Abstract
Mycobacterium avium subsp. paratuberculosis is the causative agent of Johne's disease (JD) in cattle and may be associated with Crohn's disease (CD) in humans. It is the slowest growing of the cultivable mycobacteria, and culture from clinical, veterinary, food, or environmental specimens can take 4 months or even longer. Currently, the insertion element IS900 is used to detect M. avium subsp. paratuberculosis DNA. However, closely related IS900 elements are also present in other mycobacteria, thus limiting its specificity as a target. Here we describe the use of novel primer sets derived from the sequences of two highly specific single copy genes, MAP2765c and MAP0865, for the quantitative detection of M. avium subsp. paratuberculosis within 6 h by using real-time PCR. Specificity of the target was established using 40 M. avium subsp. paratuberculosis isolates, 67 different bacterial species, and two intestinal parasites. Using the probes and methods described, we detected 27 (2.09%) M. avium subsp. paratuberculosis-positive stool specimens from 1,293 individual stool samples by the use of either IS900 or probes deriving from the MAP2765c and MAP0865 genes described here. In general, bacterial load due to M. avium subsp. paratuberculosis was uniformly low in these samples and we estimated 500 to 5,000 M. avium subsp. paratuberculosis bacteria per gram of stool in assay-positive samples. Thus, the methods described here are useful for rapid and specific detection of M. avium subsp. paratuberculosis in clinical samples.
INTRODUCTION
Mycobacterium avium subsp. paratuberculosis is the causative agent of Johne's disease (JD) in cattle, and it has been suggested that this microorganism may be associated with Crohn's disease (CD) in humans (14, 17). M. avium subsp. paratuberculosis belongs to the mycobacterial species M. avium, which is currently subdivided into three subspecies (33): M. avium subsp. avium (synonym, M. avium), M. avium subsp. paratuberculosis (synonym, M. paratuberculosis), and M. avium subsp. silvaticum (synonym, M. silvaticum). At the subspecies level, M. avium subsp. paratuberculosis can be differentiated phenotypically from M. avium subsp. avium and M. avium subsp. silvaticum by its dependence on mycobactin (51) and genotypically by the presence of multiple copies of the insertion element IS900 (2, 5, 13, 22, 45).
JD (or paratuberculosis) is a chronic, granulomatous severe form of gastroenteritis with progressive weight loss and emaciation affecting domestic and wild ruminants, e.g., cattle, sheep, goats, red deer, and rabbits worldwide (12, 30, 31). Infected livestock periodically shed M. avium subsp. paratuberculosis via feces and milk, which results in environmental distribution, where M. avium subsp. paratuberculosis can survive for extended periods (55). Milk pasteurization trials showed that high-temperature and short-duration standard pasteurization procedures do not effectively kill M. avium subsp. paratuberculosis in milk, as clinical strains of M. avium subsp. paratuberculosis have been shown to be more thermally tolerant than either M. bovis or Coxiella burnetii, the current milk pasteurization standard microorganisms (10, 44, 47, 48). Therefore, human contact can result from the consumption of inadequately pasteurized milk or raw milk or of certain other dairy products, fecally contaminated vegetables, contaminated beef, or even water (12, 14, 34, 41, 47, 48).
CD is a chronic inflammatory disease of the gastrointestinal tract in humans, affecting in particular the terminal ileum, with symptoms of general malaise, weight loss, abdominal pain, and diarrhea (4, 12, 16, 40). A hallmark of CD is the histological proof of a granulomatous inflammation, which is also characteristic of JD and other mycobacterial diseases, leading to suggestions that M. avium subsp. paratuberculosis may be associated with CD in humans (see references 12 [and references therein], 8, 15, 16, 27, and 28).
Currently, there is controversy as to whether M. avium subsp. paratuberculosis (i) is an innocent bystander that has merely colonized the intestine of Crohn's patients, (ii) could be a secondary infection but not a cause of the disease, (iii) could be the primary infectious agent and the cause of CD, (iv) acts as a superantigen, or (v) modifies the immune response in CD (6, 12, 15, 25, 32, 37, 42, 43, 50). One of the major obstacles to resolution of the debate on the role of M. avium subsp. paratuberculosis in CD and the controversial studies published is the requirement of reliable and contemporary detection and identification of M. avium subsp. paratuberculosis in complex specimens such as blood, biopsy samples, breast milk, and feces. M. avium subsp. paratuberculosis is the slowest growing of the cultivable mycobacteria, and primary culture from clinical, veterinary, food, or environmental specimens can take 4 months or even longer (12, 24, 44, 55). Moreover, the characteristics of M. avium subsp. paratuberculosis in JD and in CD seem to be totally different: in CD, M. avium subsp. paratuberculosis appears as non-acid-fast coccobacilli with the ultrastructure of spheroplasts (cell-wall-deficient forms) that do not transform into characteristic M. avium subsp. paratuberculosis organisms until after several months of incubation (4).
In this study, we aimed at establishing highly specific multiple quantitative real-time PCR (qrt-PCR) assays based on the published genome sequence of M. avium subsp. paratuberculosis strain K-10 to enable rapid and unequivocal detection and identification of M. avium subsp. paratuberculosis directly from clinical, veterinary, food, or environmental specimens as well as from pure cultures. The method developed allows the quick and reliable detection and quantification of M. avium subsp. paratuberculosis directly from stool samples within 6 h at reasonable costs.
MATERIALS AND METHODS
Human clinical samples and data.
Patient fecal and tissue specimens represented clinical routine diagnostic samples for the detection of pathogenic bacteria and parasites. Testing for the presence of M. avium subsp. paratuberculosis DNA was performed in addition to DNA detection of conventional enteropathogenic bacteria and parasites. Positive results were reported. Clinical data from patients with positive results were obtained after informed consent by record review. The study was approved by the Ethics Board of the Justus-Liebig-University of Giessen, Faculty of Medicine.
Cattle samples.
Feces and gut biopsy specimens were obtained from healthy cows and from cows with suspected paratuberculosis. The symptoms of clinical paratuberculosis are chronic diarrhea and progressive weight loss, whereas subclinically infected animals mainly exhibit decreased milk production. The guts were dissected into small pieces and subdivided into inflamed and noninflamed tissue samples based on microscopic examination.
Microorganisms and standard cultivation.
The mycobacterial and nonmycobacterial species used in the study are listed here (see Table 2). The strains were obtained from the German Resource Center for Biological Material (DSMZ) and from the strain collection of the Institute of Medical Microbiology, Justus-Liebig University of Giessen.
Table 2.
Bacterial isolates (and their sources) used in this studya
| Bacterium | Source | PCR result with indicated amplicon |
|||||
|---|---|---|---|---|---|---|---|
| I | II | III | IV | V1 | V2 | ||
| M. avium subsp. paratuberculosis type strain | DSM 44133 | + | + | + | + | + | − |
| M. avium subsp. paratuberculosis | DSM 44135 | + | + | + | + | + | − |
| M. avium subsp. paratuberculosis (n = 40) | Field isolates | + | + | + | + | + | − |
| M. avium subsp. paratuberculosis | Patient isolate | + | + | + | + | + | − |
| M. avium subsp. avium | DSM 44158 | − | − | − | − | + | − |
| M. avium subsp. silvaticum | DSM 44175 | − | − | − | − | + | − |
| Mycobacterium tuberculosis (n = 10) | Patient isolates | − | − | − | − | + | − |
| Mycobacterium marinum | Patient isolate | − | − | − | − | + | − |
| Mycobacterium kansasii | Patient isolate | − | − | − | − | + | − |
| Mycobacterium chelonae | Patient isolate | − | − | − | − | + | − |
| Mycobacterium intracellulare | Patient isolate | − | − | − | − | + | − |
| Mycobacterium abscessus | Patient isolate | − | − | − | − | + | − |
| Staphylococcus aureus | Patient isolate | − | − | − | − | + | − |
| Streptococcus agalactiae | Patient isolate | − | − | − | − | + | − |
| Lactobacillus spp. (n = 5) | Patient isolates | − | − | − | − | + | − |
| Lactococcus lactis | Patient isolate | − | − | − | − | + | − |
| Enterococcus faecalis (n = 3) | Patient isolates | − | − | − | − | + | − |
| Enterococcus faecium | Patient isolate | − | − | − | − | + | − |
| Listeria monocytogenes | Patient isolate | − | − | − | − | + | − |
| Salmonella enteritidis (n = 2) | Patient isolates | − | − | − | − | + | − |
| Salmonella typhimurium (n = 2) | Patient isolates | − | − | − | − | + | − |
| Salmonella infantis | Patient isolate | − | − | − | − | + | − |
| Salmonella typhi | Patient isolate | − | − | − | − | + | − |
| Campylobacter jejuni | Patient isolate | − | − | − | − | + | − |
| Yersinia enterocolitica | Patient isolate | − | − | − | − | + | − |
| Shigella flexneri | Patient isolate | − | − | − | − | + | − |
| Enterohemorrhagic Escherichia coli (EHEC) | Patient isolate | − | − | − | − | + | − |
| Proteus mirabilis (n = 3) | Patient isolates | − | − | − | − | + | − |
| Citrobacter diversus (n = 2) | Patient isolates | − | − | − | − | + | − |
| Enterobacter cloacae | Patient isolate | − | − | − | − | + | − |
| Enterobacter sakazakii | Patient isolate | − | − | − | − | + | − |
| Escherichia coli | Patient isolate | − | − | − | − | + | − |
| Hafnia alvei | Patient isolate | − | − | − | − | + | − |
| Helicobacter pylori | Patient isolate | − | − | − | − | + | − |
| Clostridium difficile | Patient isolate | − | − | − | − | + | − |
| Fusobacterium nucleatum | Patient isolate | − | − | − | − | + | − |
| Propionibacterium acnes | Patient isolate | − | − | − | − | + | − |
| Bacteroides fragilis | Patient isolate | − | − | − | − | + | − |
| Prevotella intermedia | Patient isolate | − | − | − | − | + | − |
| Veillonella spp. | DSM 2008 | − | − | − | − | + | − |
| Atopobium spp. | DSM 15829 | − | − | − | − | + | − |
| Entamoeba histolytica | BNI | − | − | − | − | − | + |
| Giardia lamblia | BNI | − | − | − | − | − | + |
Positive results for amplicons I through V are indicated by a plus sign, and negative results are indicated by a minus sign. n, number of isolates; DSM, strain derived from the German Resource Center for Biological Material (DSMZ); BNI, parasite DNA obtained from the Bernhard-Nocht Institute for Tropical Medicine; amplicon I, IS900; amplicon II, MAP2765c (251); amplicon III, MAP0865 (f57); amplicon IV, MAP0865; amplicon V1, universal 16S rRNA gene; amplicon V2, universal 18S rRNA gene.
M. avium subsp. paratuberculosis was cultivated on BD BBL Herrold's egg yolk agar with Mycobactin J and ANV (Becton Dickinson, Heidelberg, Germany) and in modified Middlebrook 7H9 medium for up to 4 months as previously described (12, 39). All remaining bacteria were grown under optimal conditions on appropriate media as previously recommended (3).
Acid-fast staining.
The acid-fast staining of specimens was done by the Ziehl-Neelsen procedure, and stained specimens examined under conditions of oil immersion. Dissected biopsy samples from gut tissue of cows were stained as previously described for tissue sections of bisons (20).
DNA extraction.
DNA was extracted from pure bacterial cultures with a RTP Spin Bacteria DNA kit (Invitek, Berlin, Germany). The total DNA from human and animal fecal specimens was extracted with a PSP Spin Stool DNA kit and from tissue with an Invisorb Spin Tissue minikit (Invitek) as recommended by the vendor.
Parasitic DNA from Entamoeba histolytica and from Giardia lamblia was obtained from the Bernhard-Nocht Institute for Tropical Medicine (Hamburg, Germany).
Quantitative real-time PCR (qrt-PCR) and conventional PCR.
The new primers and probes used were designed with “Primer Express” version 1.0 software (Applied Biosystems, Foster City, CA). The internal probes were labeled with the fluorescent reporter dye 5-carboxyfluoroscein (FAM) on the 5′ end and the quencher dye N′,N′,N′,N′-tetramethyl-6-carboxyrhodamine (TAMRA) on the 3′ end. The qrt-PCR was accomplished as described previously (21, 38). Protozoal universal 18S small-subunit rRNA (SSU-rRNA) detection was performed by conventional PCR using eukaryote-specific primers (Table 1) as described previously (26). The amounts of specific target sequences present in unknown samples were calculated by measuring the threshold cycle (CT) values and using standard curves generated with a series of known quantities of target sequences. The CT value represented the cycle at which the copy of the amplified target sequence intersected the threshold or baseline. Briefly, an inoculation loop of the M. avium subsp. paratuberculosis DSM 44133 type strain grown on Herrold's egg yolk was carefully resuspended in phosphate-buffered saline (PBS) and serially diluted. The total amounts of cells were counted using a Neubauer counting chamber. We determined the total cell numbers, since complex biological material may contain both viable and dead bacteria.
Table 1.
Specific primers and TaqMan probes used in this study to detect M. avium subsp. paratuberculosis
| Target (GenBank accession no.) | Oligonucleotide sequence | Reference(s) or source |
|---|---|---|
| Amplicon I IS900 (AE016958.1) | ||
| Forward | 5′-AAT GAC GGT TAC GGA GGT GGT-3′ | 21, 22 |
| Reverse | 5′-GCA GTA ATG GTC GGC CTT ACC-3′ | |
| Probe | FAM-TCC ACG CCC GCC CAG ACA GG-TAMRA | |
| Amplicon II 251 (AF445445) | ||
| Forward | 5′-GCA AGA CGT TCA TGG GAA CT-3′ | 2, 38 |
| Reverse | 5′-GCG TAA CTC AGC GAA CAA CA-3′ | |
| Probe | FAM-CTG ACT TCA CGA TGC GGT TCT TC-TAMRA | |
| Amplicon III f57 (X70277) | ||
| Forward | 5′-TAC CGA ATG TTG TTG TCA CCG-3′ | 35, this study |
| Reverse | 5′-TGG CAC AGA CGA CCA TTC AA-3′ | |
| Probe | FAM-CCG GTC CCA GGT GTG TTC GAG TTG-TAMRA | |
| Amplicon IV MAP0865 (AE016958.1) | ||
| Forward | 5′-GCG CGG CCA GTA TGG ATA TA-3′ | 19, 22, this study |
| Reverse | 5′-GAC TCA ACC CAA CGA GCT CC-3′ | |
| Probe | FAM-AGA TGC CTC TCC GAT GCT CGA TGG-TAMRA | |
| Amplicon V1 16S rRNA gene | ||
| Universal forward | 5′-TCC TAC GGG AGG CAG CAG T-3′ | 23 |
| Universal reverse | 5′-GGA CTA CCA GGG TAT CTA ATC CTG TT-3′ | |
| Universal probe | FAM-CGT ATT ACC GCG GCT GCT GGC AC-TAMRA | |
| Amplicon V2 18S rRNA gene | ||
| Universal forward | 5′-GCG GAT CCG CGG CCG CTG GTT GAT CCT GCC AGT-3′ | 26 |
| Universal reverse | 5′-GCG GAT CCG CGG CCG CGG CAG GTT CAC CTA C-3′ |
Detection of Salmonella enterica serovar enteritidis, Campylobacter jejuni, Yersinia enterocolitica, Clostridium difficile, Entamoeba histolytica, and Giardia lamblia.
The total DNA of human fecal specimens was extracted with a PSP Spin Stool kit (Invitek) as described above. S. enteritidis, C. jejuni, Y. enterocolitica, C. difficile, E. histolytica, and G. lamblia were detected by specific real-time PCRs as previously described (18, 29, 52, 53, 54).
Multiple displacement amplification (MDA) of IS900-positive fecal specimens.
MDA was performed with the extracted DNA of amplicon I (IS900)-positive but amplicon II (MAP2765c [251])-, III (MAP0865 [f57])-, and IV (MAP0865)-negative fecal samples. A commercially available GenomiPhi DNA amplification kit (Amersham Biosciences, Uppsala, Sweden), which utilizes MDA to exponentially amplify genomic DNA, was used, following the manufacturer's instructions. Briefly, 1 μl of DNA extract was added to 9 μl of sample buffer containing random hexamer primers and heated to 95°C. The chilled sample was mixed with 9 μl of reaction buffer and 1 μl of enzyme mix. The mixture was incubated for 14 h at 30°C and afterwards subjected to heat inactivation for 10 min at 65°C.
Bioinformatics.
DNA sequences were aligned with MegAlign version 5.0 software (DNASTAR Inc., Madison, WI) and compared with sequences deposited in the GenBank, EMBL, DDBJ, and PDB databases using the BLASTn basic local alignment search tool (1). The published genome of M. avium subsp. paratuberculosis strain K-10 (22; GenBank accession number AE016958.1) was analyzed and depicted with the GenomeViz bioinformatics tool (11). The standard curves to calculate the amounts of M. avium subsp. paratuberculosis in complex stool and biopsy specimens were generated with SigmaPlot 2000 version 6.00 software (SPSS GmbH Software, Munich, Germany).
RESULTS
Development of MAP0865-specific real-time PCRs.
Fragment f57 (GenBank accession no. X70277) has been previously described and used as a highly specific 620-bp-long probe for detection of M. avium subsp. paratuberculosis and diagnosis of Johne's disease (35, 49). The nucleotide-nucleotide BLAST (BLASTn) analysis we performed revealed that it was located in gene MAP0865, one of the 4,350 predicted open reading frames (ORFs) of M. avium subsp. paratuberculosis strain K-10 (Fig. 1) (22). Fragment f57 covered 48.7% of this 1,272-bp-long ORF with an identity of 100% (Fig. 1). Another BLASTn analysis using the nucleotide sequence of ORF MAP0865 with at least 3.7 million sequences deposited in databases GenBank, EMBL, DDBJ, and PDB revealed it to be unique. Consensus was detected only with deposited MAP0865 and fragment f57 sequences.
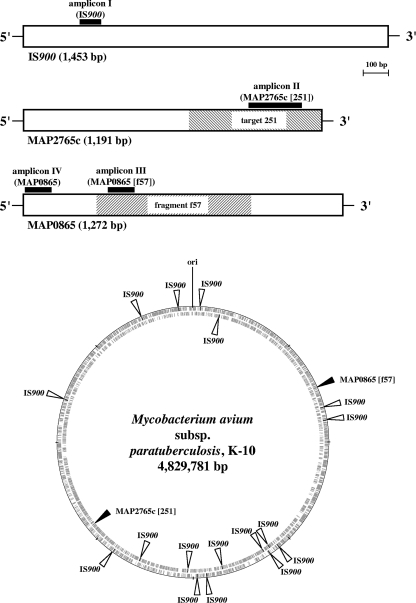
Fig. 1.
Graphic presentation of the M. avium subsp. paratuberculosis-specific chromosomal regions IS900, MAP2765c (target 251), and MAP0865 (fragment f57) and their positions in the M. avium subsp. paratuberculosis K-10 genome (GenBank accession no. AE016958.1). Outer circle, plus strand; inner circle, minus strand. The homology analyses and the graphic presentations were done using the methods BLAST, Clustal W (1), and GenomeViz (11). The black boxes represent specific TaqMan PCR amplicons I through IV, including forward and reverse primers and the internal probe labeled with FAM and TAMRA (Table 1). The amplicons were designed using Primer Express software (Applied Biosystems, Foster City, CA).
By the use of Primer Express software, two new TaqMan amplicons with primers and probes were designed for this M. avium subsp. paratuberculosis-specific MAP0865 ORF. The first one, designated amplicon III, covered both fragment f57 and ORF MAP0865, while the second, designated amplicon IV, covered only ORF MAP0865 (Fig. 1, Table 1).
Specificities of the MAP0865-specific real-time PCRs.
The specificities of the new real-time PCRs using amplicons III and IV were demonstrated by the analysis of DNA from panels of 40 M. avium subsp. paratuberculosis isolates, 17 other mycobacterial species, 13 Gram-positive bacterial species, 20 Gram-negative bacterial species (including 8 different species causing gastroenteritis), 7 obligate anaerobic bacterial species, and 2 intestinal parasite species (Table 2). The results showed that the real-time PCRs with amplicons III and IV, as well as those performed with previously developed amplicons I (IS900) and II (MAP2765c [251]), specifically amplified only the M. avium subsp. paratuberculosis DNA but not the DNA of the remaining bacteria (Table 2), indicating that the entire ORF MAP0865 is unique for M. avium subsp. paratuberculosis.
All DNA extracts gave positive results using 16S rRNA gene universal amplicon V1 for bacteria or 18S rRNA gene universal amplicon V2 for parasites (Table 2). The nucleic acid concentrations of the DNA extracts were calculated to be at ∼50 ng/μl.
Positions of element IS900, ORF MAP2765c (251), and ORF MAP0865 (f57) on the chromosome of M. avium subsp. paratuberculosis strain K-10.
A previously published genome-scale comparison of M. avium subsp. paratuberculosis with its closely related subspecies M. avium subsp. avium revealed potential new diagnostic sequences (2). Among these, target 251 with a length of 540 bp has been identified as a valuable new sequence for specific amplification of M. avium subsp. paratuberculosis DNA (38; GenBank accession no. AF445445). Nucleotide sequence alignments indicated that target 251 is located in gene MAP2765c of M. avium subsp. paratuberculosis strain K-10 (Fig. 1) (22).
Genome visualization using the GenomeViz bioinformatics tool (11) revealed a random distribution of 17 copies of IS900 on the M. avium subsp. paratuberculosis chromosome and that both ORF MAP0865 (f57) and ORF MAP2765c (251) exist as singular sequences at diametrically opposite positions (Fig. 1). Eleven copies of IS900 and MAP0865 (f57) were located on the positive strand, whereas the remaining six copies of IS900 and MAP2765c (251) were located on the negative strand (Fig. 1). This underlines the utility of IS900 as a highly sensitive PCR target for M. avium subsp. paratuberculosis due to its high copy number on the M. avium subsp. paratuberculosis chromosome. Nevertheless, ORFs MAP0865 and MAP2765c are highly specific PCR targets that can be used to identify M. avium subsp. paratuberculosis and to confirm IS900-specific PCRs.
Optimization of PCR conditions and annealing temperature of IS900-, MAP2765c (251)-, MAP0865 (f57)-, and MAP0865-specific real-time PCRs.
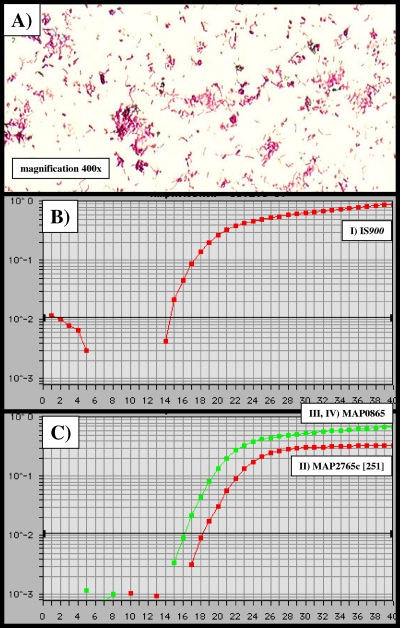
In order to determine the optimal PCR buffer conditions and the optimal primer annealing temperature for all four amplicons, we analyzed the efficiencies of the real-time PCRs with MgCl2 concentrations in the range of 1.0 to 5.0 mM and temperatures in the range of 55 to 65°C (data not shown). The DNA used was extracted from ∼108 cells of M. avium subsp. paratuberculosis, and the experiments were done in triplicate. For all amplicons (I through IV), the optimal MgCl2 concentration was 3.5 mM and the annealing temperature 57.8°C. The ideal results obtained consisted of a CT value of 14.4 for amplicon I (IS900), a CT value of 18.2 for amplicon II (MAP2765c [251]), and a CT value of 16.2 for amplicons III (MAP0865 [f57]) and IV (MAP0865) (Fig. 2). The order with respect to sensitivity and efficiency of the TaqMan-PCR was amplicon I first, amplicons III and IV second, and amplicon II third. In cases of amplicon I-positive but amplicon II- to IV-negative specimens, multiple displacement amplification (MDA) was used to exponentially amplify genomic DNA in the DNA extracts of the tested samples. The results suggest the use of MDA as a routine tool for amplification of specimens when low concentrations of M. avium subsp. paratuberculosis-specific DNA are suspected.
Fig. 2.
Temperature optimization and sensitivity of IS900-, MAP2765c-, and MAP0865-specific TaqMan PCRs. Using a DNA extract of ∼108 CFU of the M. avium subsp. paratuberculosis DSM 44133 type strain, the optimal annealing temperature for all amplicons I through IV was determined to be uniformly 57.8°C. (A) Ziehl-Neelsen staining of the M. avium subsp. paratuberculosis wild-type isolate obtained from feces of cow 7 (Table 3). (B and C) x axis, CT values; y axis, ΔRn (FAM reporter signal divided by the ROX [carboxy-X-rhodamine] passive reference signal). (B) amplicon I (IS900), CT value = 14.4; (C) amplicon II (MAP2765c [251]), CT value = 18.2; amplicons III (MAP0865 [f57]) and IV (MAP0865), CT value = 16.2.
The sensitivity of real-time PCR depends on the DNA extraction method used.
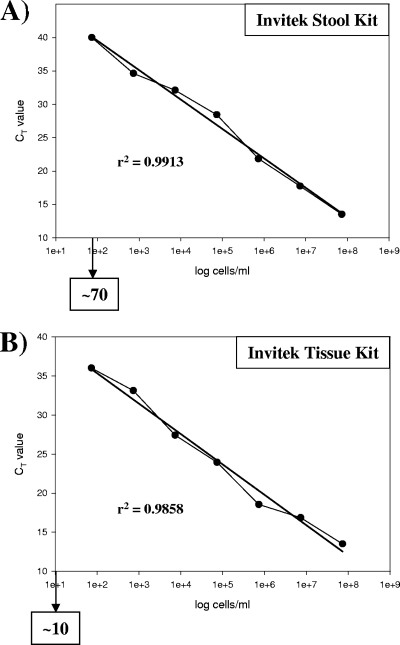
Since the real-time PCR with amplicon I (IS900) showed the highest efficiency, serial dilutions of the genomic DNA of M. avium subsp. paratuberculosis type strain DSM 44133 were used as templates to assess its sensitivity under the conditions mentioned above. The detection limit of the real-time PCR with amplicon I (IS900) was (theoretically) determined to be 1 to 10 CFU/reaction and therefore was identical to the results obtained previously (21). To identify M. avium subsp. paratuberculosis and to calculate directly the amount of the bacteria in complex biological material such as feces and tissue without cultivation, we assessed the efficiency of the DNA extraction methods with two commercially available kits: a stock solution of 7.3 × 107 M. avium subsp. paratuberculosis bacteria was serially diluted to 7.3 × 106, 7.3 × 105, 7.3 × 104, 7.3 × 103, 7.3 × 102, and 7.3 × 101 bacteria, and the DNA of the bacteria from each dilution step was extracted using commercially available kits for stool and tissue specimens (Invitek, Berlin, Germany). The stool kit had a detection limit of ≥70 cells/reaction, whereas the detection limit of the tissue kit was ≥10 cells/reaction. Both analyses confirmed very high correlation, as demonstrated by determinations of linear regression (r2) (Fig. 3).
Fig. 3.
Calculation of the detection limits for M. avium subsp. paratuberculosis by using commercially available DNA extraction kits and IS900 TaqMan PCR (amplicon I). The DNA of 1 ml of serial dilutions of the M. avium subsp. paratuberculosis DSM 44133 type strain was extracted to generate standard curves and to determine the linear regression (r2) and the detection limit (indicated by arrows) for each method. (A) Invitek stool kit (r2 = 0.9913); detection limit, ≥70 cells. (B) Invitek tissue kit (r2 = 0.9858); detection limit, ≥10 cells. The calculations were done with Sigma Plot software (SPSS Inc. Software, Munich, Germany).
Microscopic detection and cultivation of M. avium subsp. paratuberculosis in feces and gut tissue from cattle with Johne's disease.
We examined the feces and dissected gut tissue from 12 butchered cows by Ziehl-Neelsen staining and culture. Three cows were healthy, and the remaining nine were suspected to be afflicted with Johne's disease. Diseased cows 1 to 4, 6 to 9, and 12 showed acid-fast bacteria in both feces and inflamed gut tissue, and pure cultures of M. avium subsp. paratuberculosis could be obtained after 3 to 6 months of incubation (Fig. 2A, Table 3). The noninflamed gut tissue samples were uniformly negative for M. avium subsp. paratuberculosis. Healthy cows 5, 10, and 11 were negative for M. avium subsp. paratuberculosis in all samples examined (Table 3). All M. avium subsp. paratuberculosis isolates were identified by the specific real-time PCRs with amplicons I (IS900), II (MAP2765c [251]), III (MAP0865 [f57]), and IV (MAP0865). The growth of all M. avium subsp. paratuberculosis isolates was mycobactin dependent. Furthermore, the isolates were assigned to the M. avium complex by partial sequencing of the 16S rRNA genes and by performing a Genotype Mycobacterium CM/AS test (Hain Lifescience GmbH, Nehren, Germany).
Table 3.
Detection of M. avium subsp. paratuberculosis in bovine gut tissue and feces by amplicon I (IS900)-, amplicon II (MAP2765c [251])-, amplicon III (MAP0865 [f57])-, and amplicon IV (MAP0865)-specific TaqMan PCR, by acid-fast staining, and by culture (n = 12)a
| Cattle and specimen category | Acid-fast staining result | Culture result | TaqMan PCR result for indicated amplicon |
|||
|---|---|---|---|---|---|---|
| I | II | III | IV | |||
| Diseased (n = 9) | ||||||
| Noninflamed gut tissue | − | − | − | − | − | − |
| Inflamed gut tissue | + | + | + | + | + | + |
| Feces | + | + | + | + | + | + |
| Healthy (n = 3) | ||||||
| Noninflamed gut tissue | − | − | − | − | − | − |
| Feces | − | − | − | − | − | − |
Diseased cattle (n = 9) were suspected to be infected with M. avium subsp. paratuberculosis because of typical symptoms of Johne's disease (paratuberculosis).
Evaluation of the M. avium subsp. paratuberculosis-specific real-time PCRs using original bovine gut tissue and feces.
The total DNA from inflamed and noninflamed dissected gut tissue and from feces of healthy and diseased cows was extracted with commercially available kits for tissue and stool specimens (Invitek, Berlin, Germany). All of the diseased cows with positive M. avium subsp. paratuberculosis culture results were also positive for specific real-time PCR amplicons I to IV (Table 3). Using the standard curves (Fig. 3), the amount of M. avium subsp. paratuberculosis was calculated to be in the range of ∼2 × 104 to 6 × 107 bacteria per gram of inflamed gut tissue and ∼1 × 107 to 2 × 109 bacteria per gram of feces. Thus, the concentration of M. avium subsp. paratuberculosis in feces was continuously ∼30- to 500-fold higher than in inflamed parts of the intestinal tissue. All of the tissue samples from healthy M. avium subsp. paratuberculosis culture-negative cows as well as the noninflamed tissue samples of diseased cows were also negative in the M. avium subsp. paratuberculosis-specific real-time PCRs (Table 3).
Detection of M. avium subsp. paratuberculosis in stool specimens and gut tissue of patients with diarrhea.
We examined consecutive stool specimens of 1,293 hospitalized patients with mild or severe symptoms of diarrhea for the presence of S. enteritidis, C. jejuni, Y. enterocolitica, C. difficile, E. histolytica, G. lamblia, and M. avium subsp. paratuberculosis. The total DNA was extracted with an Invitek stool kit, and the M. avium subsp. paratuberculosis analysis was done with highly sensitive amplicon I (IS900). Twenty-seven patients (2.09%) gave positive results, and the concentrations of M. avium subsp. paratuberculosis were calculated in the range of ∼5 × 102 to 5 × 103 bacteria per gram of stool by the use of the corresponding standard curve (Fig. 3A). The results were confirmed by using amplicons II through IV. For 6 patients, the results for amplicons II to IV remained negative following initial amplification despite a positive amplicon I result, suggesting the presence of non-M. avium subsp. paratuberculosis mycobacterial DNA. However, following MDA, the presence of M. avium subsp. paratuberculosis was then confirmed by positive PCR results for amplicons II to IV for all 6 specimens. All of the 26 M. avium subsp. paratuberculosis-positive patients gave negative test results for S. enteritidis, C. jejuni, Y. enterocolitica, E. histolytica, and G. lamblia. Only one of these patients, who suffered from pseudomembranous colitis, subsequently tested positive for C. difficile.
Among the 1,293 patients, we identified 11 patients with chronic inflammatory bowel disease. Of these, 6 patients had clinically confirmed CD and an additional 5 were classified with ulcerative colitis (UC). Only two of the 27 M. avium subsp. paratuberculosis-positive individuals were CD patients. Additionally, a small piece of gut tissue from one CD patient also gave M. avium subsp. paratuberculosis-positive test results. The identification was confirmed by reamplification with amplicons II through IV, acid-fast staining, and culture. The remaining four CD and five UC patients gave negative results for M. avium subsp. paratuberculosis.
DISCUSSION
The most prominent target used in several studies to detect DNA of M. avium subsp. paratuberculosis by PCR is the insertion element IS900 (19, 21). The multicopy (17 copies) nature of the sequence on the M. avium subsp. paratuberculosis chromosome makes it ideal as a target sequence for the detection of M. avium subsp. paratuberculosis, since it exhibits a higher level of sensitivity compared to the use of single-copy genes as targets (22, 45). We analyzed consecutive stool specimens of 1,293 hospitalized patients by the use of target IS900. Twenty-seven (2.09%) of the cohort gave positive test results for IS900. Only two of these patients suffered from CD. The bacterial load was persistently low and was calculated in the range of 500 to 5,000 M. avium subsp. paratuberculosis bacteria per gram of stool. In addition, an analyzed section of gut tissue from one CD patient was also positive for IS900 and the bacterium could be isolated by culture. It was not possible to isolate M. avium subsp. paratuberculosis from the 26 other patients, since the analyses were performed retrospectively with stored DNA extracts.
Unfortunately, the specificity of target IS900 is not 100%, since IS900 insertion elements with close sequence homology are also present on the chromosomes of M. cookii, M. marinum, M. paraffinicum, and M. scrofulaceum isolates (7, 9, 21, 37, 40). Furthermore, polymorphisms detected in IS900 as variants of M. avium subsp. paratuberculosis have been previously described; such variants should be interpreted as suggestive of the presence of a Mycobacterium organism other than M. avium subsp. paratuberculosis until the detection has been confirmed by independent methods (38). Therefore, to enhance the specificity of M. avium subsp. paratuberculosis detection, it is indispensable to use multiple M. avium subsp. paratuberculosis-specific targets. So far, several specific targets have been used by employing different techniques: IS900 and target 251 by real-time PCR (21, 38, 44, 49), ISMap02 by a nested PCR method (46), and f57 sequences by hybridization and by PCR (35, 44, 49). Also, the completed genome sequence of M. avium subsp. paratuberculosis strain K-10 and comparative genome analysis with the closely related species M. avium subsp. avium, including experimental studies to identify M. avium subsp. paratuberculosis-specific genomic regions, revealed miscellaneous potential new diagnostic targets (2, 19, 22, 35, 38). In order to improve detection, we used several M. avium subsp. paratuberculosis-specific targets in one assay for detection and also used quantitative TaqMan real-time PCR, since the technology is highly sensitive and specific.
For a multiple real-time PCR assay, we chose IS900, target 251, and the f57 sequence, which are randomly distributed on the M. avium subsp. paratuberculosis chromosome (Fig. 1, bottom). When BLASTn analysis and bioinformatic GenomeViz software were used, target 251 and sequence f57 were found in genes MAP2765c and MAP0865 of the published M. avium subsp. paratuberculosis K-10 strain, respectively (Fig. 1, top). Computer-aided analysis of the entire MAP0865 ORF revealed that it was unique for M. avium subsp. paratuberculosis, since no corresponding sequences were detected in publicly available databases. To establish real-time PCR assays for this sequence, two new TaqMan targets were generated: amplicon III, located in the f57 sequence, and amplicon IV, located at the 5′ end of ORF MAP0865 (Fig. 1, top). Each PCR was run separately and independently in corresponding unique microtiter wells but under the same conditions with respect to PCR buffer and temperature profiles. Even though the conditions were identical, the results showed that the sensitivities of the individual PCRs were different (Fig. 2B and C). The highest sensitivity was obtained with the IS900 PCR and the lowest with the MAP2765c (251) PCR. Amplicons III (MAP0865 [f57]) and IV (MAP0865) showed identical levels of efficacy and intermediate levels of sensitivity between those of IS900 PCR and MAP2765c (251) PCR. Because of the identical efficacy results determined under the applied conditions, only one curve, representing both amplicons, is depicted (Fig. 2C). The high sensitivity of the IS900 PCR is attributed to the multicopy nature of this element, which is present as 17 copies on the M. avium subsp. paratuberculosis K-10 chromosome (Fig. 1, bottom). The sensitivities determined for amplicons II through IV were lower, since ORFs MAP2765c and MAP0865 are singular targets (Fig. 1, bottom). We also attribute the lower sensitivity of the MAP2765c PCR to its size (203 bp), which affected the efficacy of the PCR methods used. The recommended size for TaqMan-based amplicons is 70 to 150 bp (36), and both amplicons III and IV of MAP0865 represent an optimal size of 101 bp, which resulted in identical efficacy results for these independent PCRs.
The specificities of newly designed quantitative real-time PCR amplicons III and IV, along with those of the previously described specific amplicons I and II, were demonstrated using 2 M. avium subsp. paratuberculosis type strains, 40 field isolates of M. avium subsp. paratuberculosis (human, animal, and environmental origin), and 13 species of Gram-positive bacteria, 20 species of Gram-negative bacteria, 7 species of anaerobic bacteria, and 2 species of intestinal parasites (Table 2). Positive signals were obtained only for the M. avium subsp. paratuberculosis strains; all of the other microorganisms tested gave uniformly negative results. The results for the pure microorganisms were achieved with DNA extracted by an RTP Spin Bacteria kit. Since stool and tissue specimens are complex and represent difficult samples for DNA extraction and PCR with respect to potential DNA-degrading enzymes and PCR inhibitors and to standardize methodology, we used commercially available stool and tissue kits from Invitek (see Materials and Methods). DNA extraction is generally achieved within 1 h. The detection limits for M. avium subsp. paratuberculosis determined using the tissue and the stool kits for DNA extraction of tissue and stool specimens were marginally above the theoretical detection limit of 1 to 10 CFU and determined to be ∼10 CFU and ∼70 CFU, respectively (Fig. 3).
The applicability of the DNA extraction methods and the specific quantitative real-time PCR assays used was assessed by acid-fast staining and by culture using tissue and stool specimens of three healthy cattle and of nine diseased cattle with Johne's disease. The real-time PCR results were 100% identical to the results of microscopic analysis and the culture (Table 3) but could be accomplished in 6 h. The quantification obtained by qrt-PCR showed that the examined cattle shed very large amounts of M. avium subsp. paratuberculosis calculated to be in the range of approximately 1 × 107 to 2 × 109 bacteria per gram of feces, which was ∼20,000- through ∼400,000-fold higher than in human feces. Fecal samples from cattle are easily obtained and are therefore superior to other specimens such as biopsy specimens or blood samples for analysis. In particular, the results we obtained from gut tissue of infected cattle strongly depended on the biopsy specimens used. Noninflamed gut tissue from diseased cattle invariably gave negative results, whereas inflamed gut tissue gave positive results (Table 3).
The presence of M. avium subsp. paratuberculosis in human stool specimens and one biopsy specimen as detected with target IS900 was confirmed by reamplifying the samples with amplicons II through IV. This means that all samples that gave IS900-positive test results truly represented M. avium subsp. paratuberculosis. We are well aware that our human samples were single snapshots randomly collected and want to emphasize that our results for CD and UC patients are not meant to be representative. However, the results clearly demonstrate that the technology of DNA extraction and qrt-PCR utilized is appropriate for analysis of stool specimens and gut tissue for the presence of M. avium subsp. paratuberculosis and could therefore substantially contribute to the current debate about the role of M. avium subsp. paratuberculosis in CD. Further studies are necessary, and we suggest using several consecutive stool specimens, which are noninvasive compared to biopsy specimens and with which we were able to demonstrate high sensitivity and specificity for the detection of M. avium subsp. paratuberculosis.
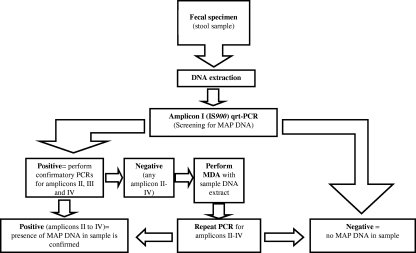
In conclusion, we demonstrate the development of qrt-PCR amplicons III and IV, which, in combination with amplicons I and II, enable unequivocal detection, identification, and quantification of M. avium subsp. paratuberculosis directly from clinical, veterinary, food, and environmental specimens as well as from pure cultures. For IS900-positive but amplicon II- to IV-negative specimens, we suggest the use of MDA and the consecutive repetition of the confirmatory PCRs. A suggested workflow is shown in Fig. 4. Fecal specimens are true alternatives for the analysis of patients and cattle because of their noninvasiveness and simplicity in collection. Our data presented here provide a basis for further structured studies of the potential role of M. avium subsp. paratuberculosis in CD and other human diseases.
Fig. 4.
Suggested workflow for the detection of M. avium subsp. paratuberculosis DNA in stool samples. MDA, multiple displacement amplification. Amplicon I = IS900; amplicon II = MAP2765c (251); amplicon III = MAP0865 (f57); amplicon IV = MAP0865.
ACKNOWLEDGMENTS
We thank Martina Klös-Langsdorf and Kirsten-Susann Bommersheim for excellent technical assistance, Isabell Krabs for the preparation of the gut tissue of slaughtered cows, Simone Ries for running qrt-PCRs during a practical course, and Egbert Tannich from the Bernhard-Nocht Institute for Tropical Medicine (Hamburg, Germany) for providing parasite DNA obtained from Entamoeba histolytica and from Giardia lamblia.
This work was supported by grants from the Bundesministerium fuer Bildung und Forschung, Germany, within the framework of the National Genome Research Network (NGFN) (contract no. 01GS0401).
Footnotes
Published ahead of print on 23 March 2011.
REFERENCES
- 1. Altschul S. F., Gish W., Miller W., Myers E. W., Lipman D. J. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403–410 [DOI] [PubMed] [Google Scholar]
- 2. Bannantine J., Baechler P. E., Zhang Q., Li L., Kapur V. 2002. Genome scale comparison of Mycobacterium avium subsp. paratuberculosis with Mycobacterium avium subsp. avium reveals potential diagnostic sequences. J. Clin. Microbiol. 40:1303–1310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Chapin K. C., Lauderdale T.-L. 2007. Reagents, stains, and media: bacteriology, p. 334–364 In Murray P. R., Baron E. J., Jorgensen J. H., Landry M. L., Pfaller M. A. (ed.), Manual of clinical microbiology, 9th ed. ASM Press, Washington, DC [Google Scholar]
- 4. Chiodini R. J. 1989. Crohn's disease and the mycobacterioses: a review and comparison of two disease entities. Clin. Microbiol. Rev. 2:90–117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Collins D. M., de Lisle G. W. 1986. Restriction endonuclease analysis of various strains of Mycobacterium paratuberculosis isolated from cattle. Am. J. Vet. Res. 47:2226–2229 [PubMed] [Google Scholar]
- 6. Committee on Diagnosis and Control of Johne's Disease 2003. Johne's disease and Crohn's disease, p. 104–120 In Diagnosis and control of Johne's disease: report for the National Research Council of the National Academy of Science. The National Academic Press, Washington, DC [Google Scholar]
- 7. Cousins D. V., et al. 1999. Mycobacteria distinct from Mycobacterium avium subsp. paratuberculosis isolated from the faeces of ruminants possess IS900-like sequences detectable IS900 polymerase chain reaction: implications for diagnosis. Mol. Cell Probes 13:431–442 [DOI] [PubMed] [Google Scholar]
- 8. Dalziel T. K. 1913. Chronic interstitial enteritis. Br. Med. J. 2:1068–1070 [Google Scholar]
- 9. Englund S., Bolske G., Johansson K. E. 2002. An IS900-like sequence found in a Mycobacterium sp. other than Mycobacterium avium subsp. paratuberculosis. FEMS Microbiol. Lett. 209:267–271 [DOI] [PubMed] [Google Scholar]
- 10. Gao A., Mutharia L., Chen S., Rahn K., Odumeru J. 2002. Effect of pasteurisation on survival of Mycobacterium paratuberculosis in milk. J. Dairy Sci. 85:3198–3205 [DOI] [PubMed] [Google Scholar]
- 11. Ghai R., Hain T., Chakraborty T. 2004. GenomeViz: visualizing microbial genomes. BMC Bioinformatics 5:198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Grant I. R. 2005. Zoonotic potential of Mycobacterium avium subsp. paratuberculosis: the current position. J. Appl. Microbiol. 98:1282–1293 [DOI] [PubMed] [Google Scholar]
- 13. Green E. P., et al. 1989. Sequence and characteristics of IS900, an insertion element identified in a human Crohn's disease isolate of Mycobacterium paratuberculosis. Nucleic Acids Res. 17:9063–9073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Harris N. B., Barletta R. G. 2001. Mycobacterium avium subsp. paratuberculosis in veterinary medicine. Clin. Microbiol. Rev. 14:489–512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hermon-Taylor J. 2001. Protagonist: Mycobacterium avium subspecies paratuberculosis is a cause of Crohn's disease. Gut 49:755–756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hermon-Taylor J., Bull T. J. 2002. Crohn's disease caused by Mycobacterium avium subsp. paratuberculosis: a public health tragedy whose resolution is long overdue. J. Med. Microbiol. 51:3–6 [DOI] [PubMed] [Google Scholar]
- 17. Hermon-Taylor J., et al. 2000. Causation of Crohn's disease by Mycobacterium avium subspecies paratuberculosis. Can. J. Gastroenterol. 14:521–539 [DOI] [PubMed] [Google Scholar]
- 18. Hoorfar J., Ahrens P., Radström P. 2000. Automated 5′ nuclease PCR assay for identification of Salmonella enterica. J. Clin. Microbiol. 38:3429–3435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hruska K., Bartos M., Kralik P., Pavlik I. 2005. Mycobacterium avium subsp. paratuberculosis in powdered infant milk: paratuberculosis in cattle—the public health problem to be solved. Vet. Med.-Czech 50:327–335 [Google Scholar]
- 20. Huntley J. F. J., Whitlock R. H., Bannantine J. P., Stabel J. R. 2005. Comparison of diagnostic detection methods for M. avium subsp. paratuberculosis in North American bison. Vet. Pathol. 42:42–51 [DOI] [PubMed] [Google Scholar]
- 21. Kim S. G., et al. 2002. Development and application of quantitative polymerase chain reaction assay based on the ABI 7700 system (TaqMan) for detection and quantification of Mycobacterium avium subsp. paratuberculosis. J. Vet. Diagn. Invest. 14:126–131 [DOI] [PubMed] [Google Scholar]
- 22. Li L., et al. 2005. The complete genome sequence of Mycobacterium avium subspecies paratuberculosis. Proc. Natl. Acad. Sci. U. S. A. 102:12344–12349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Martin F. E., Nadkami M. A., Jacques N. A., Hunter N. 2002. Quantitative microbiological study of human carious dentine by culture and real-time PCR: association of anaerobes with histopathological changes in chronic pulpitis. J. Clin. Microbiol. 40:1698–1704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. McFadden J. J., Butcher P. D., Chiodini R. J., Hermon-Taylor J. 1987. Crohn's disease-isolated mycobacteria are identical to Mycobacterium paratuberculosis, as determined by DNA probes that distinguish between mycobacterial species. J. Clin. Microbiol. 25:796–801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Mendoza J. L., Lana R., Díaz-Rubio M. 2009. Mycobacterium avium subspecies paratuberculosis and its relationship with Crohn's disease. World J. Gastroenterol. 15:417–422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Müller A., et al. 2000. Detection of Isospora belli by polymerase chain reaction using primers based on small-subunit ribosomal RNA sequences. Eur. J. Clin. Microbiol. Infect. Dis. 19:631–634 [DOI] [PubMed] [Google Scholar]
- 27. Naser S. A., Schwartz D., Shafran I. 2000. Isolation of Mycobacterium avium subsp. paratuberculosis from breast milk of Crohn's disease patients. Am. J. Gastroenterol. 95:1094–1095 [DOI] [PubMed] [Google Scholar]
- 28. Naser S. A., Ghobrial G., Romero C., Valentine J. F. 2004. Culture of Mycobacterium avium subspecies paratuberculosis from the blood of patients with Crohn's disease. Lancet 364:1039–1044 [DOI] [PubMed] [Google Scholar]
- 29. Nogva H. K., Bergh A., Holck A., Rudi K. 2000. Application of the 5′-nuclease PCR assay in evaluation and development of methods for quantitative detection of Campylobacter jejuni. Appl. Environ. Microbiol. 66:4029–4036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Olsen I., Sigurgardottir G., Djonne B. 2002. Paratuberculosis with special reference to cattle. Vet. Q. 24:12–28 [DOI] [PubMed] [Google Scholar]
- 31. Ott S. L., Wells S. J., Wagner B. A. 1999. Herd-level economic losses associated with Johne's disease on US dairy operations. Prev. Vet. Med. 40:179–192 [DOI] [PubMed] [Google Scholar]
- 32. Parrish N. M., et al. 2009. Absence of Mycobacterium avium subsp. paratuberculosis in Crohn's patients. Inflamm. Bowel Dis. 15:558–565 [DOI] [PubMed] [Google Scholar]
- 33. Pfyffer G. E. 2007. Mycobacterium: general characteristics, laboratory detection, and staining procedures, p. 543–572 In Murray P. R., Baron E. J., Jorgensen J. H., Landry M. L., Pfaller M. A. (ed.), Manual of clinical microbiology, 9th ed. American Society for Microbiology Press, Washington, DC [Google Scholar]
- 34. Pierce E. S. 2009. Possible transmission of Mycobacterium avium subspecies paratuberculosis through potable water: lessons from an urban cluster of Crohn's disease. Gut Pathog. 23:17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Poupart P., Coene M., Vanheuverswyn H., Cocito C. 1993. Preparation of a specific RNA probe for detection of Mycobacterium paratuberculosis and diagnosis of Johne's disease. J. Clin. Microbiol. 31:1601–1605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Proudnikov D., et al. 2003. Optimizing primer-probe design for fluorescent PCR. J. Neurosci. Methods 123:31–45 [DOI] [PubMed] [Google Scholar]
- 37. Quirke P. 2001. Antagonist: Mycobacterium avium subspecies paratuberculosis is a cause of Crohn's disease. Gut 49:757–760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Rajeev S., Zhang Y., Sreevatsan S., Motiwala A. S., Byrum B. 2005. Evaluation of multiple genomic targets for identification and confirmation of Mycobacterium avium subsp. paratuberculosis isolates using real-time PCR. Vet. Microbiol. 105:215–221 [DOI] [PubMed] [Google Scholar]
- 39. Schwartz D., et al. 2000. Use of short-term culture for identification of M. avium subsp. paratuberculosis in tissue from Crohn's disease patients. Clin. Microbiol. Infect. 6:303–307 [DOI] [PubMed] [Google Scholar]
- 40. Semret M., Turenne C. Y., Behr M. A. 2006. Insertion sequence IS900 revisited. J. Clin. Microbiol. 44:1081–1083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Shankar H., et al. 2010. Presence, characterization, and genotype profiles of Mycobacterium avium subspecies paratuberculosis from unpasteurized individual and pooled milk, commercial pasteurized milk, and milk products in India by culture, PCR, and PCR-REA methods. Int. J. Infect. Dis. 14:121–126 [DOI] [PubMed] [Google Scholar]
- 42. Shivananda S., et al. 1996. Incidence of inflammatory bowel disease across Europe: is there a difference between north and south? Results of the European Collaborative Study in Inflammatory Bowel Disease (EC-IBD). Gut 39:690–697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Sibartie S., et al. 2010. Mycobacterium avium subsp. paratuberculosis (MAP) as a modifying factor in Crohn's disease. Inflamm. Bowel Dis. 16:296–304 [DOI] [PubMed] [Google Scholar]
- 44. Slana I., Liapi M., Moravkova M., Kralova A., Pavlik I. 2009. Mycobacterium avium subsp. paratuberculosis in cow bulk tank milk in Cyprus detected by culture and quantitative IS900 and F57 real-time PCR. Prev. Vet. Med. 89:223–226 [DOI] [PubMed] [Google Scholar]
- 45. Soumya M. P., Pillai R. M., Antony P. X., Mukhopadhyay H. K., Rao V. N. 2009. Comparison of faecal culture and IS900 PCR assay for the detection of Mycobacterium avium subsp. paratuberculosis in bovine faecal samples. Vet. Res. Commun. 33:781–791 [DOI] [PubMed] [Google Scholar]
- 46. Stabel J. R., Bannantine J. P. 2005. Development of a nested PCR method targeting a unique multicopy element, ISMap02, for detection of Mycobacterium avium subsp. paratuberculosis in fecal samples. J. Clin. Microbiol. 43:4744–4750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Sung N., Collins M. T. 1998. Thermal tolerance of Mycobacterium paratuberculosis. Appl. Environ. Microbiol. 64:999–1005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Sung N., Collins M. T. 2000. Effect of three factors in cheese production (pH, salt and heat) on Mycobacterium avium subsp. paratuberculosis viability. Appl. Environ. Microbiol. 66:1334–1339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Tasara T., Stephan R. 2005. Development of an F57 sequence-based real-time PCR assay for detection of M. avium subsp. paratuberculosis in milk. Appl. Environ. Microbiol. 71:5957–5968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Thomas Dow C. 2008. Cows, Crohn's and more: is Mycobacterium paratuberculosis a superantigen? Med. Hypotheses 71:858–861 [DOI] [PubMed] [Google Scholar]
- 51. Thorel M. F., Krichevsky M., Levy-Frebault V. V. 1990. Numerical taxonomy of mycobactin-dependent mycobacteria, emended description of Mycobacterium avium, and description of Mycobacterium avium subsp. avium subsp. nov., Mycobacterium avium subsp. paratuberculosis subsp. nov., and Mycobacterium avium subsp. silvaticum subsp. nov. Int. J. Syst. Bacteriol. 40:254–260 [DOI] [PubMed] [Google Scholar]
- 52. van den Berg R. J., et al. 2005. Prospective multicenter evaluation of a new immunoassay and real-time PCR for rapid diagnosis of Clostridium difficile-associated diarrhea in hospitalized patients. J. Clin. Microbiol. 43:5338–5340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Verweij J. J., et al. 2004. Simultaneous detection of Entamoeba histolytica, Giardia lamblia, and Cryptosporidium parvum in fecal samples by using multiplex real-time PCR. J. Clin. Microbiol. 42:1220–1223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Vishnubhatla A., et al. 2000. Rapid 5′ nuclease (TaqMan) assay for detection of virulent strains of Yersinia enterocolitica. Appl. Environ. Microbiol. 66:4131–4135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Whittington R. J., Marshall D. J., Nicholls P. J., Marsh I. B., Reddacliff L. A. 2004. Survival and dormancy of Mycobacterium avium subsp. paratuberculosis in the environment. Appl. Environ. Microbiol. 70:2989–3004 [DOI] [PMC free article] [PubMed] [Google Scholar]