with oral or intravenous corticosteroids may be given empirically, but these treatments are not without toxicity or immunosuppressive penalties. A worsening status, common early in the course of therapy, may precipitate a shift to pentamidine or a combination of primaquine and clindamycin, again with significant toxicities (3, 5, 34). Transfer to a high-dependency unit or ventilatory support may be required. Empiricism may be necessary, but ruling PCP in or out as a serious diagnostic consideration pays many management dividends for the patient and the clinician.
The diagnosis of PCP is established by microscopy of lung tissue, bronchial lavage, or other deep respiratory samples (3, 9, 15). PCP primarily affects the alveoli, and deep pulmonary samples are necessary for adequate microscopy. In the only comparative study of the diagnostic performance of different microscopic techniques in PCP, immunofluorescence (IF) was nonstatistically superior in sensitivity (91%) to staining with Calcofluor white (74%) and silver (77%), which were themselves more sensitive than Diff-Quik (48%) (26). Many fewer organisms are usually present in non-AIDS patients with PCP, which adversely affects diagnostic performance (34).
The first reports of PCR for the detection of fungal infections were for Pneumocystis carinii and Candida albicans, both in August 1990 (2, 38). There are many published papers indicating the utility of standard and, more recently, real-time PCR to diagnose Pneumocystis pneumonia (1–4, 8, 10, 11, 14, 16, 18, 22, 38, 39). Many laboratories have developed their own assays and use them routinely. However, lack of standardization and clinical validation of the laboratory-developed assays is the primary reason why the EORTC/MSG group decided to omit all fungal PCR from the definitions of invasive fungal diseases (7).
In addition, several papers attest to the fact that PCR diagnosis of PCP is more sensitive than microscopy (4, 10, 11, 13, 22, 24). The generally higher sensitivity of PCR assays than of microscopy may be disproportionately important for non-AIDS patients and may result in fewer missed clinical diagnoses.
We report here the first prospective multicenter evaluation of a commercially launched real-time PCR assay for Pneumocystis, primarily in non-AIDS patients. The four clinical centers were located in Europe and the United States. Subjects were enrolled in the study if they were at risk for infection with Pneumocystis jirovecii. Clinical diagnosis was provided for all enrolled subjects in accordance with local routine diagnostic procedures at the four institutions, and the diagnostic comparator method of choice was the Merifluor direct fluorescent antigen (DFA) immunofluorescence test. Respiratory samples were collected, and DNA was extracted at all sites using a standardized extraction system and was subsequently tested with the MycAssay Pneumocystis kit on a SmartCycler real-time PCR platform.
MATERIALS AND METHODS
Materials required.
The following equipment or materials were provided or required by the clinical sites to conduct this study: MycAssay Pneumocystis kits (Myconostica Ltd., Manchester, United Kingdom), the MycXtra fungal DNA extraction kit (Myconostica Ltd.), the BD BBL MycoPrep specimen digestion/decontamination kit (BD Diagnostic Systems, Oxford, United Kingdom), the SmartCycler real-time PCR platform (Cepheid, Sunnyvale, CA), the Vortex adaptor plate (Myconostica Ltd.), Vortex Genie 2 (Scientific Industries, NY), and a dedicated minicentrifuge and the Merifluor-Pneumocystis assay (Meridian Bioscience, Inc., Cincinnati, OH).
Subjects enrolled.
Subjects were enrolled in the study for a period of 14 months (November 2008 to December 2009) if they met the following inclusion criteria: (i) either HIV/AIDS with bilateral infiltrates and/or hypoxemia (moderate probability of PCP) or bronchoalveolar lavage (BAL) to diagnose pulmonary infiltrates of uncertain etiology, or bronchoscopy for other reasons (e.g., lung cancer, solid-organ transplantation, hematology patients, intensive-care unit [ICU] patients, interstitial lung disease), or the availability of induced-sputum samples from non-HIV/AIDs patients (sputum if induced sputum was not available) (all low probability of PCP) and (ii) informed consent according to local and national guidelines given in those institutions in which an institutional review board (IRB) or ethical committee required it. Patients were excluded if they were less than 18 years of age, unless parental consent had been given, or if the sample collection procedure was unduly risky.
One hundred thirty-one unique subjects were enrolled in the study. Of these, 21 were excluded from analysis, either because they withdrew consent (n = 1) or because there was an insufficient amount of sample for PCR testing (n = 20), leaving 110 unique study subjects with analyzable data. At one site, 16 subjects were enrolled in the study multiple times. For the purpose of the primary analysis, data from the first visit of each unique patient for whom a PCR sample was available were transferred to the analysis database. Subsequent visits for the repeat subjects were located in a separate database for analysis. Taking all repeat visits into consideration, 132 samples were available for testing using the PCR assay. Data from the total number of samples were analyzed in order to look at the assay failure rate.
The study was reviewed and approved by each institution's institutional review board or ethical committee, and patients gave written informed consent in accordance with their local requirements.
Samples.
The majority of samples tested (101 [92%]) were obtained from a directed BAL procedure required for routine clinical purposes. Two of the remaining specimens were sputa, one of which was induced, and there were 7 other lower respiratory tract (LRT) specimens. These were recorded as a specimen from bronchial washing following lavage, bronchial aspirates, bronchial secretions, and two bronchial biopsy specimens. The volume of sample available for DNA extraction ranged from <1 ml to 60 ml. Following collection, the samples were transferred to the molecular testing laboratories, where they were either (i) extracted within 72 h (n = 80), and the DNA extracts stored at −20°C or −70°C, or (ii) stored at −20°C or −70°C for as long as 19 days prior to DNA extraction.
Extraction procedure.
All samples were extracted on site by the local laboratory. Fungal DNA was extracted from the samples using the MycXtra fungal DNA extraction kit according to the manufacturer's instructions. A minimum volume of 2 ml of sample is recommended. DNA was extracted directly from liquid BAL samples, while viscous samples were preprocessed using either the N-acetyl-cysteine–sodium hydroxide (NALC-NaOH) reagent in the BD BBL MycoPrep specimen digestion/decontamination kit (BD Diagnostic Systems, Oxford, United Kingdom) according to the manufacturer's instructions or a local NALC procedure for acid-fast bacillus (AFB) decontamination. DNA extracts were shipped on dry ice to Myconostica for molecular testing.
Molecular testing.
MycAssay Pneumocystis is a commercially available qualitative real-time PCR test utilizing molecular beacons (36) for the detection of P. jirovecii. The target sequence is the Pneumocystis mitochondrial ribosomal large subunit (mLSU). The primers and molecular beacon avoid the polymorphisms described in this region (35), ensuring that detection is not affected by known genetic heterogeneity in the Pneumocystis population (5). Human DNA is not detected. The kit also contains an internal amplification control (IAC) sequence, a DNA fragment not present in P. jirovecii, to confirm amplification. The P. jirovecii target sequence is labeled with a 6-carboxyfluorescein (FAM) beacon, and the IAC sequence is labeled with a HEX beacon. The Cepheid SmartCycler real-time PCR system was used to monitor the fluorescence emitted by each beacon within the reaction mixture. On this platform, the assay limit of blank, defined as the experimentally determined negative cutoff value used to distinguish positive from negative results, is a cycle threshold (CT) of 39. A CT lower than this is positive (20). The CT values can be interpreted in a semiquantitative manner, since there is an approximate relationship between the CT and the fungal burden.
Failure of the IAC renders a negative assay unreportable. The assay limit of detection (LoD), the lowest DNA concentration at which ≥95% of results are positive (i.e., CT below the limit of blank), was calculated in accordance with NCCLS procedure EP17-A (21) and was determined to be <35 copies per 10 μl of target DNA sample.
Upon receipt from the clinical sites, DNA extracts were stored at −80°C at Myconostica Ltd. until the PCR testing was complete. Storage time for the extracts ranged from 9 months to 6 weeks. All samples were thawed, batched, and assayed on the same day. The PCR results were not returned to the clinician or patient, nor were they used for establishing a clinical diagnosis.
Merifluor DFA immunofluorescence testing.
The comparator diagnostic test was the Merifluor DFA immunofluorescence test. The manufacturer's instructions were followed.
Other diagnostic tests.
Other diagnostic tests, including Calcofluor white (3 sites) and Gomori methenamine silver (GMS) (1 site) microscopy, serum lactate dehydrogenase (LDH) levels, C-reactive protein (CRP), plasma viscosity (PV), and erythrocyte sedimentation rate (ESR), were conducted in accordance with local requirements. Where these results were available, the data were captured on the study forms.
Clinical risk factors and final diagnosis.
At study entry, in addition to assigning a pretest probability of moderate or low risk for PCP infection, the enrolling clinician was asked to assign the clinical condition or conditions that contributed to the subject's at-risk status for infection.
Clinical diagnosis for each subject enrolled in the study was performed by the enrolling clinician, in accordance with his or her local diagnostic procedures and paradigms, up to 10 weeks after the subject had been enrolled and the sample had been taken for analysis. Information taken into account in reaching a clinical diagnosis of PCP included radiological appearance, hypoxemia, serum LDH level, identification of other pathogens, and local microscopy results for P. jirovecii. Additional clinical information was requested for 3 months after the study for subjects whose PCR result was positive but whose clinical diagnosis did not confirm PCP infection.
Data collection and analysis.
Clinical and laboratory data were collected on dedicated data collection forms for each subject enrolled in the study. The data were entered by a double data entry convention into a study database. Data queries were generated as required and were followed to closure. An audit of the accuracy of the database entries relative to the original data collection forms was conducted by two independent persons prior to database locking and analysis.
RESULTS
Subject disposition.
Data from 110 enrolled subjects were eligible for analysis. The demographics of the study subjects, by site, are recorded in Table 1. The population studied was predominantly not HIV infected (92%), and by the study entry criteria, their probability of infection was considered to be low. The risk categories assigned at the time of study entry were combined into groups for analysis, based on the predisposition for PCP. The analysis groups were as follows: (i) HIV/AIDS, (ii) lung transplant (one subject with a dual organ transplant [lung/kidney] was included in this category), (iii) all other organ transplants and allogeneic hematopoietic stem cell transplants (HSCT), (iv) leukemia, other hematological disorders, and autologous HSCT, (v) other solid tumors, and (vi) other nonmalignant conditions.
Table 1.
Subject enrollment, disposition, and demographics by sitea
| Characteristic | Valueb |
|||
|---|---|---|---|---|
| Site 1 | Site 2 | Site 3 | Site 4 | |
| Enrollment | ||||
| Unique subjects | 18 | 29 | 22 | 62 |
| Subjects entering into primary analysisc | 18 | 22 | 22 | 48 |
| Subjects with repeat visits | 0 | 0 | 0 | 16 |
| No. of samples assayed | 18 | 22 | 22 | 70 |
| Gender | ||||
| Male | 7 | 10 | 13 | 29 |
| Female | 11 | 12 | 9 | 19 |
| Race | ||||
| Caucasian | 12 | 4 | 0 | 48 |
| Black | 6 | 13 | 0 | 0 |
| Other (nonwhite) | 0 | 5 | 0 | 0 |
| No data | 0 | 0 | 22 | 0 |
| Mean (range) age (yr) | 57 (26–78) | 54 (21–75) | 52 (26–74) | 56 (23–73) |
| Pretest probability of PCP | ||||
| Moderate | 0 | 3 | 4 | 0 |
| Low | 18 | 19 | 18 | 48 |
| Sample type | ||||
| BAL | 18 | 20 | 20 | 43 |
| Sputum | 0 | 1 | 1 | 0 |
| Other LRT | 0 | 1 | 1 | 5 |
| At-risk group | ||||
| HIV/AIDS | 0 | 3 | 6 | 0 |
| Lung transplant | 8 | 0 | 1 | 31 |
| Other transplants | 1 | 7 | 3 | 0 |
| Leukemia | 1 | 1 | 7 | 0 |
| Other solid tumors | 2 | 1 | 1 | 7 |
| Other nonmalignant conditions | 6 | 10 | 4 | 10 |
The testing and enrollment sites for the study were Duke University School of Medicine, Durham, NC; Albert Einstein College of Medicine and Montefiore Medical Center, Bronx, NY; Centre Hospitalier Universitaire Vaudois and University of Lausanne, Lausanne, Switzerland; and Medizinische Universität and Landeskrankenhaus Natters, Innsbruck, Austria.
Except where otherwise indicated, values are numbers of patients.
Subjects were excluded from analysis if they withdrew consent or if there was inadequate sample left for PCR testing.
MycAssay Pneumocystis.
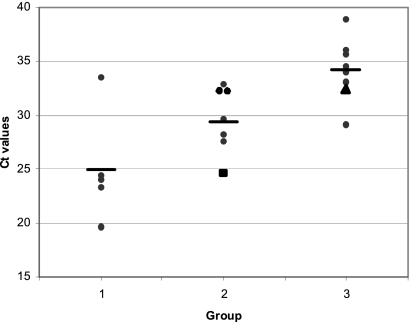
Thirteen of the 14 subjects with a final clinical diagnosis of PCP were positive by the MycAssay Pneumocystis kit (Table 2), giving an assay sensitivity of 93%. Five of these 13 subjects (group 1) had HIV/AIDS, with a moderate probability of PCP infection; they recorded an average CT of 25 (range, 19.6 to 33.5). The remaining 8 positive subjects (group 2), all with a low probability of infection, recorded an average CT of 29.4 (range, 24.6 to 32.9) (Fig. 1). A third group, of 9 subjects who were PCP negative by clinical diagnosis and PCR positive, recorded an average CT of 34.2 (range, 29.1 to 38.9). One-factor analysis of variance (ANOVA) showed a statistically significant difference among the three groups (P < 0.001). Posthoc multiple-comparison tests (Scheffe's test) adjusting for multiple testing gave a statistical difference (P < 0.001) between groups 1 and 3, with a less significant difference (P = 0.031) between groups 2 and 3. Comparison of the results in groups 1 and 2 demonstrated some limited evidence of a difference that did not reach significance (P = 0.10).
Table 2.
Sensitivity and specificity of MycAssay Pneumocystis relative to clinical diagnosisa
| MycAssay Pneumocystis result | No. of patients with the following clinical diagnosis: |
|
|---|---|---|
| PCP | Condition other than PCP | |
| Positive | 13 | 9 |
| Negative | 1 | 87 |
A total of 110 patients were tested.
Fig. 1.
CT values for all study subjects with positive PCR results. The graph shows the results for subjects in group 1 (confirmed PCP and HIV/AIDS) (mean CT, 25), group 2 (confirmed PCP and no HIV/AIDS) (mean CT, 29.4), and group 3 (diagnoses other than PCP) (mean CT, 34.2) at the end of the study. Horizontal lines indicate means. Analysis by Scheffe's test yielded the following P values for comparisons between groups: 0.10 for groups 1 and 2, <0.001 for groups 1 and 3, and 0.031 for groups 2 and 3. In group 2, the subject with a CT of 24.6 (represented by the large square) was negative by immunofluorescence and microscopy. In group 3, the triangle represents the subject who went on to develop PCP.
One subject had an assigned clinical diagnosis of PCP; however, the PCR was negative. This subject was a 41-year-old female with HIV/AIDS, with a CD4+ cell count of 33 × 106/liter, on corticosteroids, and admitted to the ICU. The IF test result was positive, and the LDH level was 249 U/liter (normal). The sample obtained was induced sputum, with a volume of <1 ml, which is below the recommended extraction volume of 2 ml. A comment written on the relevant data collection form queried the quality of the sample.
One 63-year-old male lung transplant recipient with a clinical diagnosis of PCP was PCR positive (CT, 24.6) and negative by IF. He was on corticosteroids and other immunosuppressants and had an elevated LDH level of 527 U/liter and a CRP level of 20 mg/liter. Microscopy and IF were both negative.
Nine subjects were positive by PCR, without clinical evidence of PCP, and negative by IF (Tables 3 and 4; Fig. 1). All were considered to have a low probability of PCP infection at enrollment. Five of the subjects had lung transplants, 2 had renal transplants, and 2 were enrolled with other nonmalignant clinical conditions. Eight were microscopy negative, and one was microscopy positive for yeasts. Four of the nine had normal chest X-ray or chest computed tomography (CT) results reported at the time of testing. LDH levels ranged from 154 to 423 U/liter and CRP levels from <0.3 to 150 mg/liter. Clinical follow-up data for these nine subjects were requested for a period of 3 months post-study entry; one of these subjects developed PCP infection a month after sampling.
Table 3.
Comparison of Merifluor immunofluorescence test with MycAssay Pneumocystis resultsa
| MycAssay Pneumocystis result | No. of patients with the indicated result |
|||
|---|---|---|---|---|
| Clinically diagnosed with PCP (n = 14) |
Clinically diagnosed with a condition other than PCP (n = 69) |
|||
| IF+ | IF− | IF+ | IF− | |
| Positive | 12 | 1 | 0 | 7 |
| Negative | 1 | 0 | 0 | 62 |
A total of 83 patients were tested.
Table 4.
Details for 9 PCR-positive patients with assigned clinical diagnoses other than PCP at study exita
| Patient no. | CT | Age (yr) | Sex | Risk factor(s) | Episode diagnosis | Test result |
PCP medicationc | Follow-up | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Merifluor IFb | Microscopy | LDH (U/liter) | PO2 (mm Hg) | FIO2 (%) | CxR | CT | ||||||||
| 1 | 32.4 | 64 | F | Lung Tx, corticosteroids | Lung infection | − | − | 220 | 68 | 21 | Normal | Normal | None at time of testing | Developed PCP within 3 mo of study exit |
| 2 | 35.6 | 62 | M | Renal Tx, corticosteroids | Candida colonization and presumptive BOOP | − | − | 423 | 50, 75 | 20, 60 | Air space opacities | Patchy air space opacities and small mediastinal lymph nodes | Posttesting: cotrimoxazole (Septrin or Bactrim) | No relevant development |
| 3 | 34 | 55 | M | Renal Tx, corticosteroids | Pneumonia | − | − | 251 | 93% | RA (21) | Bilateral infiltrates | Patchy foci of consolidation, calcified bilateral hilar lymph nodes | None at time of testing | No relevant development |
| 4 | 38.9 | 53 | F | Lung Tx, corticosteroids | P. aeruginosa-A. fumigatus colonization and infections | − | − | No data | No data | No data | No data | Bibasal lung infiltrate and single nodule | None at time of testing; voriconazole, >30 days | No relevant development |
| 5 | 33.1 | 69 | M | Lung Tx, corticosteroids | PLTS | − | − | 204 | 70 | 21 | Normal | No data | None at time of testing | No relevant development |
| 6 | 34.1 | 67 | M | Lung Tx, corticosteroids | PLTS | − | − | 187 | 81 | 21 | Normal | Normal | None at time of testing | No relevant development |
| 7 | 36.1 | 56 | F | Lung Tx, corticosteroids | Viral pneumonia/pneumonitis | − | − | 312 | 90 | 21 | Multiple consolidations | No data | None at time of testing; posaconazole, ∼4 days | No relevant development |
| 8 | 29.1 | 62 | M | Other (NMC) | Lung carcinoma | No data | − | 154 | 59 | 21 | Infiltrates, right upper lobe | Right midzone, massive infiltrates | None at time of testing | No relevant development |
| 9 | 34.5 | 47 | F | Other (NMC) | Hemoptysis | No data | − | 167 | No data | No data | Normal | No data | None at time of testing | No relevant development |
CT, cycle threshold; IF, immunofluorescence; Tx, transplant; BOOP, bronchiolitis obliterans organizing pneumonia; LDH, lactate dehydrogenase; NMC, nonmalignant conditions; PLTS, post-lung transplant surveillance; PO2, partial pressure of oxygen; FIO2, fractional inspired oxygen; CxR, chest X ray; CT, computed tomography; RA, room air.
−, negative.
Voriconazole and posaconazole have no effect on P. jirovecii.
The PCR test demonstrated a sensitivity, specificity, positive predictive value (PPV), and negative predictive value (NPV) of 93%, 91%, 59%, and 99%, respectively; positive and negative likelihood ratios of 9.9 and 0.08; and a diagnostic odds ratio of 125.6. These sensitivity and specificity numbers are comparable to, or better than, those published for other real-time PCR assays (10, 17, 29).
Merifluor DFA-IF test.
IF data were available for 83 of the 110 subjects. Thirteen were IF positive and 70 were IF negative, giving a sensitivity and specificity of 93% and 100%, respectively. All IF-positive subjects had a confirmed final diagnosis of PCP. The PCR results for this subset (83 of the 110 subjects) were compared to the Merifluor IF results for the clinically positive PCP population (n = 14) and the clinically negative PCP population (n = 69) (Table 3). Overall the sensitivity, specificity, PPV, and NPV obtained were similar to the results obtained for the total study population (93%, 90%, 65%, and 98%, respectively).
Pretest probability.
Subjects with HIV/AIDS and the presence of bilateral ground glass effects on radiography or hypoxemia are considered to have a moderate probability of infection with P. jirovecii. Seven of the 9 subjects with HIV/AIDS presented with the criteria for moderate probability, and 6 (86%) were assigned a clinical diagnosis of PCP at study exit.
Eight of 103 (8%) subjects considered to have a low probability of infection were assigned a clinical diagnosis of PCP at study exit. Of these 8, all had been on corticosteroids or another immunosuppressive drug, and none were receiving prophylactic trimethoprim-sulfamethoxazole. They were evenly distributed across all 5 of the “at-risk categories” detailed above, excluding HIV/AIDS. Seven had evidence of bilateral ground glass effects on radiography. One patient, without HIV/AIDS, had a CD4+ count of <100 ×106/liter. Of the remaining 95 subjects, half were not on steroids or immunosuppressive agents. Of the 48 who were receiving steroids or similar drugs, none had evidence of bilateral ground glass effects and 18 were receiving prophylactic trimethoprim-sulfamethoxazole.
Assay robustness.
Two of the 132 (1.5%) samples tested in the study were reported “Negative/Fail” and therefore invalid because the IAC was out of range, suggesting PCR inhibition. In both cases, the sample matrix was BAL fluid, and the final diagnoses were not PCP. Under these circumstances, the results were reported as “inhibitor present, negative test result cannot be reported.”
Repeat sample analysis.
One site enrolled 16 subjects in the study multiple times, for a total of 42 samples. Although the repeat samples were not part of the primary analysis, other analyses were done to look at the pattern of PCR positivity with sequential visits, with respect to date of diagnosis and therapy regimens. The visit schedules covered a 3- to 4-month window. Nine of the 16 patients had a single follow-up visit (where a respiratory sample was available for testing).
Two of the 16 subjects were diagnosed with PCP. Both subjects tested positive by PCR and have been included in the primary study analysis. For one subject, for whom data from three samples were available, the first PCR sample was positive 1 month before the clinical diagnosis of PCP was made (Table 4, patient 1). The third sample was negative by PCR and had been collected during a course of therapy. For the other subject, for whom three samples were available for analysis, both PCP diagnosis and PCR were positive at the first visit. Subsequent samples were negative by PCR and were collected during the course of therapy.
Nine of the 16 subjects, contributing 24 samples to the study, had a final clinical diagnosis other than PCP at the time of the initial visit and on all subsequent visits. All 24 samples tested negative by PCR.
Five of the 16 subjects, contributing 12 samples to the study, had a final clinical diagnosis other than PCP at the time of the initial visit and all subsequent visits; however, 6 samples tested positive by PCR. For one individual, a post-lung transplant surveillance subject, 2 consecutive samples taken in a 14-day window tested positive by PCR. These subjects were not treated for PCP infection, and none developed PCP during the 3-month follow-up period.
DISCUSSION
The timely diagnosis and treatment of P. jirovecii infection remain a challenge to the clinician and mycology laboratories, where the gold standard has been visualization of characteristic P. jirovecii cysts and/or trophozoites in lung tissue biopsy specimens. The advent of the AIDS epidemic led to the use of less stringent diagnostic criteria, since many patients were too ill to undergo biopsy. Microscopy of BAL and other respiratory samples became the norm for laboratory diagnosis. The comparative study of the diagnostic performance of microscopy demonstrated that immunofluorescence was nonstatistically superior to Calcofluor white and Gomori methenamine silver (GMS) staining, with a sensitivity of about 90% (26).
Twenty years on, the literature still debates different PCR protocols: standard versus real-time versus touchdown PCR and the diagnostic benefits of quantitative versus qualitative PCR. Standardization of the approach, and clinical validation, for a PCR protocol and diagnostic algorithm would facilitate the use and acceptance of PCR in patient management. The results reported in this study show the diagnostic value of a validated commercial real-time PCR assay when used for patients at risk for PCP infection. PCR was 100% concordant with a positive clinical diagnosis. In this study, the sensitivity of PCR was at least equivalent to that of immunofluorescence. If the small-volume induced-sputum sample of questionable quality were excluded, PCR would outperform immunofluorescence. The sensitivity, specificity, and likelihood ratios reported here are comparable to other reports in the literature for real-time PCR assays.
The negative IF result for the subject who was PCP positive by diagnosis and strongly positive by PCR is interesting. The CT value was very low at 24.6, suggesting a severe infection. The subject, who underwent lung transplantation in 2006 and PCP prophylaxis for 1 year posttransplantation, suffered late-onset PCP at the time of testing for this study. The failure of microscopy and immunofluorescence to detect PCP infection with a majority of trophozoites and few cysts has been discussed in the literature (28, 31, 32). An imbalance in the cyst-to-trophozoite ratio has been reported for patients studied while receiving prophylaxis (32).
The average CT of 25 for the PCP-with-HIV/AIDS group (group 1) was lower than that (29.4) for the non-HIV/AIDS PCP group (group 2). This finding was expected, since the PCP burden in the HIV/AIDS population is generally found to be higher. The average CT (34.2) for the non-PCP PCR-positive group (group 3) demonstrated overlap with group 2, with weak significance (P = 0.031) confirming the difficulty of setting a clinical cutoff for non-HIV/AIDS populations who may have low-level PCP infection or colonization. Consequently, the PPV and NPV will differ in different patient populations.
The probability of PCP infection is well documented for the immunocompromised HIV/AIDS population and was used in this study to distinguish between moderate and low pretest probabilities of disease. Increasingly, PCP infection is found in other immunocompromised populations, without HIV/AIDS. The numerical risk of PCP infection in these groups is less well documented. A subset (17/48) of the non-HIV/AIDS patients enrolled in this study at one site were considered at moderate risk by their enrolling physicians. All were lung transplant patients who had stopped their prophylaxis treatment 1 year after the transplant. None progressed to PCP infection during the time course of this study. Chronic steroid use is associated with a higher rate of colonization and mortality in the non-HIV population (5), as is the presence of underlying lung disorders, including chronic obstructive pulmonary disorders (13).
Many papers have compared PCR diagnosis of PCP to microscopy (10, 17, 28, 29). The definition of the cutoff in each paper has usually been that most consistent with the microscopy results, and the different molecular targets and methods prevent direct comparison between many of the papers. The generally higher sensitivity of PCR assays than of microscopy could be of most value for non-AIDS patients, in whom the fungal load is generally lower. In this study, 1 of the 9 (11%) patients who were PCR positive and PCP negative at study exit developed PCP within 6 weeks of the initial diagnosis despite normal chest X-ray and CT results and negative microscopy results at the time of the study. The remaining 8 patients, for whom details are given in Table 4, did not progress to PCP infection during the 3 months of follow-up, and it is likely that the P. jirovecii amplified from the clinical samples of these patients is the result of colonization (6, 12, 19, 25, 37). Since microscopy and immunofluorescence are part of the diagnostic algorithm, a negative test is diagnostic for the absence of infection but may also be due to sampling error, low fungal burdens, or few P. jirovecii cysts. A positive PCR result, if microscopy is negative, could be confirmed by the β-1,3-glucan test, which is commonly positive in the blood of patients with PCP (23, 25, 33) but lacks specificity for the disease. In clinical settings where a positive PCR result is not supported by other clinical signs and symptoms, clinicians will have to decide whether to treat; with a highly sensitive PCR assay, this may lead to overtreatment. The very high NPV is helpful for excluding the diagnosis.
Of the 16 subjects studied more than once, 2 were diagnosed with PCP infection. For both these subjects, once the clinical diagnosis was made and PCP therapy started, the PCR results were negative. Although the sample size was too small to allow one to draw conclusions, this does suggest that a PCR assay loses sensitivity once the infection is being treated and is therefore unlikely to be of diagnostic use during prolonged periods of treatment. It may be possible to monitor the response to therapy with real-time PCR assays, as others have reported (8, 35). Five patients with lung transplants contributed 12 samples, of which at least 1 sample was PCR positive and negative by microscopy. One subject was receiving PCP prophylactic therapy. The most likely explanation of these PCR-positive test results is low-level colonization with P. jirovecii and carriage of the organism in the patient with the lung transplant. Transmission of infection between individuals, via environmental contamination, is a known risk (5, 15, 27). If colonization can be confirmed for an asymptomatic subject, this opens the possibility of treatment and targeted reduction of the reservoir of P. jirovecii.
ACKNOWLEDGMENTS
This study was funded by Myconostica Ltd.
We are indebted to Julie Morris for the statistical analyses.
The following authors are or were employees and/or shareholders of Myconostica at the time the study was conducted: M. Hughes, S. A. Follett, X. Cui, F. Leung, G. Morgan, and A. Moody. D. S. Perlin is a member of the scientific advisory board for Myconostica Ltd. and a shareholder. D. W. Denning is a Board director of Myconostica Ltd. and holds founder shares in F2G Ltd. and Myconostica Ltd., both University of Manchester spin-off companies. D. W. Denning has received grant support from F2G as well as from the Fungal Research Trust, the Wellcome Trust, the Moulton Trust, The Medical Research Council, The Chronic Granulomatous Disease Research Trust, the National Institute of Allergy and Infectious Diseases, the National Institute of Health Research, the European Union, AstraZeneca, and Basilea. He has continued to act as an advisor/consultant to F2G and Myconostica, as well as other companies over the past 5 years, including Basilea, Vicuron (now Pfizer), Pfizer, Schering Plough, Nektar, Daiichi, Astellas, Gilead, and York Pharma. He has been paid for talks on behalf of Schering, Astellas, Novartis, Merck, Dainippon, and Pfizer.
Footnotes
Published ahead of print on 2 March 2011.
The authors have paid a fee to allow immediate free access to this article.
REFERENCES
- 1. Arcenas R. C., et al. 2006. A real-time polymerase chain reaction assay for detection of Pneumocystis from bronchoalveolar lavage fluid. Diagn. Microbiol. Infect. Dis. 54:169–175 [DOI] [PubMed] [Google Scholar]
- 2. Buchman T. G., Rossier M., Merz W. G., Charache P. 1990. Detection of surgical pathogens by in vitro DNA amplification. Part I. Rapid identification of Candida albicans by in vitro amplification of a fungus-specific gene. Surgery 108:338–347 [PubMed] [Google Scholar]
- 3. Carmona E. M., Limper A. H. 24 August 2010. Update on the diagnosis and treatment of Pneumocystis pneumonia. Ther. Adv. Respir. Dis. doi:10.1177/1753465810380102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Chen S. C., Halliday C. L., Meyer W. 2002. A review of nucleic acid-based diagnostic tests for systemic mycoses with an emphasis on polymerase chain reaction-based assays. Med. Mycol. 40:333–357 [DOI] [PubMed] [Google Scholar]
- 5. Cushion M. T. 2010. Are members of the fungal genus Pneumocystis (a) commensals; (b) opportunists; (c) pathogens; or (d) all of the above? PLoS Pathog. 6:e1001009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Davis J. L., et al. 2008. Pneumocystis colonisation is common among hospitalised HIV infected patients with non-Pneumocystis pneumonia. Thorax 63:329–334 [DOI] [PubMed] [Google Scholar]
- 7. De Pauw B., et al. 2008. Revised definitions of invasive fungal disease from the European Organization for Research and Treatment of Cancer/Invasive Fungal Infections Cooperative Group and the National Institute of Allergy and Infectious Diseases Mycoses Study Group (EORTC/MSG) Consensus Group. Clin. Infect. Dis. 46:1813–1821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Durand-Joly I., et al. 2005. Molecular diagnosis of Pneumocystis pneumonia. FEMS Immunol. Med. Microbiol. 45:405–410 [DOI] [PubMed] [Google Scholar]
- 9. Festic E., Gajic O., Limper A. H., Aksamit T. R. 2005. Acute respiratory failure due to Pneumocystis pneumonia in patients without human immunodeficiency virus infection: outcome and associated features. Chest 128:573–579 [DOI] [PubMed] [Google Scholar]
- 10. Flori P., et al. 2004. Comparison between real time PCR, conventional PCR and different staining techniques for diagnosing Pneumocystis jirovecii pneumonia from bronchoalveolar lavage specimens. J. Med. Microbiol. 53:603–607 [DOI] [PubMed] [Google Scholar]
- 11. Gupta R., et al. 2007. Improved detection of Pneumocystis jirovecii infection in a tertiary care reference hospital in India. Scand. J. Infect. Dis. 39:571–576 [DOI] [PubMed] [Google Scholar]
- 12. Hauser P. M., Blanc D. S., Bille J., Nahimana A., Francioli P. 2000. Carriage of Pneumocystis carinii by immunosuppressed patients and molecular typing of the organisms. AIDS 14:461–462 [DOI] [PubMed] [Google Scholar]
- 13. Huang L., Morris A., Limper A. H., Beck J. M. 2006. An official ATS Workshop summary: recent advances and future directions in Pneumocystis pneumonia (PCP). Proc. Am. Thorac. Soc. 3:655–664 [DOI] [PubMed] [Google Scholar]
- 14. Huggett J. F., et al. 2008. Development and evaluation of a real-time PCR assay for detection of Pneumocystis jirovecii DNA in bronchoalveolar lavage fluid of HIV-infected patients. Thorax 63:154–159 [DOI] [PubMed] [Google Scholar]
- 15. Krajicek B. J., Limper A. H., Thomas C. F., Jr 2008. Advances in the biology, pathogenesis and identification of Pneumocystis pneumonia. Curr. Opin. Pulm. Med. 14:228–234 [DOI] [PubMed] [Google Scholar]
- 16. Larsen H. H., et al. 2002. Development and evaluation of a quantitative, touch-down, real-time PCR assay for diagnosing Pneumocystis carinii pneumonia. J. Clin. Microbiol. 40:490–494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Linssen C. F. M., et al. 2006. Inter-laboratory comparison of three different real time PCR assays for the detection of Pneumocysis jirovecii in bronchoalveolar lavage fluid samples. J. Med. Microbiol. 55:1229–1235 [DOI] [PubMed] [Google Scholar]
- 18. Lipschik G. Y., et al. 1992. Improved diagnosis of Pneumocystis carinii infection by polymerase chain reaction on induced sputum and blood. Lancet 340:203–206 [DOI] [PubMed] [Google Scholar]
- 19. Maskell N. A., et al. 2003. Asymptomatic carriage of Pneumocystis jirovecii in subjects undergoing bronchoscopy: a prospective study. Thorax 58:594–597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Myconostica Ltd 9 November 2010. Instructions for use for MycAssay Pneumocystis (IVD SmartCycler), version 3.2. Myconostica Ltd., Manchester, United Kingdom [Google Scholar]
- 21. NCCLS 2004. Protocols for determination of limits of detection and limits of quantitation; approved guideline. NCCLS document EP17-A, vol. 24, no. 34 NCCLS, Wayne, PA [Google Scholar]
- 22. Olsson M., Stralin K., Holmberg H. 2001. Clinical significance of nested polymerase chain reaction and immunofluorescence for detection of Pneumocystis carinii pneumonia. Clin. Microbiol. Infect. 7:492–497 [DOI] [PubMed] [Google Scholar]
- 23. Persat F., et al. 2008. Contribution of the (1→3)-β-d-glucan assay for diagnosis of invasive fungal infections. J. Clin. Microbiol. 46:1009–1013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Pinlaor S., et al. 2004. PCR diagnosis of Pneumocystis carinii on sputum and bronchoalveolar lavage samples in immuno-compromised patients. Parasitol. Res. 94:213–218 [DOI] [PubMed] [Google Scholar]
- 25. Pisculli M. L., Sax P. E. 2008. Use of a serum β-glucan assay for diagnosis of HIV-related Pneumocystis jirovecii pneumonia in patients with negative microscopic examination results. Clin. Infect. Dis. 46:1928–1930 [DOI] [PubMed] [Google Scholar]
- 26. Procop G. W., et al. 2004. Detection of Pneumocystis jiroveci in respiratory specimens by four staining methods. J. Clin. Microbiol. 42:3333–3335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Rabodonirina M., et al. 2004. Molecular evidence of interhuman transmission of Pneumocystis pneumonia among renal transplant recipients hospitalized with HIV-infected patients. Emerg. Infect. Dis. 10:1766–1773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ribes J. A., Limper A. H., Espy M. J., Smith T. F. 1997. PCR detection of Pneumocystis carinii in bronchoalveolar lavage specimens: analysis of sensitivity and specificity. J. Clin. Microbiol. 35:830–835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Rohner P., Jacomo V., Studer R., Schrenzel J., Graf J-D. 2009. Detection of Pneumocystis jirovecii by two staining methods and two quantitative PCR assays. Infection 37:261–265 [DOI] [PubMed] [Google Scholar]
- 30. Su Y. S., Lu J. J., Perng C. L., Chang F. Y. 2008. Pneumocystis jirovecii pneumonia in patients with and without human immunodeficiency virus infection. J. Microbiol. Immunol. Infect. 41:478–482 [PubMed] [Google Scholar]
- 31. Sukura A. 1995. Trophozoite-to-cyst ratio increases during recovery from Pneumocystis carinii pneumonia in rats. APMIS 103:300–306 [PubMed] [Google Scholar]
- 32. Tamburrini E., et al. 1996. Imbalance between Pneumocystis carinii cysts and trophozoites in bronchoalveolar lavage fluid from patients with pneumocystosis receiving prophylaxis. J. Med. Microbiol. 45:146–148 [DOI] [PubMed] [Google Scholar]
- 33. Tasaka S., et al. 2007. Serum indicators for the diagnosis of Pneumocystis pneumonia. Chest 131:1173–1180 [DOI] [PubMed] [Google Scholar]
- 34. Thomas C. F., Jr., Limper A. H. 2004. Pneumocystis pneumonia. N. Engl. J. Med. 350:2487–2498 [DOI] [PubMed] [Google Scholar]
- 35. Tsolaki A. G., Miller R. F., Wakefield A. E. 1999. Oropharyngeal samples for genotyping and monitoring response to treatment in AIDS patients with Pneumocystis carinii pneumonia. J. Med. Microbiol. 48:897–905 [DOI] [PubMed] [Google Scholar]
- 36. Tyagi S., Kramer F. R. 1996. Molecular beacons: probes that fluoresce upon hybridization. Nat. Biotechnol. 14:303–308 [DOI] [PubMed] [Google Scholar]
- 37. Vidal S., et al. 2006. Pneumocystis jirovecii colonisation in patients with interstitial lung disease. Clin. Microbiol. Infect. 12:231–235 [DOI] [PubMed] [Google Scholar]
- 38. Wakefield A. E., et al. 1990. Detection of Pneumocystis carinii with DNA amplification. Lancet 336:451–453 [DOI] [PubMed] [Google Scholar]
- 39. Wakefield A. E., Guiver L., Miller R. F., Hopkin J. M. 1991. DNA amplification on induced sputum samples for diagnosis of Pneumocystis carinii pneumonia. Lancet 337:1378–1379 [DOI] [PubMed] [Google Scholar]