Abstract
Information about the genotype of varicella-zoster virus (VZV) is useful to monitor outbreaks of vaccine strains. However, in South Korea, where varicella vaccine was introduced in 1988, there are limited data about the genotype of VZV. VZV was isolated from vesicular lesions of patients with herpes zoster or varicella in South Korea between January 2007 and June 2009. DNAs were purified from single-passage isolates. The genotype was determined by sequence analysis of open reading frame (ORF) 22. The PstI restriction enzyme site in ORF 38 and the BglI restriction enzyme site in ORF 54 were evaluated by restriction enzyme analysis. Forty-four patients with herpes zoster and nine patients with varicella were enrolled. The median age of patients with herpes zoster was 59.5 (range, 10 to 77) years, and the median age of patients with varicella was 8 (range, 6 to 9) years. In sequence analysis of ORF 22, all isolates were genotype J, irrespective of the age group. In restriction enzyme analysis, 51 of 54 (94.3%) isolates contained a PstI site in ORF 38, and all isolates contained a BglI site in ORF 54. Our data suggest that genotype J has been circulating since the 1940s in South Korea.
INTRODUCTION
Varicella-zoster virus (VZV) is a human herpesvirus that causes chicken pox upon primary infection. Following primary infection, VZV establishes latency in sensory nerve ganglia. When VZV-specific cell-mediated immunity declines, VZV reactivates and manifests as herpes zoster (8). In the prevaccination era, varicella affected almost everyone, with 90% of cases occurring before the age of 15 years (1, 18). In the early 1970s, varicella vaccine, which was composed of the Oka strain of live, attenuated VZV, was developed in Japan (24, 25). Varicella vaccine was first licensed for use in European countries and Japan in the 1980s (2, 19). Since the introduction of varicella vaccine in the United States in 1995, the incidence of varicella and varicella-related deaths has decreased significantly (17, 23).
Since varicella vaccine was a live, attenuated vaccine, varicella-like rashes after vaccination were a problem. Postlicensure safety surveillance revealed 3,640 cases of vesicular rash (37.4/100,000 doses). The genotyping of VZV was useful to distinguish vaccine strains from wild strains (13, 20). In earlier studies, vaccine strains were identified through restriction fragment length polymorphism (RFLP) analysis of the PstI restriction enzyme site in open reading frame (ORF) 38 and the BglI restriction enzyme site in ORF 54 (11). RFLP analysis showed that wild isolates from the United Kingdom and from the United States were PstI+ in ORF 38 and BglI− in ORF 54. The Oka vaccine strain (vOka) was PstI− in ORF 38 and BglI+ in ORF 54 (10, 11). The PstI− BglI+ strain was believed to be unique to Oka varicella vaccine and no longer circulating. Therefore, RFLP seemed to distinguish wild-type VZV and vaccine strains. However, Loparev et al. reported that three wild-type VZV strains with the PstI− BglI+ profile of the Japanese Oka vaccine strain genotype were isolated in Hawaii in 2002 (14). Therefore, RFLP analysis of ORF 38 and ORF 54 could not distinguish vaccine strains from wild-type strains.
In 2000, RFLP analysis of the SmaI restriction enzyme site in ORF 62 was developed and distinguished the Oka vaccine strain from wild-type strains (13). In 2004, a novel strategy for VZV genotyping based on the sequencing of ORF 22 was developed, and VZV strains from different areas of the world are classified into three major genotypes, E (European), J (Japanese), and M (mosaic) (14). The major reference strains are Dumas and Oka parental. The Dumas strain was found in the Netherland in the 1970s and fully sequenced in 1986. It was the first fully sequenced VZV strain and the representative of the E genotype (5, 6, 15).
In South Korea, varicella vaccine was licensed in 1988 and has been included in national immunization recommendations since 2005 (19). The representative vaccines licensed in Korea are Varilix, Varivax, and Sudavax (3). Varilix and Varivax are made from the Oka strain. Sudavax is made from the MAV/06 strain, which originated from a Korean varicella patient (9, 7, 16). However, until recently, there was limited study to determine the genotype of circulating VZV in South Korea. We previously reported the genotype of VZV isolated from elderly zoster patients (4). The VZV isolates from 22 adults over 60 years old were all of the J genotype. To evaluate whether the genotype changed over time, in the current study, we included VZV isolated from zoster patients younger than 60 years of age, as well as from children with varicella. We aimed to determine the genotype of VZV in South Korea.
(This study was presented in part at the 2nd Vaccine Congress, Boston, MA, 2008.)
MATERIALS AND METHODS
Patients.
The specimens for VZV genotyping were prospectively collected from vesicular lesions of ambulatory or hospitalized patients with clinically diagnosed varicella or herpes zoster from February 2007 to June 2009. All specimens were from clinically diagnosed cases seen in three secondary or tertiary care hospitals and one pediatrics clinic. We obtained data, including patients' ages, sex, underlying diseases, involved dermatome, and extent of rashes. The age groups were classified at intervals of 10 years.
Isolation and genotyping of VZV. (i) Isolation of VZV.
A sterile swab and a syringe were used to aspirate and scrub an unroofed vesicle. Specimens were placed in sterile tubes. Viruses were propagated in MRC-5 human lung fibroblasts at 37°C with 5% CO2 until there were ≥25% cytopathic effects. Single-passage isolates were collected and stored at −80°C until subsequent processes were performed.
(ii) Genotyping of VZV isolates. (a) DNA preparation.
DNAs were purified from the lysates of VZV-infected MRC-5 cells using the QIAamp DNA Mini Kit (Qiagen, Hilden, Germany) according to the manufacturer's instructions.
(b) Evaluation of ORF 38 (PstI), ORF 54 (BglI), and ORF 62 (SmaI) single-nucleotide polymorphisms.
To characterize the Oka vaccine strain and distinguish it from wild-type VZV, conventional PCR targeting ORF 38, ORF 54, and ORF 62 were performed, followed by restriction endonuclease digestion with PstI, BglI, and SmaI, respectively (11, 13). Nla (5′-GGA ACC CCT GCA CCA TTA AA-3′)/Fok (5′-TCC CTT CAT GCC CGT TAC AT-3′) and Pst A (5′-TTG AAC AAT CAC GAA CCG TT-3′)/Pst B (5′-CGG GTG AAC CGT ATT CTG AG-3′) were used to amplify ORFs 54 and 38, respectively. DNA amplification reactions were performed with an MJ Research PTC-100 Thermal Cycler (MJ Research, Waltham, MA) in 50-μl reaction volumes, using AccuPower PCR PreMix (Bioneer, Daejeon, South Korea; 2.5 U Top DNA polymerase, 1.5 mM MgCl2, and a 250 μM concentration of each deoxynucleotide triphosphate), a 100 pM concentration of each primer, and 5 to 10 μl of VZV DNA extracts under the conditions previously described (11). The amplification products of ORF 38 and ORF 54 were digested at 37°C with PstI (Promega, Madison, WI) and BglI (Promega, Madison, WI), respectively (11). PKVL6U (5′-TTC CCA CCG CGG CAC AAA CA-3′)/PKVL1L (5′-GGT TGC TGG TGT TGG ACG CG-3′) were used to amplify ORF 62 (13). The amplification products of ORF 62 were digested at 25°C with SmaI (Promega, Madison, WI).
(c) Phylogenetic analysis of ORF 22.
To identify genotypes E, J, and M, conventional PCR of a 447-bp fragment (positions 37837 to 38264) of VZV ORF 22 was done. The PCR forward primer, p22R1f (5′-GGG TTT TGT ATG AGC GTT GG-3′, positions 37837 to 37856), and the reverse primer, p22R1r (5′-CCC CCG AGG TTC GTA ATA TC-3′, positions 38383 to 38356), were used to amplify VZV ORF 22. DNA amplification reactions were performed under the conditions previously described (14).
Six polymorphic foci, positions 37902, 38019, 38055, 38081, 38177, and 38229, were sequenced as previously described (14). All VZV genomic locus numbers used in this study are based on the nucleotide sequence of the Dumas strain (GenBank accession number XO4370). Automated DNA sequencing was performed with an ABI Prism 3730XL DNA analyzer (Applied Biosystems, Foster City, CA). Sequencing reactions of PCR products were performed by using the ABI Prism Big Dye Terminator Cycle Sequencing Ready Reaction Kit V.3.1 (Applied Biosystems, Foster City, CA). Primary DNA sequence assembly and analysis were performed with CLC Combined Workbench 3.6.1 (CLC Bio, Aarhus, Denmark).
To create the phylogenetic trees of ORF 22, the reference strains of genotypes E, M1, M2, and J were used. Dumas, CA123 (GenBank accession number DQ457052), DR (GenBank accession number DQ452050), and the Oka parental strain (pOka; GenBank accession number AB097933) were used as reference strains for genotypes E, M1, M2, and J, respectively (15). We also included VariVax and VarilRix for J; strain 8 for M2; and strains 11, 22, 36, 49, BC, 32 P5, 32 P22, 32 P72, KEL, SD, NH293, and HJO for E in the phylogenetic analysis (15). We used a neighbor-joining method to create the phylogenetic trees.
Nucleotide sequence accession numbers.
Genome sequences from this study were submitted to GenBank with accession numbers FJ425229 to FJ425272.
RESULTS
Patients' characteristics.
Forty-four patients with herpes zoster and nine patients with varicella were included. The mean age of the patients with herpes zoster was 59.5 (range, 10 to 77) years, and the mean age of the patients with varicella was 8 (range, 6 to 9) years (Table 1). Among patients with herpes zoster, the most common underlying diseases were hematologic malignancies and human immunodeficiency virus infection. Disseminated zoster cases were 6.8% (3/44). The most common dermatomes that herpes zoster involved were T1 to T6.
Table 1.
Baseline characteristics of the 53 patients with herpes zoster or varicella
| Characteristic | Value |
|
|---|---|---|
| Herpes zoster (n = 44) | Varicella (n = 9) | |
| Clinical characteristics | ||
| Age (yr) [median (range)] | 59.5 (10–77) | 8 (6–9) |
| Male [n (%)] | 30 (68.2) | 4 (44.4) |
| Underlying disease [n (%)] | ||
| Solid cancer | 7 (15.9) | 0 |
| Hematologic malignancy | 9 (20.5) | 0 |
| HIV | 9 (20.5) | 0 |
| Chronic kidney disease | 2 (4.5) | 0 |
| Rheumatologic disease | 1 (2.3) | 0 |
| Others | 5 (11.4) | 0 |
| None | 6 (13.6) | 9 (100) |
| Characteristics of herpes zoster | ||
| Disseminated infection [n (%)] | 3 (6.8) | NAa |
| Involved dermatomes [n (%)] | NA | |
| Cranial nerves | 6 (13.6) | |
| C1–C8 | 7 (15.9) | |
| T1–T6 | 10 (22.7) | |
| T7–T12 | 6 (13.6) | |
| L1–L5 | 6 (13.6) | |
NA, not applicable.
Genotype of the VZV isolates.
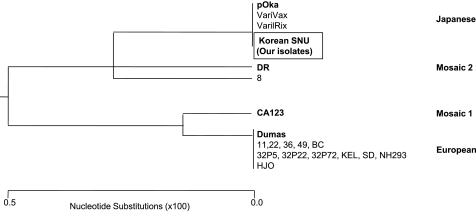
In RFLP analysis of ORF 38 and ORF 54, the PstI restriction enzyme site in ORF 38 was found in 94.3% (50/53) and the BglI restriction enzyme site in ORF 54 was found in 100% (53/53) of isolates (Table 2). In RFLP analysis of ORF 62, no SmaI+ isolate, which was representative of the Oka vaccine strain, was found. To distinguish genotypes E, M, and J, we performed sequence analysis of ORF 22. The sequences were compared to the reference strains Dumas, CA123, DR, and pOka, representing E, M1, M2, and J, respectively. Sequence analysis showed changes in three positions: A to G in position 37902, T to C in position 38081, and G to A in position 38177. The findings showed no difference between pOka and our isolates (Korean SNU) (Table 3). The phylogenetic tree of ORF 22 showed that our isolates clustered with the pOka strain (Fig. 1).
Table 2.
Nucleotide polymorphisms of ORF 22 and RFLP analysis of ORF 38 (PstI) and ORF 54 (BglI) of 53 isolates of varicella-zoster virus from Korea
| Age group (yr) | Patients [n (%)] | Residue at ORF 22 position: |
PstI+ [n (%)] | BglI+ [n (%)] | |||||
|---|---|---|---|---|---|---|---|---|---|
| 37902 | 38019 | 38055 | 38081 | 38177 | 38229 | ||||
| 0–9 | 9a (17.0) | G | G | C | C | A | A | 8 (88.9) | 9 (100) |
| 10–19 | 2 (3.8) | G | G | C | C | A | A | 1 (50) | 2 (100) |
| 20–29 | 2 (3.8) | G | G | C | C | A | A | 2 (100) | 2 (100) |
| 30–39 | 4 (7.5) | G | G | C | C | A | A | 4 (100) | 4 (100) |
| 40–49 | 5 (9.4) | G | G | C | C | A | A | 5 (100) | 5 (100) |
| 50–59 | 7 (13.2) | G | G | C | C | A | A | 7 (100) | 7 (100) |
| 60–69 | 16 (30.2) | G | G | C | C | A | A | 16 (100) | 16 (100) |
| 70–79 | 4 (7.5) | G | G | C | C | A | A | 4 (100) | 4 (100) |
| Unknown | 4 (7.5) | G | G | C | C | A | A | 3 (75) | 4 (100) |
Varicella immunization was introduced in 1988 in South Korea. Therefore, the 9 patients under 10 years of age with varicella might have been vaccinated against varicella. Unfortunately, we could not confirm if they had been vaccinated, because the records of vaccination were not available.
Table 3.
Comparison of nucleotide polymorphisms of Korean isolates with reference strains
Fig. 1.
Phylogenetic tree based on sequencing analysis of ORF 22 of VZV by the neighbor-joining method. The reference strains for each genotype are from reference 15. Our isolates are shown as “Korean SNU” in the box. The major reference strains of genotypes J, E, and M are shown in boldface.
DISCUSSION
This was the largest study for the genotypic analysis of VZV in South Korea. Our study demonstrated that Korean isolates belong to genotype J. In previous genotypic studies of VZV from Southeast Asian countries, genotype M was the most frequent genotype (14). In Bangladesh, genotype M1 was found, and in Nepal and India, genotypes M1 and M2, respectively, were found (14, 15). In early studies with three isolates from south China, the isolates were genotype M2, and three isolates from north China were genotype E (14). In a study of the middle eastern region of China, the VZV isolates were genotype J. Therefore, the genotypes of VZV isolates in China differed according to the area where VZV was collected (12). In a previous study, we performed genotyping of VZV isolates from 22 elderly patients with herpes zoster and showed that the genotypes of the VZV isolates were all J (4). The 53 isolates in this study were collected in different areas of South Korea at latitudes between 35.0°N and 38.0°N in a temperate climate. Our isolates were revealed as genotype J. This observation is consistent with a previous report from a similar location, such as the middle eastern region of China, with a latitude of 31.85°N and a typical temperate climate (12). However, in a genotypic analysis of 130 VZV samples in the United States, 3% (4/130) were revealed to be genotype J (22). Therefore, genotype J is frequently found in isolates from the area near Japan and is rarely found in isolates from other areas.
This result has a significant implication. In cases of vesicular rashes after varicella vaccination in countries without genotype J, genotype J could be a suspect for vaccine strain infection. However, in Japan and South Korea, where the wild-type strains are mostly genotype J, in cases of varicella-like rash after vaccination, genotypic analysis of ORF 22 could not distinguish vaccine strain infections from wild-type strain infections. To distinguish vOka and the wild-type strain, ORF 62-based PCR and RFLP analysis with SmaI should be used.
In our results, 94.3% (50/53) of isolates were PstI+ in ORF 38. Of the 3 isolates that were PstI− in ORF 38, one was isolated from a 9-year-old girl with varicella and another from a herpes zoster patient, a 19-year-old male with multiple cerebral infarctions. For the other isolate, we could not get clinical information. To distinguish vOka and the wild-type strain, we also performed RFLP analysis of ORF 62 with SmaI. All 3 of the isolates were SmaI− in ORF 62, suggesting the isolates were wild-type VZV rather than vOka.
In previous studies in the United Kingdom and Germany, the genotypes of VZV were changing (10, 21). In the United Kingdom, RFLP analysis revealed that 10.5% of isolates contained a BglI restriction enzyme in ORF 54 in 1970 and the 1980s; however, isolates containing a BglI restriction enzyme in ORF 54 increased to 18.2% in the 1990s. In Germany, 6% of isolates were BglI+ in ORF 54 in late 1997 to 2001 and 21% in 2006 (20, 21). The increase in isolates that were BglI+ in ORF 54 was attributed to immigrants from tropical areas, such as Asia and Africa (10, 21). In our study, all isolates were BglI+ in ORF 54, which was consistent with a previous study showing that isolates that were BglI+ in ORF 54 were frequently found in Asia (14).
Primary infection with VZV usually occurs during childhood, around 10 years of age (8). Herpes zoster develops by the reactivation of VZV, and the VZV isolated from zoster patients is the VZV acquired during childhood. Therefore, the VZV isolated from the zoster patients aged 70 circulated approximately 60 years ago, in the 1940s. Our study showed that all 53 isolates were genotype J and over 90% of isolates were PstI+ in ORF 38 and BglI+ in ORF 54. Therefore, we could conclude that the genotype of the circulating VZV strain had not changed during the last 7 decades.
In South Korea, varicella vaccination was introduced in 1988 and has been recommended as a universal childhood immunization since 2005. Therefore, many children have been vaccinated against varicella during the last 10 years. In the current study, all 9 isolates from children under 10 years of age were from varicella, whereas the remaining 44 isolates were from zoster. It is notable that 1 of 9 isolates (11.1%) from children under 10 years of age was PstI− in ORF 38. In contrast, one isolate (2.5%) from the 40 patients over 10 years of age, excluding one isolate from an unknown age group, was PstI− in ORF 38. These findings suggest that the introduction of varicella vaccination might have an impact on the genotypes of circulating VZV in South Korea.
In conclusion, all the VZV isolates were identified as a single strain of Japanese type, irrespective of the ages of the patients, suggesting that genotype J has been circulating since the 1940s in South Korea. Further studies are needed to monitor the possible impact of vaccination on the genotypes of circulating VZV in South Korea.
ACKNOWLEDGMENT
We are grateful to Son Yongkyu for collecting varicella zoster virus at his clinic.
Footnotes
Published ahead of print on 16 March 2011.
REFERENCES
- 1. Centers for Disease Control and Prevention 1999. Prevention of varicella. Update recommendations of the Advisory Committee on Immunization Practices (ACIP). MMWR Recommend. Rep. 48:1–5 [PubMed] [Google Scholar]
- 2. Centers for Disease Control and Prevention 1996. Prevention of varicella: recommendations of the Advisory Committee on Immunization Practices (ACIP). MMWR Recommend. Rep. 45:1–36 [PubMed] [Google Scholar]
- 3. Choi E. H. 2008. Varicella vaccine. Hanyang Med. Rev. 28:30–36 [Google Scholar]
- 4. Choi Y. J., Kim K. H., Oh M.-d. 2010. Genotype of varicella zoster virus isolated from Korean elderly patients with herpes zoster. Infect. Chemother. 42:162–170 [Google Scholar]
- 5. Davison A. J., Scott J. E. 1986. The complete DNA sequence of varicella-zoster virus. J. Gen. Virol. 67:1759–1816 [DOI] [PubMed] [Google Scholar]
- 6. Dumas A. M., Andrew J. D., Geelen J. L. M. C., Maris W., Noordaa J. V. D. 1980. Infectivity and molecular weight of varicella-zoster virus DNA. J. Gen. Virol. 47:233–235 [Google Scholar]
- 7. GlaxoSmithKline 2010. Varilrix package leaflet: information for the user. GlaxoSmithKline, Rixensart, Belgium [Google Scholar]
- 8. Gnann J. W., Whitley J. R. J. 2002. Herpes zoster. N. Engl. J. Med. 347:340–346 [DOI] [PubMed] [Google Scholar]
- 9. Green Cross Company 2010. Suduvax package insert. Green Cross Company, Yongin, South Korea [Google Scholar]
- 10. Hawrami K., et al. 1997. Molecular epidemiology of varicella-zoster virus in East London, England, between 1971 and 1995. J. Clin. Microbiol. 35:2807–2809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. LaRussa P., et al. 1992. Restriction fragment length polymorphism of polymerase chain reaction products from vaccine and wild-type varicella-zoster virus isolates. J. Virol. 66:1016–1020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Liu J., Wang M., Gan L., Yang S., Chen J. 2009. Genotyping of clinical varicella-zoster virus isolates collected in China. J. Clin. Microbiol. 47:1418–1423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Loparev V. N., Argaw T., Krause P. R., Takayama M., Schmid D. S. 2000. Improved identification and differentiation of varicella-zoster virus (VZV) wild-type strains and an attenuated varicella vaccine strain using a VZV open reading frame 62-based PCR. J. Clin. Microbiol. 38:3156–3160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Loparev V. N., et al. 2004. Global identification of three major genotypes of varicella-zoster virus: longitudinal clustering and strategies for genotyping. J. Virol. 78:8349–8358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Loparev V. N., et al. 2007. Identification of five major and two minor genotypes of varicella-zoster virus strains: a practical two-amplicon approach used to genotype clinical isolates in Australia and New Zealand. J. Virol. 81:12758–12765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Merck and Co., Inc 2010. Varivax prescribing information. Merck and Co., Inc., Whitehouse Station, NJ [Google Scholar]
- 17. Nguyen H. Q., Jumaan A. O., Seward J. F. 2005. Decline in mortality due to varicella after implementation of varicella vaccination in the United States. N. Engl. J. Med. 352:450–458 [DOI] [PubMed] [Google Scholar]
- 18. Roush S. W., McIntyre L., Baldy L. M. (ed.). 2008. Manual for the surveillance of vaccine-preventable diseases, 4th ed. Centers for Disease Control and Prevention, Atlanta, GA [Google Scholar]
- 19. Sadzot-Delvaux C., et al. 2008. Varicella vaccination in Japan, South Korea, and Europe. J. Infect. Dis. 197(Suppl. 2):S185–S190 [DOI] [PubMed] [Google Scholar]
- 20. Sauerbrei A., et al. 2003. Molecular characterisation of varicella-zoster virus strains in Germany and differentiation from the Oka vaccine strain. J. Med. Virol. 71:313–319 [DOI] [PubMed] [Google Scholar]
- 21. Sauerbrei A., Wutzler P. 2007. Different genotype pattern of varicella-zoster virus obtained from patients with varicella and zoster in Germany. J. Med. Virol. 79:1025–1031 [DOI] [PubMed] [Google Scholar]
- 22. Sergeev N., Rubtcova E., Chizikov V., Schmid D. S., Loparev V. N. 2006. New mosaic subgenotype of varicella-zoster virus in the U. S. A.: VZV detection and genotyping by oligonucleotide-microarray. J. Virol. Methods 136:8–16 [DOI] [PubMed] [Google Scholar]
- 23. Seward J. F., et al. 2002. Varicella disease after introduction of varicella vaccine in the United States, 1995–2000. JAMA 287:606–611 [DOI] [PubMed] [Google Scholar]
- 24. Takahashi M., et al. 2008. Development of varicella vaccine. J. Infect. Dis. 197:S41–S44 [DOI] [PubMed] [Google Scholar]
- 25. Takahashi M., et al. 1974. Live vaccine used to prevent the spread of varicella in children in hospital. Lancet ii:1288–1290 [DOI] [PubMed] [Google Scholar]