Abstract
Identification of Streptococcus pneumoniae in the nasopharynx is critical for an understanding of transmission, estimates of vaccine efficacy, and possible replacement disease. Conventional nasopharyngeal swab (NPS) culture and serotyping (the WHO protocol) is likely to underestimate multiple-serotype carriage. We compared the WHO protocol with methods aimed at improving cocolonization detection. One hundred twenty-five NPSs from an infant pneumococcal-carriage study, containing ≥1 serotype by WHO culture, were recultured in duplicate. A sweep of colonies from one plate culture was serotyped by latex agglutination. DNA extracted from the second plate was analyzed by S. pneumoniae molecular-serotyping microarray. Multiple serotypes were detected in 11.2% of the swabs by WHO culture, 43.2% by sweep serotyping, and 48.8% by microarray. Sweep and microarray were more likely to detect multiple serotypes than WHO culture (P < 0.0001). Cocolonization detection rates were similar between microarray and sweep, but the microarray identified the greatest number of serotypes. A common serogroup type was identified in 95.2% of swabs by all methods. WHO methodology significantly underestimates multiple-serotype carriage compared to these alternate methods. Sweep serotyping is cost-effective and field deployable but may fail to detect serotypes at low abundance, whereas microarray serotyping is more costly and technology dependent but may detect these additional minor carried serotypes.
INTRODUCTION
Streptococcus pneumoniae remains one of the most important bacterial pathogens globally (23). The introduction of pneumococcal conjugate vaccines has resulted in a profound reduction in invasive pneumococcal disease (IPD) caused by serotypes covered by the vaccines, but increases in the numbers of cases due to nonvaccine serotypes have been observed (25). Conjugate vaccines have also been shown to prevent nasopharyngeal carriage of the vaccine serotypes, although an increase in the carriage of nonvaccine serotypes has also been documented (20). To plan future vaccine strategy, there is a need to better understand the dynamics of pneumococcal colonization of the nasopharynx (a critical early step in the pathogenesis of IPD [3]), particularly in relation to carriage of, and competition between, multiple serotypes. Nonvaccine serotypes carried at low density may become important disease-causing serotypes once vaccine serotypes are removed from, or suppressed within, the nasopharyngeal niche (18). Longitudinal studies of pneumococcal colonization allow detailed exploration of carriage dynamics (7, 10, 12, 15), but standard approaches based on agar plate culture and conventional serotyping techniques may not be optimal for the detection of multiple-serotype carriage (22). Gratten et al. serotyped up to six colonies from nasal-swab culture plates and found multiple-serotype carriage in 29.5% of Papua New Guinean children (9). They went on to serotype at least 50 colonies from 10 selected nasal-swab cultures and concluded that the minor carried serotype accounted for 4 to 27% of the total pneumococcal population. A review of published data on multiple carriage concluded that, to detect a minor carried serotype, it would be necessary to serotype at least five colonies to have a 95% chance of detecting the serotype if it accounted for 50% of the total pneumococcal population, and one would need to examine 299 colonies if the serotype was present at a relative abundance of 1% (14). This approach would clearly not be cost- or time effective using conventional Quellung serotyping. Several alternative approaches have been described for the detection of multiple-serotype carriage. Researchers in The Gambia have developed a latex agglutination technique in which colonies from the primary culture plate are suspended in saline and serotyped by latex agglutination. Using this extremely cost-effective method, up to 10.4% of pneumococcal acquisitions were found to be of multiple serotypes in a longitudinal infant cohort study (12). However, there are no published studies comparing this technique with conventional pneumococcal-serotyping methodologies for detection of multiple-serotype colonization. Several other methods have demonstrated increased detection of multiple carriage compared to standard culture, including swab enrichment culture followed by either Quellung typing (16) or multiplex PCR (8) from the broth, immunoblotting (4) or multiplex PCR (26) from the primary culture plate, multiplex PCR direct from the nasopharyngeal swab–skim milk-tryptone-glucose-glycerol (NPS-STGG) transport medium specimen (2), pneumolysin noncoding region (plyNCR) PCR followed by terminal restriction fragment length polymorphism (RFLP) direct from the swab (6), and microarray-based detection and serotyping (5, 13). However, in general, these methods are either expensive, time-consuming, limited in the number of serotypes detected, or a combination of these factors, and therefore are not well suited for use in large-scale carriage studies, particularly those carried out in less developed countries, where there is the largest burden of pneumococcal disease (23).
The objectives for this study were first to determine the amount by which the standard culture method for detection of pneumococcal nasopharyngeal carriage underestimates cocolonization by multiple serotypes and, secondly, to assess the utility of the two alternate methods, one molecular (for reference laboratory use) and the other culture based (for field laboratory use), for detection of pneumococcal cocolonization in an infant carriage study.
(This work was presented in part at the 7th International Symposium on Pneumococci and Pneumococcal Diseases, March 2010, abstract 292.)
MATERIALS AND METHODS
NPSs collected during a longitudinal infant pneumococcal-carriage study were used in the current work. In this cohort, 965 Burmese/Karen refugee infants, born in Maela camp on the Thailand-Myanmar border between October 2007 and November 2008, were followed from birth to 24 months of age. NPSs were obtained from each infant at monthly intervals throughout the follow-up period. The study infants received WHO Expanded Program on Immunisation (EPI) immunizations, but pneumococcal conjugate vaccines were not available during the study period. Maela camp is a crowded refugee camp: approximately 40,000 people inhabit an area of 4 km2. Respiratory infections are a common cause of morbidity and mortality in this population: interim data from the infant cohort described in this paper found the incidence of clinical pneumonia to be 0.75 episodes per child-year in the first year of life (29).
Detection of pneumococcal colonization by the WHO culture protocol.
Swabs were collected and processed according to the current WHO pneumococcal-carriage detection protocol (22). Briefly, a Dacron-tipped nasopharyngeal swab (Medical Wire and Equipment, Corsham, United Kingdom) was inserted into the nasopharynx, rotated 180°, and removed. Using sterile scissors, the tip was cut off into a 2-ml cryovial (Simport, Beloeil, Quebec, Canada) containing 1 ml STGG transport medium. The NPS-STGG specimens were transported to the laboratory in a cool box, frozen at −80°C on the day of collection, and subsequently cultured in batches. Ten microliters of fully thawed STGG was streaked onto Columbia CNA agar with 5% sheep blood (bioMérieux, Marcy l'Etoile, France), and the plates were incubated overnight at 36°C in 5% CO2. Two alpha-hemolytic colonies, or more if clear morphological variation was observed, were subsequently subcultured onto sheep blood agar (Clinical Diagnostics, Bangkok, Thailand). S. pneumoniae was identified by colonial morphology and 5-μg (6-mm) optochin disc susceptibility (Oxoid, Basingstoke, United Kingdom). Suspected pneumococcal isolates with an intermediate optochin disc zone diameter (7 to 13 mm) were tested for bile solubility to confirm identity (27). Where colonial morphology and the optochin zone were identical in both subcultured colonies, only one of the isolates was serotyped. If any variation was seen, all isolates were serotyped. Serotyping was performed by latex agglutination (1, 17), and equivocal results were confirmed by Quellung serotyping (28). Nontypeable (NT) pneumococci were provisionally identified when weak agglutination with pool B and serogroup 19 latex antisera, but no agglutination with group 19 factor antisera (19b, 19c, and 19f; 7h), was observed (Shoklo Malaria Research Unit [SMRU], unpublished data). These isolates were confirmed as NT pneumococci by bile solubility and demonstration of the absence of capsular swelling with Omniserum (SSI Diagnostica, Copenhagen, Denmark).
Detection of pneumococcal colonization by sweep serotyping or microarray.
One hundred twenty-five NPS-STGG specimens known to contain at least one pneumococcal serotype by the WHO culture method were selected from the entire swab collection. Laboratory personnel were blinded to the original WHO culture result.
The specimens were thawed and mixed, and 10-μl aliquots of STGG were recultured on two CNA-blood agar plates. Both plates were incubated overnight at 36°C in 5% CO2. Growth was assessed after overnight culture, and those with growth only in the primary streak were reincubated for a further 24 h. If growth was still poor at 48 h, the specimen was recultured using 50-μl aliquots of STGG. The entire growth from each plate was removed using a sterile cotton swab (Medical Wire and Equipment, Corsham, United Kingdom) and suspended in aliquots of sterile saline. The first aliquot (0.5 to 2 ml saline adjusted to an organism density of 0.5 McFarland standard) was used for sweep serotyping, and genomic DNA was extracted from the second aliquot using a Gram-positive bacterium protocol (QIAamp minikit; Qiagen, Hilden, Germany). Briefly, the organism suspension was centrifuged to pellet the bacteria; this pellet was resuspended in 180 μl of an enzyme lysis solution (20 mg/ml lysozyme, 20 mM Tris-HCl, pH 8.0, 2 mM EDTA, 1.2% Triton) and incubated at 37°C for 30 min. Qiagen buffer AL (200 μl) and 20 μl proteinase K were added, and the mixture was incubated for 30 min at 56°C, followed by 15 min at 95°C. Following these lysis steps, the standard manufacturer's extraction protocol was followed, and DNA was eluted into 200 μl Qiagen buffer AE. The DNA extract was stored at −80°C prior to being tested by the Bacterial Microarray Group at St. George's, University of London, London, United Kingdom (BμG@S; http://bugs.sgul.ac.uk).
Sweep serotyping by latex agglutination was performed on the saline suspension: the suspension was first tested with antiserum pools A to I, followed by all appropriate group, type, and factor antisera, as previously described (12). NT pneumococci were provisionally identified by the latex agglutination reactions described previously.
Molecular serotyping was performed on the DNA extracts using the BμG@S SP-CPS v1.4.0 microarray, and the output was analyzed using a Bayesian hierarchical model as described previously (5). In addition to serotype detection, the array was used to determine the relative serotype proportions in specimens containing multiple serotypes (13).
To confirm a single highly discrepant result, a swab in which sweep serotyping identified two serotypes but the microarray detected nine serotypes, intensive study of the NPS-STGG specimen by conventional culture and serotyping was undertaken. To ensure single colonies would be well separated, five 2-μl aliquots of the thawed STGG were recultured onto CNA-blood agar plates and incubated overnight as described previously. A total of 300 randomly selected alpha-hemolytic colonies were subcultured onto plain blood agar plates (with an optochin disc) and subsequently serotyped by latex agglutination. Finally, sequential multiplex PCR was attempted on the DNA extract from this specimen culture to provide a second confirmatory molecular-serotyping result (24).
Statistical analysis.
Data were entered into an Access 2003 database (Microsoft, Redmond, WA) and systematically checked for errors. Statistical analyses were carried out using STATA 10.1 (StataCorp, College Station, TX). Continuous data (nonnormally distributed) were analyzed by the Wilcoxon matched-pairs signed-rank test, and proportions were compared by the chi-squared test. Comparisons of multiple-serotype carriage detection by method were done using McNemar's test. Statistical significance was inferred from two-sided P values of ≤0.05.
At the time of the study, it was not possible to resolve serotypes 6A and 6C using antisera; therefore, these serotypes were considered identical (and labeled 6A/C). In a small number of specimens, microarray testing identified multiple nontypeable pneumococci, based on variation of the cps locus components; however, they were considered to be a single serotype when methods were compared.
Study ethics.
Written informed consent was obtained from the mothers of all infants prior to enrollment in the study. Ethical approval was granted by the Ethics Committee of The Faculty of Tropical Medicine, Mahidol University, Thailand (MUTM 2009-306), and the Oxford Tropical Research Ethics Committee, Oxford University, United Kingdom (031-06).
RESULTS
Swabs and study participants.
One hundred twenty-five nasopharyngeal swabs from 42 infants, aged between 26 and 516 days, were included. Swabs from up to 12 different sampling time points per infant were examined.
Detection of multiple-serotype colonization.
Multiple pneumococcal serotypes were identified in 14/125 (11.2%) NPSs by the WHO culture method. A single serotype was identified in the other 111/125 (88.8%) swabs. The proportion of swabs in which multiple-serotype carriage was identified increased using either sweep (54/125; 43.2%) or microarray (61/125; 48.8%) serotyping. Sweep serotyping was 3.9 times (95% confidence interval [CI], 2.4 to 6.2) and microarray serotyping 4.4 times (95% CI, 2.8 to 6.9) more likely to detect multiple pneumococcal serotype colonization than the WHO culture method (both P < 0.0001).
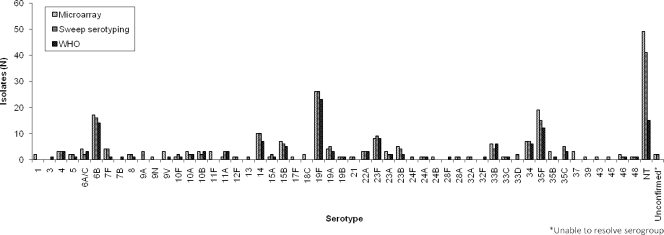
Considering all 125 swabs together, the microarray identified significantly more pneumococci (222 pneumococci; 47 serotypes) than either sweep serotyping (195 pneumococci; 39 serotypes) or WHO culture (139 pneumococci; 35 serotypes) (P < 0.0001) (Fig. 1). The number of serotypes identified per swab ranged from one to nine, as summarized in Table 1. Sweep serotyping detected significantly fewer serotypes per swab than the microarray (P = 0.04), although this difference became nonsignificant (P = 0.06) if a single outlying swab was excluded. In this particular swab, nine serotypes were identified by microarray but only two serotypes by sweep serotyping and by further examination of 300 discrete alpha-hemolytic colonies by conventional culture (246/300 colonies were identified as S. pneumoniae, and only these were serotyped), although multiplex PCR also indicated the presence of at least four serotypes, which included those identified by sweep serotyping plus others detected by microarray. In two instances, the sweep and microarray serotyping methods could not resolve a serogroup (i.e., could not serotype to the “type” level). By all three methods, a common six serotypes accounted for 56.9 to 60.0% of the total, but the rank order of these serotypes varied by method (Table 2).
Fig. 1.
Distribution of serotypes identified by each detection method.
Table 1.
Number of pneumococcal serotypes identified per nasopharyngeal swab by each of the three detection methods
| Total no. of serotypes detected | WHO culture [n (%)] | Sweep serotype [n (%)] | Microarray [n (%)] |
|---|---|---|---|
| 1 | 111 (88.8) | 71 (56.8) | 64 (51.2) |
| 2 | 14 (11.2) | 41 (32.8) | 36 (28.8) |
| 3 | 0 (0) | 10 (8.0) | 20 (16.0) |
| 4 | 0 (0) | 3 (2.4) | 3 (2.4) |
| ≥5 | 0 (0) | 0 (0) | 2 (1.6) |
Table 2.
Pneumococcal serotypes most frequently identified by each detection method
| Serotype | WHO culture [n (%)] | Sweep serotype [n (%)] | Microarray [n (%)] |
|---|---|---|---|
| 19F | 23 (16.6) | 26 (13.3) | 26 (11.7) |
| NT | 15 (10.8) | 41 (21.0) | 49 (22.1) |
| 6B | 14 (10.1) | 16 (8.2) | 17 (7.7) |
| 35F | 12 (8.6) | 15 (7.7) | 19 (8.6) |
| 23F | 8 (5.8) | 9 (4.6) | 8 (3.6) |
| 14 | 7 (5.0) | 10 (5.1) | 10 (4.5) |
| Proportion of total serotypes identified | 79/139 (56.9) | 117/195 (60.0) | 129/222 (58.2) |
Both sweep and microarray serotyping identified a significantly larger proportion of NT pneumococci than WHO culture (22.1% [microarray] and 21.0% [sweep] versus 10.8% [WHO]; P = 0.007). However, even when these NT pneumococci were excluded from the analyses, sweep or microarray serotyping still identified a significantly greater number of pneumococci and multiple-serotype cocolonization episodes than the WHO culture method. The microarray detected 173 typeable pneumococci (46 serotypes) and multiple-serotype carriage in 41/125 (32.8%) NPSs, sweep serotyping detected 154 typeable pneumococci (38 serotypes) and multiple-serotype carriage in 31/125 (24.8%) NPSs, and WHO culture detected 124 typeable pneumococci (34 serotypes) and multiple-serotype carriage in 11/125 (8.8%) NPSs (all P < 0.0001).
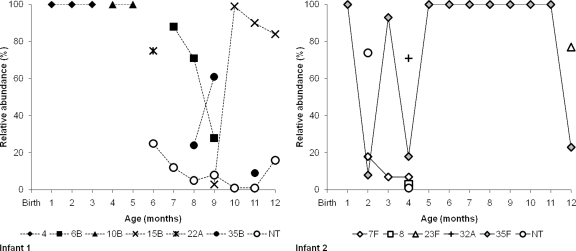
There was a nonsignificant trend in the proportion of PCV13 serotypes versus nonvaccine serotypes detected by the three methods: PCV13 serotypes accounted for 46.8% (WHO culture), 39.5% (sweep serotyping), and 38.3% (microarray serotyping) of all of the serotypes detected (P = 0.25). Considering only the microarray data, in swabs where a single serotype was identified (i.e., completely “dominant”), the serotype was a PCV13 type in 35/64 (54.7%), but in swabs where multiple serotypes were detected, the “dominant” serotype was a PCV13 type in only 21/61 (34.4%; P = 0.02). The ability of the microarray to determine the relative abundance of each serotype allowed detailed exploration of the temporal flux of relative serotype levels in two representative infants, for whom a total of 24 swabs were examined (Fig. 2).
Fig. 2.
Temporal changes in pneumococcal carriage over the first year of life in two infants (microarray data of serotypes detected and their relative abundances).
Serotype result agreement between detection methods.
Overall, at least one common serotype was found by all three detection methods in 109/125 specimens (87.2%; 95% CI, 81.1 to 93.1%). Relaxing agreement to the serogroup type (SGT) level, there was at least one common SGT identified by all three methods in 119/125 specimens (95.2%; 95% CI, 91.4 to 99.0%) and SGT agreement between two of the three methods in the remaining six specimens.
In 44 specimens, a single pneumococcal serotype was identified by all three detection methods. In 42/44 (95.5%; 95% CI, 89.0 to 100%), all three methods identified the same serotype. In one swab, WHO and microarray both identified a type 9V pneumococcus, but sweep serotyping determined the serotype to be 9A. In the other noncongruent specimen, both WHO culture and sweep serotyping identified a nontypeable pneumococcus, but the microarray detected type 6B capsule genes.
Limits of detection of sweep serotyping and microarray.
In order to assess the limits of the sweep serotyping assay, serial dilutions of overnight cultures of S. pneumoniae (six clinical isolates; serotypes 6A, 6B, 14, 19F, 19A, and 23F) were made in normal saline and serotyped using latex antisera as described previously. The organism density in the saline suspensions was determined by the plate count technique (21). Reproducible serotype results could be obtained at organism densities of 107 to 108 CFU/ml (i.e., 0.5 McFarland standard). In vitro mixtures of these suspensions confirmed that, at an overall organism density of 0.5 McFarland standard (visual), serotypes present at ≥25% of the total pneumococcal population could be reliably detected by latex agglutination (data not shown).
In the 61 swabs in which multiple serotypes were detected by microarray, the relative proportion of the minority serotype (defined as the least abundant serotype detected) ranged from 1% to 46% (median, 7%; interquartile range [IQR], 3 to 14%) (Table 3). Microarray serotyping detected additional serotypes compared to sweep serotyping in 38 specimens; in 30/38 (78.9%), the predominant additional serotype detected was NT, and in 21/38 (55.3%), the only additional serotype was NT. In the 17/38 specimens in which the microarray detected additional typeable pneumococci, 9/17 (53.0%) serotypes were present at <10%, 5/17 (29.4%) were present at 10 to 19%, and only 3/17 (17.6%) were present at ≥20% of the total pneumococcal population.
Table 3.
Relative proportion of minority serotypes detected by microarray compared with the total number of pneumococcal serotypes detected
| Relative proportion (%) of minority serotype | Value for total no. of serotypes detected of: |
Total | ||||
|---|---|---|---|---|---|---|
| 2 | 3 | 4 | 5 | 9 | ||
| <10 | 12 | 15 | 3 | 1 | 1 | 32 |
| 10–19 | 11 | 5 | 0 | 0 | 0 | 16 |
| 20–29 | 6 | 0 | 0 | 0 | 0 | 6 |
| 30–39 | 3 | 0 | 0 | 0 | 0 | 3 |
| 40–49 | 4 | 0 | 0 | 0 | 0 | 4 |
| Total | 36 | 20 | 3 | 1 | 1 | 61 |
DISCUSSION
We have demonstrated that culture and serotyping of nasopharyngeal-swab specimens using the current WHO protocol significantly underestimates the frequency of multiple pneumococcal serotype carriage. Sweep serotyping identified 1.4 times, and microarray 1.6 times, as many pneumococci as the standard culture method. These alternate methods also revealed greater diversity of carriage serotypes, reflected by the greater proportion of nonvaccine serotypes identified by each method. As expected, the microarray identified the largest number of carried serotypes and was able to detected minor serotypes present at very low relative abundance. The microarray data confirmed the previously described finding that the minority serotype in a multiple-carriage episode is present at low abundance: we documented a median relative abundance of 7% (range, 1 to 46%) for the minority serotype compared with 4 to 27% found in the Papua New Guinea study using Quellung typing of multiple colonies (9). The ability of microarray and sweep serotyping to confidently document the frequency of nontypeable pneumococcal carriage is also useful, since these organisms are suspected of being an important reservoir of antimicrobial resistance, although they are, as yet, not an important cause of invasive disease (19). Identification of nontypeable pneumococci can be problematic given the phenotypic and genetic similarities to closely related streptococcal species, such as Streptococcus mitis and Streptococcus pseudopneumoniae. However, characterization of 168 NT isolates from the study population by multilocus sequence typing (MLST) concluded that 162/168 (96%) could be assigned a sequence type, and only six isolates were found to be nonpneumococcal streptococci by multilocus sequence analysis (http://www.emlsa.net; unpublished data). This result adds confidence to the finding of frequent NT pneumococcal detections in the current evaluation.
The agreement between the three methods, in terms of detection of a common serotype, was excellent, with at least one common SGT identified in 95.2% of swabs. The two discrepancies observed in 44 swabs in which a single serotype was identified by all methods were minor: a factor discrepancy (9A versus 9V) between methods in one and a potential nonexpressed capsule in the other (microarray identified 6B capsule genes, but both WHO and sweep found a nontypeable pneumococcus). A single result was unsatisfactory: a swab in which the microarray identified nine serotypes but sweep serotyping identified only two and WHO culture one serotype. This specimen was recultured, and 246 discrete pneumococcal colonies were individually serotyped by latex agglutination; the sweep serotyping result was confirmed by this approach, and additional serotypes were not encountered. In spite of this, the microarray result may well be correct and is supported to some extent by multiplex PCR results. It is likely that to detect several very low-abundance serotypes it would be necessary to serotype even more than the 300 colonies that were attempted here. Furthermore, the presence of conserved capsule polysaccharide genes in other non-alpha-hemolytic or optochin-resistant alpha-hemolytic Streptococcus spp. in the primary-plate DNA extract may have resulted in signal on the array, possibly considered a false positive but reflecting the total gene pool and capacity for capsule polysaccharide production in the sample.
The WHO culture method will detect the predominant serotype but statistically cannot be expected to detect low-abundance serotypes (14). Increasing the number of colonies serotyped by random selection from the primary culture plate has been shown to improve detection of multiple colonization (11), but the cost and time required for the additional serotyping renders this difficult to sustain in the setting of a large carriage study. These factors also limit the utility of immunoblotting or broth enrichment methods in such studies (4, 16).
Sweep serotyping is cheap, quick, and technically straightforward and was not significantly inferior to the microarray method in detecting multiple-carriage episodes, although it was not efficient at detecting pneumococcal serotypes present at very low abundance, i.e., <25% of the total. Further work is planned to improve the detection of the minor carried type. If this can be improved, sweep serotyping may ultimately become a gold standard method for detection, but not relative quantitation, of multiple serotypes in nasopharyngeal samples.
Molecular techniques, especially when applied directly to the swab, reduce the cost and time of multiple-serotype detection compared to Quellung typing, but currently, multiplex PCR can detect only a limited number of serotypes and may therefore miss serotypes that are ultimately important for replacement disease after vaccination (3, 8, 24). The significant advantages of the microarray method used in the current study are the ability to detect all known pneumococcal serotypes and to estimate their relative abundances in a mixed population. In the setting of a longitudinal carriage study, these additional data permit detailed observations of the interactions between pneumococcal serotypes and, indeed, between pneumococci and other bacterial or viral species over time.
Both sweep and microarray serotyping enable researchers to better interrogate nasopharyngeal-swab specimens with regard to detection of multiple pneumococcal serotype carriage. In the current study, the microarray was more sensitive than sweep serotyping and provided useful data regarding relative serotype abundance. However, microarray-based molecular serotyping is currently restricted to a small number of research laboratories, and the cost per test is relatively high, although additional simultaneous analyses can be conducted. Sweep serotyping is extremely cost-effective (10 μl neat pneumococcal antiserum is used to produce 6 ml of latex reagent) and can be performed in any microbiology laboratory capable of serotyping S. pneumoniae by Quellung, which makes it a highly useful technique for pneumococcal-carriage studies, particularly in resource-poor settings.
ACKNOWLEDGMENTS
We are grateful to all of the Maela carriage study participants, the staff at the SMRU Maela camp clinic and Mae Sot microbiology laboratory, and Richard Adegbola (MRC Laboratories, The Gambia) for sharing the latex antiserum preparation method.
This work was supported by the Wellcome Trust (grants 083735 [P.T.], 077166/Z/05 [F.N. and C.T.], and 086547 [J.H. and K.G.]).
We report no potential conflicts of interest.
Footnotes
Published ahead of print on 16 March 2011.
The authors have paid a fee to allow immediate free access to this article.
REFERENCES
- 1. Adegbola R. A., et al. 2006. Serotype and antimicrobial susceptibility patterns of isolates of Streptococcus pneumoniae causing invasive disease in The Gambia 1996–2003. Trop. Med. Int. Health 11:1128–1135 [DOI] [PubMed] [Google Scholar]
- 2. Antonio M., Hakeem I., Sankareh K., Cheung Y. B., Adegbola R. A. 2009. Evaluation of sequential multiplex PCR for direct detection of multiple serotypes of Streptococcus pneumoniae from nasopharyngeal secretions. J. Med. Microbiol. 58:296–302 [DOI] [PubMed] [Google Scholar]
- 3. Bogaert D., De Groot R., Hermans P. W. 2004. Streptococcus pneumoniae colonisation: the key to pneumococcal disease. Lancet Infect. Dis. 4:144–154 [DOI] [PubMed] [Google Scholar]
- 4. Bronsdon M. A., et al. 2004. Immunoblot method to detect Streptococcus pneumoniae and identify multiple serotypes from nasopharyngeal secretions. J. Clin. Microbiol. 42:1596–1600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Brugger S. D., Frey P., Aebi S., Hinds J., Muhlemann K. 2010. Multiple colonization with S. pneumoniae before and after introduction of the seven-valent conjugated pneumococcal polysaccharide vaccine. PLoS One 5:e11638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Brugger S. D., Hathaway L. J., Muhlemann K. 2009. Detection of Streptococcus pneumoniae strain cocolonization in the nasopharynx. J. Clin. Microbiol. 47:1750–1756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Coles C. L., et al. 2001. Pneumococcal nasopharyngeal colonization in young South Indian infants. Pediatr. Infect. Dis. J. 20:289–295 [DOI] [PubMed] [Google Scholar]
- 8. da Gloria Carvalho M., et al. 2010. Revisiting pneumococcal carriage by use of broth enrichment and PCR techniques for enhanced detection of carriage and serotypes. J. Clin. Microbiol. 48:1611–1618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Gratten M., et al. 1989. Multiple colonization of the upper respiratory tract of Papua New Guinea children with Haemophilus influenzae and Streptococcus pneumoniae. Southeast Asian J. Trop. Med. Public Health 20:501–509 [PubMed] [Google Scholar]
- 10. Gray B. M., Converse G. M., III, Dillon H. C., Jr 1980. Epidemiologic studies of Streptococcus pneumoniae in infants: acquisition, carriage, and infection during the first 24 months of life. J. Infect. Dis. 142:923–933 [DOI] [PubMed] [Google Scholar]
- 11. Hare K. M., Morris P., Smith-Vaughan H., Leach A. J. 2008. Random colony selection versus colony morphology for detection of multiple pneumococcal serotypes in nasopharyngeal swabs. Pediatr. Infect. Dis. J. 27:178–180 [DOI] [PubMed] [Google Scholar]
- 12. Hill P. C., et al. 2008. Nasopharyngeal carriage of Streptococcus pneumoniae in Gambian infants: a longitudinal study. Clin. Infect. Dis. 46:807–814 [DOI] [PubMed] [Google Scholar]
- 13. Hinds J., et al. 2009. Molecular serotyping of Streptococcus pneumoniae: a microarray-based tool with enhanced utility for isolate typing, novel serotype discovery, non-typeable investigation, multiple carriage detection, and direct analysis of nasopharyngeal swabs, abstr. A-O5. Europneumo 2009: 9th Eur. Meet. Mol. Biol. Pneumococcus, Bern, Switzerland [Google Scholar]
- 14. Huebner R. E., Dagan R., Porath N., Wasas A. D., Klugman K. P. 2000. Lack of utility of serotyping multiple colonies for detection of simultaneous nasopharyngeal carriage of different pneumococcal serotypes. Pediatr. Infect. Dis. J. 19:1017–1020 [DOI] [PubMed] [Google Scholar]
- 15. Hussain M., et al. 2005. A longitudinal household study of Streptococcus pneumoniae nasopharyngeal carriage in a UK setting. Epidemiol. Infect. 133:891–898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kaltoft M. S., Skov Sorensen U. B., Slotved H. C., Konradsen H. B. 2008. An easy method for detection of nasopharyngeal carriage of multiple Streptococcus pneumoniae serotypes. J. Microbiol. Methods 75:540–544 [DOI] [PubMed] [Google Scholar]
- 17. Lafong A. C., Crothers E. 1988. Simple latex agglutination method for typing pneumococci. J. Clin. Pathol. 41:230–231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Lipsitch M. 1997. Vaccination against colonizing bacteria with multiple serotypes. Proc. Natl. Acad. Sci. U. S. A. 94:6571–6576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Marsh R., et al. 2010. The nonserotypeable pneumococcus: phenotypic dynamics in the era of anticapsular vaccines. J. Clin. Microbiol. 48:831–835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Mbelle N., et al. 1999. Immunogenicity and impact on nasopharyngeal carriage of a nonavalent pneumococcal conjugate vaccine. J. Infect. Dis. 180:1171–1176 [DOI] [PubMed] [Google Scholar]
- 21. Miles A. A., Misra S. S., Irwin J. O. 1938. The estimation of the bactericidal power of the blood. J. Hyg. 38:732–749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. O'Brien K. L., Nohynek H. 2003. Report from a WHO working group: standard method for detecting upper respiratory carriage of Streptococcus pneumoniae. Pediatr. Infect. Dis. J. 22:133–140 [DOI] [PubMed] [Google Scholar]
- 23. O'Brien K. L., et al. 2009. Burden of disease caused by Streptococcus pneumoniae in children younger than 5 years: global estimates. Lancet 374:893–902 [DOI] [PubMed] [Google Scholar]
- 24. Pai R., Gertz R. E., Beall B. 2006. Sequential multiplex PCR approach for determining capsular serotypes of Streptococcus pneumoniae isolates. J. Clin. Microbiol. 44:124–131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Pilishvili T., et al. 2010. Sustained reductions in invasive pneumococcal disease in the era of conjugate vaccine. J. Infect. Dis. 201:32–41 [DOI] [PubMed] [Google Scholar]
- 26. Rivera-Olivero I. A., Blommaart M., Bogaert D., Hermans P. W., de Waard J. H. 2009. Multiplex PCR reveals a high rate of nasopharyngeal pneumococcal 7-valent conjugate vaccine serotypes co-colonizing indigenous Warao children in Venezuela. J. Med. Microbiol. 58:584–587 [DOI] [PubMed] [Google Scholar]
- 27. Spellerberg B., Brandt C. 2007. Streptococcus, p. 423 In Murray P. R., Baron E. J., Jorgensen J. H., Landry M. L., Pfaller M. A. (ed.), Manual of Clinical Microbiology, 9th ed., vol. 1 ASM Press, Washington, DC [Google Scholar]
- 28. SSI Diagnostica 2006. SSI Diagnostica pneumococcal antisera kit insert, 2nd ed. Statens Serum Institut, Copenhagen, Denmark [Google Scholar]
- 29. Turner C., et al. 2010. Pneumococcal carriage and clinical pneumonia episodes in under one year old refugee children on the Thailand-Myanmar border, poster 194. 7th Int. Symp. Pneumococci Pneumococcal Dis., Tel Aviv, Israel [Google Scholar]