Abstract
The polysaccharide capsule (CPS) of Campylobacter jejuni is the major serodeterminant of the Penner serotyping scheme. There are 47 Penner serotypes of C. jejuni, 22 of which fall into complexes of related serotypes. A multiplex PCR method for determination of capsule types of Campylobacter jejuni which is simpler and more affordable than classical Penner typing was developed. Primers specific for each capsule type were designed on the basis of a database of gene sequences from the variable capsule loci of 8 strains of major serotypes sequenced in this study and 10 published sequences of other serotypes. DNA sequence analysis revealed a mosaic nature of the capsule loci, suggesting reassortment of genes by horizontal transfer, and demonstrated a high degree of conservation of genes within Penner complexes. The multiplex PCR can distinguish 17 individual serotypes in two PCRs with sensitivities and specificities ranging from 90 to 100% using 244 strains of known Penner type.
INTRODUCTION
Campylobacter jejuni is one of the major causes of human bacterial diarrheal disease (29), with an estimated annual incidence of 400 million worldwide. The disease is zoonotic, with wild and domesticated poultry representing major reservoirs (13, 47). The symptoms of campylobacteriosis can range from a mild, watery diarrhea to dysentery, often with abdominal pain and fever (1). Moreover, there are several serious postinfectious complications associated with C. jejuni, including reactive arthritis (38), Guillain-Barré syndrome (2), and irritable bowel syndrome (35, 44).
The polysaccharide capsule (CPS) represents an interface between the bacterium and the environment and contributes to virulence. Noncapsulated mutants of C. jejuni were demonstrated to have reduced abilities to invade INT407 cells in vitro (6), to colonize chickens (5, 12), and to cause diarrheal disease in ferrets (6). CPS also contributes to resistance to normal human serum (6). The CPS is phase variable in expression, presumably due to slip-strand mismatch repair in genes essential for CPS synthesis, a mechanism of variation that is common in C. jejuni (14, 31, 45). The ability to turn CPS expression on and off suggests that there may be advantages to the pathogen to express this polysaccharide coat only at specific times during its life cycle. Importantly, a prototype CPS conjugate vaccine against C. jejuni 81-176 provided 100% efficacy against diarrheal disease in a nonhuman primate model, further implicating this structure as both a virulence determinant and a protective antigen (27).
The genes for CPS biosynthesis are located in one of the more hypervariable regions of the C. jejuni genome. The high degree of variability of CPS genes is consistent with CPS being the major serodeterminant of the Penner or heat-stable (HS) serotyping scheme (19). However, in some serotypes lipooligosaccharide (LOS) has been shown to play a role in Penner serospecificity (39). The Penner scheme is a passive slide hemagglutination assay that includes 47 C. jejuni serotypes (33, 34). Twenty-two of the 47 serotypes fall into complexes of what appear to be structurally related CPS types (39). Currently, the DNA sequences of 10 C. jejuni hypervariable CPS loci (HS1, HS19, HS23, HS36, HS23/36, HS41, HS2, HS6, HS4/13/64, HS53) have been reported (9, 10, 18, 31, 32), and these loci range in size from 15 to 34 kb. The genes in the variable CPS loci include genes encoding sugar biosynthetic enzymes, putative glycosyl transferases, and genes of unknown function. Genes involved in heptose biosynthesis are conserved in the CPS loci of many strains. These genes include hddC, a putative heptose transferase; gmhA2, a phosphoheptose isomerase; hddA, a putative d-glycero-d-manno-heptose 7-phosphate kinase; and dmhA, a GDP-mannose 4,6-dehydratase responsible for the conversion of heptose to deoxyheptose. The genes for biosynthesis of O-methyl phosphoramidate (MeOPN), an unusual modification on some C. jejuni capsules, were initially identified and characterized in C. jejuni NCTC 11168 (23). These highly conserved MeOPN biosynthesis genes have been found in about 70% of the strains sequenced to date. Similarly, McNally et al. identified two distinct MeOPN transferases in NCTC 11168 responsible for addition of MeOPN to distinct sugar moieties in the NCTC 11168 CPS (23). There is variability in the predicted protein sequences of MeOPN transferases among sequenced strains, consistent with attachment of MeOPN to different sugars in different CPS structures. The MeOPN transferases also appear to undergo phase variation in expression, which contributes to the observed nonstoichiometric levels of MeOPN on CPS. Phase variations affecting capsule structure appear to modulate the Penner serotype within the HS23/36 complex. HS23 and HS36 strains and strains that serotype as both HS23 and HS36 (e.g., C. jejuni strain 81-176) all express a capsule composed of repeating units of α-d-galactose, β-d-GlcNAc, and d-glycero-d-altro-heptose or deoxyheptose variants with and without methyl groups (4). The difference in serotype (HS23, HS36, or HS23/36) is due to phase variations in the genes encoding heptose synthesis and the MeOPN transferase (4, 18).
The Penner serotyping scheme was used extensively for epidemiological studies for many years, but the complexities and expense of producing the antisera specific to 47 C. jejuni serotypes have limited its usefulness. Other methods have largely replaced Penner typing for general epidemiological studies, such as multilocus sequence typing (MLST), pulsed-field gel electrophoresis (PFGE), and ribotyping. Given the recent data demonstrating that a CPS conjugate vaccine can protect against C. jejuni diarrhea in a primate model (27), we are interested in evaluating the feasibility of a multivalent CPS conjugate approach to prevent C. jejuni infections. Critical to this end is the determination of the valency that would be required for an effective vaccine in areas of the world where C. jejuni is endemic or hyperendemic. To address this, we have developed a multiplex PCR method that can rapidly determine CPS types. The primers were designed on the basis of published CPS loci from 10 serotypes and another 8 serotypes that were sequenced as part of this work. Here, we report the sequences of the CPS loci of these 8 serotypes and describe a multiplex PCR system that can accurately predict 17 major CPS types of C. jejuni. We also provide new information on the relationship of serotypes within some additional Penner complexes.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The strains used for sequence analysis and for development of the multiplex PCR are shown in Table 1. Collections of clinical isolates from Thailand and Egypt were used to validate the multiplex PCR. The Thai isolates were obtained from cases and asymptomatic controls in an acute diarrhea study among foreign travelers at Bumrungrad Hospital, Bangkok, Thailand, during 2001 and 2002, as well as C. jejuni isolates collected from U.S. soldiers with acute diarrhea and from asymptomatic controls during Operation Cobra Gold exercises at different sites in Thailand from 1998 to 2003. The Thai isolates were serotyped by Helen Tabor at the Canadian Reference Lab in Winnipeg, Manitoba, Canada. The Egyptian isolates were from a longitudinal village-based study of diarrhea in rural Egyptian children (41) and were serotyped by Eva Nielsen at the Danish Veterinary Laboratory, Copenhagen, Denmark. C. jejuni strains were routinely cultured at 37°C under microaerobic conditions (5% O2, 10% CO2, and 85% N2) on Mueller-Hinton (MH) agar plates.
Table 1.
Strains used in this study
| Strain | Penner type | Reference or source |
|---|---|---|
| ATCC 43429 | HS1 (type) | 34 |
| ATCC 43430 | HS2 (type) | 34 |
| NCTC 11168 | HS2 | 31 |
| ATCC 43431 | HS3 (type) | 34 |
| ATCC 43432 | HS4 (type) | 34 |
| BH-01-0142 | HS3/13/50 | 37 |
| GC8486 | HS4/13/64 | 36 |
| ATCC 43433 | HS5 (type) | 34 |
| 81116 | HS6 | 30 |
| ATCC 43436 | HS8 (type) | 34 |
| ATCC 43438 | HS10 (type) | 34 |
| ATCC 43441 | HS13 (type) | 34 |
| ATCC 43442 | HS15 (type) | 34 |
| ATCC 43444 | HS17 (type) | 34 |
| ATCC 43446 | HS19 (type) | 34 |
| ATCC 43448 | HS22 (type) | 34 |
| 81-176 | HS23/36 | 20 |
| ATCC 43478 | HS28 (type) | 34 |
| ATCC 43460 | HS41 (type) | 34 |
| ATCC 43461 | HS42 (type) | 34 |
| ATCC 43463 | HS44 (type) | 34 |
| ATCC 43465 | HS50 (type) | 34 |
| RM1221 | HS53 | 25 |
| ATCC 49302 | HS64 (type) | 34 |
DNA purification.
C. jejuni genomic DNA was extracted from 16-h cultures following the method described by Sambrook et al. (43).
CPS sequencing strategies.
Due to the large size of the CPS regions (15 to 35 kb), primers were designed in the highly conserved heptose genes hddA (hddA-L, 5′-GAAAGAGAAGATTTAGGCATAGTAGG; hddA-R, 5′-TCCATGATTTAACCCCCTCTT) and dmhA (dmhA-L, 5′-GGATTTACAGGGCAAGTTGG; dmhA-R, 5′-TTCTTGTAACAAAAGTGCGAATG). If the strains to be sequenced produced a positive amplification with primers hddA-L/R and dmhA-L/R, these genes were used as anchors for long-range PCR. In this case, primer kpsC-R (5′-GGATTTTCTTTTATGGCATCTTTT) was used with primer hddA-L and primer dmhA-R was used with primer kpsF-F (5′-AGTCGATGCTGATGCTATGG). This two-step PCR increases the probability of amplification by lowering the size of the PCR product. PCRs were performed using a MasterAmp Extra-Long PCR kit from Epicentre or LongAmp Taq DNA polymerase from New England BioLabs, following the manufacturers' protocols. PCRs with primers kpsC-R/hddA-L were performed at an annealing temperature of 55°C for 40 s and dmhA-R/kpsF-F at an annealing temperature of 56°C for 40 s. Both reactions were performed with a denaturing step at 94°C for 40 s and amplification of 10 min at 72°C for 25 cycles. PCR products were used to construct a genomic library using the TOPO cloning system (Invitrogen), following the manufacturer's protocol. Sequencing reactions were performed using an ABI Prism BigDye Terminator cycle sequencing kit and were purified and sequenced on an ABI Prism 3100 genetic analyzer (Applied Biosystems, Foster City, CA). Sequences were assembled using Sequencher (version 4.8) software (Gene Codes). Additional primers were designed to fill gaps or to correct potential errors or low-quality sequences, as needed. Artemis software (42) was used to annotate the CPS locus.
PCR primer design.
A database of 18 CPS loci was created to identify unique regions of each serotype, and PCR primers were designed using the online software MuPlex MultiObjective multiplex PCR (40). Primers were designed with the following parameters: length between 18 and 30 residues, 20 to 50% GC content, and melting temperature ranging from 57 to 63°C. Primer sequences were further verified for absence of dimerization or hairpin formation using the web-enabled AutoDimer interface (46). The primer sets were designed in two mixes, alpha and beta, to produce amplicons that differ by at least 20 bp from the other amplicons in the same mix.
Multiplex PCR.
PCRs were performed using AmpliTaq DNA polymerase FS (Perkin-Elmer–Applied Biosystems) and a 2720 thermal cycler. Alpha and beta mixes are used to obtain a final concentration of 3.25 ng/μl of each primer in the PCR. All PCRs were performed using the following parameters: 94°C for 30 s, 56°C for 30 s, and 72°C for 45 s for a total of 29 cycles. The PCR products were analyzed by gel electrophoresis on 15-cm-long 2% agarose gels in 0.5× TBE (Tris-borate-EDTA) buffer at 175 V for 75 min. The sizes of the PCR products and corresponding serotype were determined by comparison with a 100-bp molecular size standard (New England BioLabs).
Nucleotide sequence accession numbers.
The accession numbers for the CPS sequences described in this paper are shown in Table 2.
Table 2.
Summary of sequenced C. jejuni CPS loci
| Penner type | Size (bp) | GenBank accession no. | GC (%) | No. of genes | No. of MeOPN transferases | Heptose | Deoxyheptose | No. of sugar transferases | CPS structure available |
|---|---|---|---|---|---|---|---|---|---|
| HS1 | 15,180 | BX545859a | 26.8 | 11 | 1 | No | No | 3 | 22 |
| HS2 | 34,180 | AL111168.1b | 26.5 | 28 | 2 | Yes | No | 8 | 18 |
| HS3 | 26,371 | HQ343268f | 27.3 | 23 | 1 | Yes | Yes | 10 | 3 |
| HS3/13/50 | 26,371 | HQ343267f | 27.3 | 23 | 1 | Yes | Yes | 10 | Monteiro et al. (unpublished) |
| HS4 | 22,836 | HQ343269f | 28.0 | 18 | 2 | Yes | Yes | 4 | Monteiro et al. (unpublished) |
| HS4/13/64 | 23,423 | AASY01000000c | 28.0 | 18 | 2 | Yes | Yes | 4 | 9 |
| HS6 | 26,729 | NC_009839d | 27.6 | 23 | 0 | No | No | 8 | 28 |
| HS8 | 22,063 | HQ343270f | 27.1 | 18 | 0 | Yes | Yes | 5 | Monteiro et al. (unpublished) |
| HS10 | 27,307 | HQ343271f | 27.1 | 25 | 1 | Yes | Yes | 4 | Monteiro et al. (unpublished) |
| HS15 | 23,868 | HQ343272f | 28.3 | 22 | 1 | Yes | Yes | 5 | Monteiro et al. (unpublished) |
| HS17 | 22,064 | HQ343273f | 27.1 | 18 | 0 | Yes | Yes | 5 | Monteiro et al. (unpublished) |
| HS19 | 16,727 | BX545860a | 26.1 | 13 | 1 | No | No | 4 | 21 |
| HS23 | 24,627 | AY332625a | 27.0 | 21 | 1 | Yes | Yes | 7 | 4, 18 |
| HS36 | 24,625 | AY332624a | 26.9 | 21 | 1 | Yes | Yes | 7 | 4, 18 |
| HS23/36 | 24,625 | BX545858a | 27.1 | 21 | 1 | Yes | Yes | 7 | 4 |
| HS41 | 34,118 | BX545857a | 27.2 | 30 | 0 | Yes | Yes | 2 | 16 |
| HS42 | 23,268 | HQ343274f | 26.9 | 21 | 0 | Yes | Yes | 7 | Monteiro et al. (unpublished) |
| HS53 | 18,272 | CP000025.1e | 27.0 | 15 | 0 | Yes | Yes | 7 | 11 |
RESULTS AND DISCUSSION
Selection of strains for additional CPS locus sequencing.
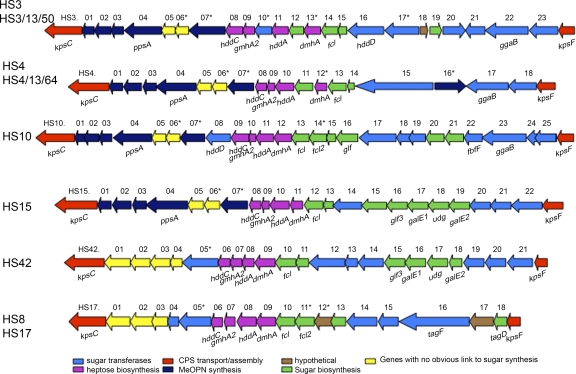
Strains were selected for CPS sequencing on the basis of limited information on serotypes common in the Armed Forces Research Institute of Medical Sciences (AFRIMS) collection. This included 5 serotypes for which no CPS sequence information existed: HS10, HS15, HS42, and the complex composed of HS8 and HS17. We compared these sequences to those of other CPS loci for the presence of conserved genes (e.g., heptose, fucose, and MeOPN biosynthesis genes). We also examined the sequence of the CPS loci of strains from two major Penner complexes, HS3 and HS4. In this case we compared the sequences of the type strains of HS3 and HS4 to other published sequences from strains with mixed serotypes within each complex in an attempt to clarify the relationship among strains within these complexes. The overall data are summarized in Fig. 1 and Table 2. Actual open reading frames (ORFs) for each stain sequenced are shown in Table S1 in the supplemental material.
Fig. 1.
Schematic of variable CPS loci from representative Penner serotypes. CPS loci were sequenced from the strains listed in Table 1. Gene names were attributed if the predicted protein showed >80% sequence identity with other known C. jejuni proteins. Genes are color coded as shown in the key to the figure on the basis of best homology to any predicted protein in databases by BLAST analyses. Genes containing homopolymeric tracts of >8 G or C residues are marked with an asterisk.
CPS loci of HS3 type strain and an HS3/13/50 strain.
The CPS loci of the HS3 type strain and an HS3/13/50 strain (BH-01-142) are very similar. Most of the predicted proteins encoded by the genes in the CPS loci of HS3 and HS3/13/50 show 100% identity; only minimal amino acid variation (≥99.3% identity) was seen in the predicted products of two genes (see Table S1 in the supplemental material). In all previously sequenced CPS loci, all of the genes were found on the same strand (the negative strand, based on the genome sequence of NCTC 11168). However, three genes in HS3 and HS3/13/50, HS3.18, HS3.19, and HS3.20, are encoded on the other strand. Both strains contain genes for MeOPN synthesis and a single predicted MeOPN transferase gene that contains a homopolymeric tract capable of phase variation (Fig. 1; see Table S1 in the supplemental material). Both strains contain genes for heptose and deoxyheptose synthesis.
The structure of the CPS of the HS3 type strain is a repeating disaccharide of Gal and ld-ido-Hep [ld-ido-Hep-(1 → 4)-Gal-(1 → 3)]n (3). The structure of the CPS of the HS3/13/50 strain is being determined (M. A. Monteiro, personal communication), but composition analyses have indicated that this CPS is composed of Gal, 6-deoxy-ido-Hep (6d-ido-Hep), and small amounts of ld-ido-Hep (37). Moreover, 31P nuclear magnetic resonance detected the presence of MeOPN on the CPS of HS3/13/50 (37); MeOPN was not reported on the CPS of the HS3 type strain, likely due to differences in techniques used in this earlier study. Since the LOS cores of these two strains are identical (3, 37), it is likely that the presence of 6d-ido-Hep in HS3/13/50 CPS is responsible for the mixed serotype.
Comparison of CPS loci of HS4 type strain and an HS4/13/64 strain.
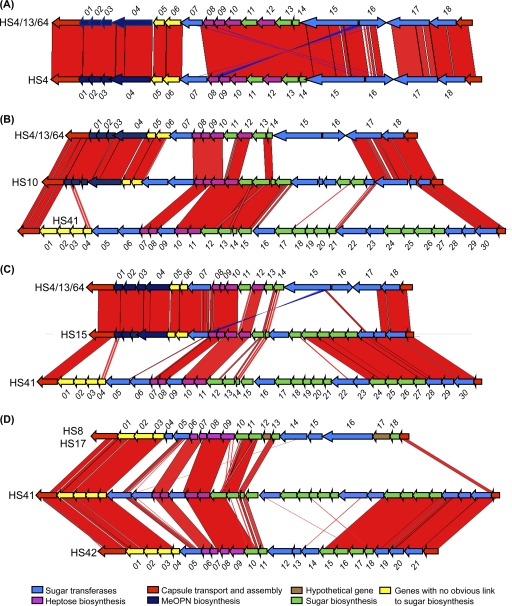
The CPS locus of the HS4 type strain was very similar to that of a strain that serotyped as HS4/13/64 (CG8486) (9) (Fig. 2A). In both strains, all genes except one, HS4.16, were encoded on the negative strand. The polysaccharide structure of the CPS of the type strain of HS4 has not been determined, but that of HS4/13/64 has been shown to be a repeating disaccharide of 4-substituted N-acetyl-β-d-glucopyranosamine and 3-substituted 6-deoxy-β-d-ido-heptopyranose, with MeOPN attached to both the O-2 and O-7 positions of the heptopyranose residue (9). At the DNA level, both the HS4 type strain and HS4/13/64 have genes for heptose, deoxyheptose, and MeOPN synthesis, as well as two putative MeOPN transferase genes (Fig. 1; see Table S1 in the supplemental material). The predicted proteins encoded by the CPS genes of HS4/13/64 and the HS4 type strain show >92% identity, with the exception of the two putative MeOPN transferases, HS4.07 and HS4.16 (Fig. 2A). HS4/13/64 contains two MeOPN transferases (Cj8486_1464c and Cj8486_1475c), consistent with the presence of MeOPN on either the O-2 or O-7 position of the heptose (9). In contrast, in the type strain of HS4, one of these MeOPN transferases (HS4.07) is truncated, suggesting that the HS4 CPS contains MeOPN in a single position. Structural studies to examine this are under way (M. A. Monteiro, personal communication).
Fig. 2.
Comparison of sequences of CPS loci of C. jejuni strains. (A) Comparison of the sequences of the CPS loci of C. jejuni HS4/13/64 (strain GC8486) and the HS4 type strain; (B) comparison of the CPS loci of C. jejuni HS4/13/64 (strain GC8486), the HS10 type strain, and a strain of HS41 (strain 176.83) (17); (C) comparison of the CPS loci of C. jejuni HS4/13/64 (strain GC8486), the HS15 type strain, and an HS41 strain (strain 176.83); (D) comparison of the CPS loci of strains C. jejuni HS41 (strain 176.83) and the type strains of HS8, HS17, and HS42. Vertical bars represent identical regions between loci. The red bars correspond to regions that are oriented similarly, and blue bars indicate regions oriented in opposite directions. The outermost genes in red represent kpsC (left) and kpsF (right), two conserved genes in CPS synthesis that bracket the hypervariable CPS loci. The comparison was made using the Artemis comparison tool (ACT).
CPS sequencing of HS10.
The HS10 locus appeared to be a mosaic of HS41 (18) and the HS4 complex (9) (Fig. 2B). The sequence revealed the presence of MeOPN, heptose, deoxyheptose, and fucose biosynthesis genes. HS10 contains 8 unique genes, including a single MeOPN transferase gene.
CPS sequence of HS15.
The HS15 CPS locus also appeared, like that of HS10, to be a mosaic between HS41 and the HS4 complex, as shown in Fig. 2C. The genes from kpsC to hddA show a high degree of similarity with those in the HS4/13/64 CPS locus, including one of the two MeOPN transferases (see Table S1 in the supplemental material). The last seven genes at the right end of the HS15 CPS locus are conserved with the corresponding region of HS41. The CPS of HS41 contains arabinose (16), and the similarity of HS41 to HS15 in this region might suggest that the CPS of HS15 also contains arabinose. The CPS structure of HS15 is being determined (M. A. Monteiro, personal communication).
HS42 CPS sequencing.
The HS42 CPS locus is related to the HS41 locus (Fig. 1 and Fig. 2D; see Table S1 in the supplemental material), including the putative genes for arabinose incorporation into CPS. The major differences are that 4 unique putative glycosyl transferases that are not found in HS41 are present in HS42 and that HS42 lacks 12 genes found in HS41. The capsule locus of HS42, like that of HS41, lacks genes for MeOPN synthesis but contains conserved genes involved in heptose and deoxyheptose biosynthesis.
HS17 and HS8 CPS loci.
HS8 and HS17 form another Penner complex, and DNA sequence analysis of their CPS loci indicates that they are virtually identical to each other and are a mosaic of other CPS types. Both strains contain genes involved in heptose and deoxyheptose synthesis but lack genes for MeOPN. The only major difference between the CPS loci of HS8 and HS17 is in orf16, which encodes a putative CDP glycerol-phosphotransferase. In HS8, this ORF encodes a predicted protein of 1,619 amino acid residues, and in HS17 the protein is 1,578 amino acids. The truncation occurs at a homopolymeric tract of 6 T residues in HS8, whereas the homopolymeric tract is 7 T residues in HS17, which results in a premature truncation in HS17. The significance of this difference is not known, but it may reflect slip-strand mismatch repair (17).
There is considerable sequence identity of genes located between kpsC and gmhA2 in HS41 and those in HS8 and HS17 (Fig. 2D), although the HS41.03 and HS41.04 ORFs are fused together in HS8 and HS17 (HS8.02). This difference is not due to a homopolymeric tract. HS8.07 to HS8.13 includes heptose and fucose synthesis genes. HS8.16, HS8.17, and HS8.18 appear to be related to the HS1 locus (see Table S1 in the supplemental material).
Design of multiplex PCR.
Table 2 summarizes data for all sequenced CPS loci, including the 8 sequenced as part of this study. A database containing sequences of all available CPS loci was created to identify unique regions of each serotype. This data set included a partial sequence of the type strain of HS44, which forms a complex with HS1. The full sequence of HS44 will be presented with the HS44 CPS structure (M. A. Monteiro et al., unpublished data). Two primer sets were designed for the HS4 complex, and these are based on differences in MeOPN transferases among the sequenced strains in this complex (see above). This decision was based on an assumption that the differences among the members of the HS4 complex are due to differences in the position of MeOPN on the polysaccharide, a hypothesis that is being investigated (Monteiro et al., unpublished). These primer sets, named Mu_HS4 and Mu_8486, respectively, were designed in HS4.07 and Cj8486_1475, both putative MeOPN transferases. Since some HS4 strains contain two MeOPN transferases, a strain can theoretically be positive with both Mu_8486 and Mu_HS4, and two such clinical isolates were identified (see below). Since the CPS loci of HS8 and HS17 are so similar (see above), a single primer set was designed for this complex. Although CPS has been shown not to be the serodeterminant of the HS6 serotype (18), the CPS gene sequences in this strain do not match any of the other published sequences, and a primer set was designed for this CPS type as well. Primers were designed in regions that were found to be unique to each particular serotype (Table 3), as described in Materials and Methods. Each primer set was tested on the strain from which it was designed and the 23 strains shown in Table 1 to confirm specificity for these serotypes. Primer sets were judged to be satisfactory if they produced the expected size PCR product on their Penner serotype DNA template or related complexes and were negative for the other tested serotypes. Data are shown in Table S2 in the supplemental material.
Table 3.
Multiplex PCR primers for determination of CPS type
| Mix | Product size (bp) | Penner type recognized | Gene in which primer was designed | Forward sequence | Reverse sequence |
|---|---|---|---|---|---|
| Alpha | |||||
| Mu_HS2 | 62 | HS2 | Cj1437c | CAGCATTGGAGGATTTACAATATAT | CATCCTAGCACAACTCACTTCA |
| Mu_HS3 | 149 | HS3 | HS3.17 | GGTAAGGTTGATTCTGGGTTTAAT | AGATTAGGCCAAGCAATGATAA |
| Mu_HS4 | 370 | HS4 A | HS4.07 | TATATTTGGTTAGGGATCCA | CCTAACATATCATACACTACGGT |
| Mu_HS6 | 185 | HS6 | C8J_1331 | CATACATTTGCTTTCAGATTCTTTAC | ACACGCCTATTGTTGTTGTTC |
| Mu_HS10 | 229 | HS10 | HS10.08 | TCTTATGCAGCACGCTGAT | CAAATTCAATCGACTAGCCACT |
| Mu_HS15C | 325 | HS15 and HS31 | HS15.12 | ACAGGTAATAAAATGTGCGAGTTT | ATGCATCTGCAACATCATCC |
| Mu_HS41 | 279 | HS41 | HS41.22c | CTTACATATGCTGGTAGAGATGATATG | TGCAATCTCTAAAGCCCAAG |
| Mu_HS53 | 251 | HS53 | CJE1602 | AGGCAAGCAGGAATTGTTT | TTAATTGCTCTTTGGCAATCTT |
| Beta | |||||
| Mu_HS1D | 607 | HS1 complex | HS1.08 | TTGGCGGTAAGTTTTTGAAGA | GCAAGAGAAACATCTCGCCTA |
| Mu_HS17 | 342 | HS8 and HS17 | HS17.16 | TTCACGTGGAGGATTATTGG | TTGAACATTTCATGTGTATTCCCTA |
| Mu_8486 | 652 | HS4 B | Cj8486_1475 | GTGGACATGGAACTGGGACT | AAAACGTTTAAAGTCAGTGGAAA |
| Mu_HS23 | 161 | HS23/36 | CJJ81176_1435 | GCTTGGGAGATGAATTTACCTTTA | GCTTTATATCTATCCAGTCCATTATCA |
| Mu_HS42E | 441 | HS42 | HS42.14 | ATGGTAAAACCGGCATTTCA | ATGCTTCAGTTCCACCCAAA |
| Mu_HS44 | 148 | HS44 | Not annotated | AGAAGATGCACTAGGCTCTAG | GCTATCTAATTCCATCCCTG |
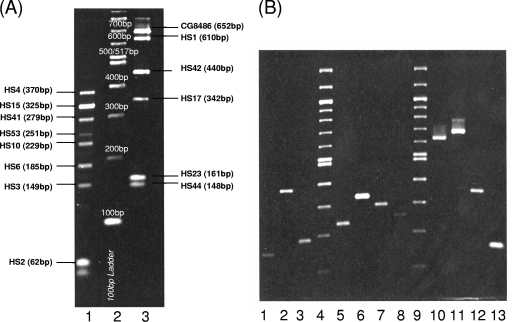
Next the primer sets were grouped into two mixes on the basis of the sizes of the products (Table 3). The alpha mix contained primers that distinguish HS2, the HS3 complex, HS6, HS10, HS15, HS41, HS53, and part of the HS4 complex (HS4 and HS13, termed HS4A). The beta mix contains primers that distinguish the HS1 complex (including HS1 and HS1/44), the HS23/36 complex, the HS8 complex (HS8 and HS17), HS42, HS44, and part of the HS4 complex (HS4/13/64 or CG8486-like termed HS4B). Following amplification, the alpha and beta mix PCRs were run separately on a 2% agarose gel in parallel with a 100-bp ladder to decipher the capsule type. Amplicon sizes ranged from 62 bp for HS2 to 652 bp for HS4B (Fig. 3). Expected sizes are listed in Table 3.
Fig. 3.
Representative multiplex PCR with alpha and beta mixes separated on 2% agarose. (A) Lane 1, mixture of PCR products amplified with the alpha primer sets on HS4A, HS15, HS41, HS53, HS10, HS6, HS3, and HS2 DNAs; lane 2, 100-bp NEB DNA standard; lane 3, mixture of PCR products amplified with the beta primer sets on HS4B, HS1, HS42, HS17, HS23, and HS44 DNAs. Amplicons were separated on 2% agarose as described in Materials and Methods. The position of each CPS-specific amplicon is shown. (B) Typical PCR products obtained with primer mixes alpha and beta. Lane 1, alpha primer sets on HS3 DNA; lane 2, alpha primer sets on HS4 DNA; lane 3, alpha primer sets on HS6 DNA; lane 4, 100-bp NEB DNA standard; lane 5, alpha primer sets on HS10 DNA; lane 6, alpha primer sets on HS15 DNA; lane 7, alpha primer sets on HS41 DNA; lane 8, alpha primer sets on HS53 DNA; lane 9, 100-bp NEB DNA standard; lane 10, PCR beta primer sets on HS1 DNA; lane 11, beta primer sets on HS4/13/64 DNA; lane 12, beta primer sets on HS8 DNA; lane 13, beta primer sets on HS23 DNA.
Validation of multiplex PCR.
To validate the multiplex PCR, the alpha and beta mixes were tested on 244 serotyped strains from Thailand and Egypt. The results are summarized in Table 4. These strains included 184 strains that typed as one of the CPS types included in the multiplex, 37 strains that typed as serotypes that were not included in the multiplex, and 23 strains that were not typeable in the Penner scheme.
Table 4.
Validation of CPS multiplex PCR with 244 strains of known Penner type
| Primer set | No. of strains with the following result: |
%a |
||||||
|---|---|---|---|---|---|---|---|---|
| Total | True positive | False positive | False negative | True negative | Accuracy | Sensitivity | Specificity | |
| HS1/44 complex | 25 | 23 | 0 | 2 | 219 | 99.18 | 92.00 | 100.00 |
| HS2 | 30 | 30 | 0 | 0 | 214 | 100.00 | 100.00 | 100.00 |
| HS3 complex | 26 | 24 | 1 | 2 | 217 | 98.77 | 92.31 | 99.54 |
| HS4 complex | 20 | 18 | 0 | 2 | 224 | 99.18 | 90.00 | 100.00 |
| HS6 | 1 | 1 | 5 | 0 | 238 | 97.95 | 100.00 | 97.94 |
| HS8/17 complex | 10 | 10 | 0 | 0 | 234 | 100.00 | 100.00 | 100.00 |
| HS10 | 14 | 13 | 2 | 1 | 228 | 98.77 | 92.86 | 99.13 |
| HS15 | 19 | 19 | 6 | 0 | 219 | 97.54 | 100.00 | 97.33 |
| HS23/36 complex | 13 | 13 | 1 | 0 | 230 | 99.59 | 100.00 | 99.57 |
| HS41 | 2 | 2 | 0 | 0 | 242 | 100.00 | 100.00 | 100.00 |
| HS42 | 8 | 8 | 0 | 0 | 236 | 100.00 | 100.00 | 100.00 |
| HS53 | 16 | 16 | 1 | 0 | 227 | 99.59 | 100.00 | 99.56 |
Accuracy was calculated as follows: (true positive + true negative)/(true positive + true negative + false positive + false negative). Sensitivity was calculated as follows: (true positive)/(true positive + false negative). Specificity was calculated as follows: (true negative)/(true negative + false positive).
Overall, the multiplex method had a specificity and accuracy of >97% and a sensitivity of >90% for the 184 strains that serotyped as 1 of the 17 serotypes covered in the multiplex (Table 4). The method detected 100% of strains of HS2 (30/30), HS8/17 (10/10), HS15 (19/19), HS23/36 (13/13), HS41 (2/2), HS53 (16/16), and HS6 (1/1). There were two false-negative results with each of the HS1/44 primers (2/25), the HS3 complex primers (2/26), and the HS4 complex primers (2/20). The HS10 primers resulted in one false-negative result (1/14) and two false-positive results (one HS1/44 and one HS44 serotype). The HS6 primers picked up five strains, two of which belonged to the HS3 complex, and the HS15 primers picked up 6 false-positive results, all of which were HS31 (see below).
There were 37 strains tested that belonged to 15 serotypes not included in the multiplex. A total of 27 of these 37 strains were negative with the multiplex primers, but 6 HS31 strains reacted with the HS15 primers, as discussed above. This might suggest that HS31 and HS15 share some genes and that primers may need to be redesigned to distinguish these serotypes. There were also individual strains that typed as HS32, HS35, and HS59 that reacted with the HS6 primers, and one HS37 strain reacted with the HS3 primers. Again, HS32, HS35, HS37, and HS59 strains have not been characterized, and the significance of these results will require additional investigation. Of the 23 strains that were untypeable in the Penner scheme, 8 reacted with the HS6 primers, 2 reacted with the HS15 primers, and 1 each reacted with the HS2 and HS10 primers. These data suggest that some strains are untypeable in the Penner scheme because the phase-variable CPS was not being expressed in the colonies selected for serology.
Conclusions.
The potential of a CPS conjugate vaccine against this pathogen offers a novel solution to the problem of C. jejuni-induced diarrhea for travelers and residents of areas where it is endemic, but determination of major CPS types in areas of endemicity is critical to development of such a vaccine. Classical Penner serotyping is based primarily on CPS, although other structures (e.g., LOS) have been shown to confer serospecificity in some cases (18, 39). Penner typing is time and labor-intensive and is currently performed in only a few labs worldwide. The availability of a multiplex PCR method to determine CPS type overcomes many of the complexities of the Penner scheme, and the method also does not require that the phase-variable CPS be expressed at the time of testing. In this study, we demonstrated the possibility of specifically recognizing CPS types by a multiplex PCR. Although the present system can discriminate some of the complexes observed within the original Penner scheme (e.g., HS4 and HS4/13/64), it has not been designed to break all of the complexes into individual serotypes but rather can distinguish related CPS types. Thus, the differences among the members of the HS23/36 complex are based on changes in phase-variable genes and cannot be discriminated by PCR. Similarly, the minor differences in sequence between HS8 and HS17 could not be distinguished.
Including this study, the CPS loci from 18 different Penner serotypes have been sequenced. Collectively, these data, including those for the 8 loci reported here, reveal a mosaic nature of the CPS genes, which is likely due to horizontal transfer among strains of this naturally transformable organism. Thus, we have demonstrated that HS10 and HS15 are mosaics of HS4 and HS41 and that HS8 and HS17 are mosaics of HS41, HS42, and HS1. We view this multiplex system to be dynamic and expect it to evolve as additional CPS loci are sequenced. The current data suggest that strains within a given complex are highly related and that the differences among some serotypes are due to phase variation of a limited number of genes. This is consistent with older data that showed serotype variations within Penner complexes (7, 24, 26, 39).
The multiplex system, in conjunction with structural analyses of additional CPS polysaccharides, will help refine our understanding of the relationship of strains within the Penner scheme and help define strains that may be immunologically cross-reactive. In another mucosal pathogen for which CPS is a protective antigen, Streptococcus pneumoniae, some CPS structures are associated with more severe disease (8, 15). The ability to rapidly determine CPS type may also help determine if specific C. jejuni CPS structures are also associated with more severe disease.
Supplementary Material
ACKNOWLEDGMENTS
We thank Eva Nielsen and Helen Tabor for performing Penner serotyping of strains, Stephen J. Savarino for providing the Egyptian strains, Piyarat Pootong and Panida Nopthai for technical assistance, and Mario A. Monteiro for helpful comments on the manuscript.
This work was supported by U.S. Naval Medical Research and Development Command Work Unit 6000.RAD1.DA3.A0308.
The opinions or assertions contained herein are the private views of the authors and are not to be construed as official or as reflecting the views of the U.S. government.
P.G., O.S., and C.M. are employees of the U.S. government, and this work was prepared as part of their official duties.
Footnotes
Supplemental material for this article may be found at http://jcm.asm.org/.
Published ahead of print on 16 March 2011.
REFERENCES
- 1. Allos B. M. 2001. Campylobacter jejuni infections: update on emerging issues and trends. Clin. Infect. Dis. 32:1201–1206 [DOI] [PubMed] [Google Scholar]
- 2. Ang C. W., Jacobs B. C., Laman J. D. 2004. The Guillain-Barre syndrome: a true case of molecular mimicry. Trends Immunol. 25:61–66 [DOI] [PubMed] [Google Scholar]
- 3. Aspinall G. O., Lynch C. M., Pang H., Shaver R. T., Moran A. P. 1995. Chemical structures of the core region of Campylobacter jejuni O:3 lipopolysaccharide and an associated polysaccharide. Eur. J. Biochem. 231:570–578 [PubMed] [Google Scholar]
- 4. Aspinall G. O., McDonald A. G., Pang H. 1992. Structures of the O chains from lipopolysaccharides of Campylobacter jejuni serotypes O:23 and O:36. Carbohydr. Res. 231:13–30 [DOI] [PubMed] [Google Scholar]
- 5. Bachtiar B. M., Coloe P. J., Fry B. N. 2007. Knockout mutagenesis of the kpsE gene of Campylobacter jejuni 81116 and its involvement in bacterium-host interactions. FEMS Immunol. Med. Microbiol. 49:149–154 [DOI] [PubMed] [Google Scholar]
- 6. Bacon D. J., et al. 2001. A phase-variable capsule is involved in virulence of Campylobacter jejuni 81-176. Mol. Microbiol. 40:769–777 [DOI] [PubMed] [Google Scholar]
- 7. Bradbury W. C., Pearson A. D., Marko M. A., Congi R. V., Penner J. L. 1984. Investigation of a Campylobacter jejuni outbreak by serotyping and chromosomal restriction endonuclease analysis. J. Clin. Microbiol. 19:342–346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Brueggemann A. B., et al. 2003. Clonal relationships between invasive and carriage Streptococcus pneumoniae and serotype- and clone-specific differences in invasive disease potential. J. Infect. Dis. 187:1424–1432 [DOI] [PubMed] [Google Scholar]
- 9. Chen Y. H., Poly F., Pakulski Z., Guerry P., Monteiro M. A. 2008. The chemical structure and genetic locus of Campylobacter jejuni CG8486 (serotype HS:4) capsular polysaccharide: the identification of 6-deoxy-d-ido-heptopyranose. Carbohydr. Res. 343:1034–1040 [DOI] [PubMed] [Google Scholar]
- 10. Fouts D. E., et al. 2005. Major structural differences and novel potential virulence mechanisms from the genomes of multiple Campylobacter species. PLoS Biol. 3:e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Gilbert M., Mandrell R. E., Parker C. T., Li J., Vinogradov E. 2007. Structural analysis of the capsular polysaccharide from Campylobacter jejuni RM1221. Chembiochem 8:625–631 [DOI] [PubMed] [Google Scholar]
- 12. Grant A. J., et al. 2005. Signature-tagged transposon mutagenesis studies demonstrate the dynamic nature of cecal colonization of 2-week-old chickens by Campylobacter jejuni. Appl. Environ. Microbiol. 71:8031–8041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Griffiths P. L., Park R. W. 1990. Campylobacters associated with human diarrhoeal disease. J. Appl. Bacteriol. 69:281–301 [DOI] [PubMed] [Google Scholar]
- 14. Guerry P., et al. 2002. Phase variation of Campylobacter jejuni 81-176 lipooligosaccharide affects ganglioside mimicry and invasiveness in vitro. Infect. Immun. 70:787–793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hanage W. P., et al. 2005. Invasiveness of serotypes and clones of Streptococcus pneumoniae among children in Finland. Infect. Immun. 73:431–435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hanniffy O. M., et al. 1999. Chemical structure of a polysaccharide from Campylobacter jejuni 176.83 (serotype O:41) containing only furanose sugars. Carbohydr. Res. 319:124–132 [DOI] [PubMed] [Google Scholar]
- 17. Hendrixson D. R. 2006. A phase-variable mechanism controlling the Campylobacter jejuni FlgR response regulator influences commensalism. Mol. Microbiol. 61:1646–1659 [DOI] [PubMed] [Google Scholar]
- 18. Karlyshev A. V., et al. 2005. Analysis of Campylobacter jejuni capsular loci reveals multiple mechanisms for the generation of structural diversity and the ability to form complex heptoses. Mol. Microbiol. 55:90–103 [DOI] [PubMed] [Google Scholar]
- 19. Karlyshev A. V., Linton D., Gregson N. A., Lastovica A. J., Wren B. W. 2000. Genetic and biochemical evidence of a Campylobacter jejuni capsular polysaccharide that accounts for Penner serotype specificity. Mol. Microbiol. 35:529–541 [DOI] [PubMed] [Google Scholar]
- 20. Korlath J. A., Osterholm M. T., Judy L. A., Forfang J. C., Robinson R. A. 1985. A point-source outbreak of campylobacteriosis associated with consumption of raw milk. J. Infect. Dis. 152:592–596 [DOI] [PubMed] [Google Scholar]
- 21. McNally D. J., et al. 2006. The HS:19 serostrain of Campylobacter jejuni has a hyaluronic acid-type capsular polysaccharide with a nonstoichiometric sorbose branch and O-methyl phosphoramidate group. FEBS J. 273:3975–3989 [DOI] [PubMed] [Google Scholar]
- 22. McNally D. J., et al. 2005. The HS:1 serostrain of Campylobacter jejuni has a complex teichoic acid-like capsular polysaccharide with nonstoichiometric fructofuranose branches and O-methyl phosphoramidate groups. FEBS J. 272:4407–4422 [DOI] [PubMed] [Google Scholar]
- 23. McNally D. J., et al. 2007. Commonality and biosynthesis of the O-methyl phosphoramidate capsule modification in Campylobacter jejuni. J. Biol. Chem. 282:28566–28576 [DOI] [PubMed] [Google Scholar]
- 24. Melby K., Storvold G., Congi R. V., Penner J. L. 1985. Serotyping of Campylobacter jejuni isolated from sporadic cases and outbreaks in northern Norway. Acta Pathol. Microbiol. Immunol. Scand. B 93:83–86 [DOI] [PubMed] [Google Scholar]
- 25. Miller W. G., et al. 2000. Detection on surfaces and in Caco-2 cells of Campylobacter jejuni cells transformed with new gfp, yfp, and cfp marker plasmids. Appl. Environ. Microbiol. 66:5426–5436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Mills S. D., Kuzniar B., Shames B., Kurjanczyk L. A., Penner J. L. 1992. Variation of the O antigen of Campylobacter jejuni in vivo. J. Med. Microbiol. 36:215–219 [DOI] [PubMed] [Google Scholar]
- 27. Monteiro M. A., et al. 2009. Capsule polysaccharide conjugate vaccine against diarrheal disease caused by Campylobacter jejuni. Infect. Immun. 77:1128–1136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Muldoon J., et al. 2002. Structures of two polysaccharides of Campylobacter jejuni 81116. Carbohydr. Res. 337:2223–2229 [DOI] [PubMed] [Google Scholar]
- 29. Oberhelman R. A., Taylor D. N. 2000. Campylobacter infections in developing countries, p. 139–154 In Campylobacter, 2nd ed. American Society for Microbiology, Washington, DC [Google Scholar]
- 30. Palmer S. R., et al. 1983. Water-borne outbreak of campylobacter gastroenteritis. Lancet i:287–290 [DOI] [PubMed] [Google Scholar]
- 31. Parkhill J., et al. 2000. The genome sequence of the food-borne pathogen Campylobacter jejuni reveals hypervariable sequences. Nature 403:665–668 [DOI] [PubMed] [Google Scholar]
- 32. Pearson B. M., et al. 2007. The complete genome sequence of Campylobacter jejuni strain 81116 (NCTC11828). J. Bacteriol. 189:8402–8403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Penner J. L., Hennessy J. N. 1980. Passive hemagglutination technique for serotyping Campylobacter fetus subsp. jejuni on the basis of soluble heat-stable antigens. J. Clin. Microbiol. 12:732–737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Penner J. L., Hennessy J. N., Congi R. V. 1983. Serotyping of Campylobacter jejuni and Campylobacter coli on the basis of thermostable antigens. Eur. J. Clin. Microbiol. 2:378–383 [DOI] [PubMed] [Google Scholar]
- 35. Pimentel M., et al. 2008. A new rat model links two contemporary theories in irritable bowel syndrome. Dig. Dis. Sci. 53:982–989 [DOI] [PubMed] [Google Scholar]
- 36. Poly F., et al. 2007. Genome sequence of a clinical isolate of Campylobacter jejuni from Thailand. Infect. Immun. 75:3425–3433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Poly F., et al. 2008. Characterization of two Campylobacter jejuni strains for use in volunteer experimental-infection studies. Infect. Immun. 76:5655–5667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Pope J. E., Krizova A., Garg A. X., Thiessen-Philbrook H., Ouimet J. M. 2007. Campylobacter reactive arthritis: a systematic review. Semin. Arthritis Rheum. 37:48–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Preston M. A., Penner J. L. 1989. Characterization of cross-reacting serotypes of Campylobacter jejuni. Can. J. Microbiol. 35:265–273 [DOI] [PubMed] [Google Scholar]
- 40. Rachlin J., Ding C., Cantor C., Kasif S. 2005. MuPlex: multi-objective multiplex PCR assay design. Nucleic Acids Res. 33:W544–W547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Rao M. R., et al. 2001. Pathogenicity and convalescent excretion of Campylobacter in rural Egyptian children. Am. J. Epidemiol. 154:166–173 [DOI] [PubMed] [Google Scholar]
- 42. Rutherford K., et al. 2000. Artemis: sequence visualization and annotation. Bioinformatics 16:944–945 [DOI] [PubMed] [Google Scholar]
- 43. Sambrook J., Fritsch E. F., Maniatis T. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 44. Spiller R. C. 2007. Role of infection in irritable bowel syndrome. J. Gastroenterol. 42(Suppl. 17):41–47 [DOI] [PubMed] [Google Scholar]
- 45. Szymanski C. M., et al. 2003. Detection of conserved N-linked glycans and phase-variable lipooligosaccharides and capsules from campylobacter cells by mass spectrometry and high resolution magic angle spinning NMR spectroscopy. J. Biol. Chem. 278:24509–24520 [DOI] [PubMed] [Google Scholar]
- 46. Vallone P. M., Butler J. M. 2004. AutoDimer: a screening tool for primer-dimer and hairpin structures. Biotechniques 37:226–231 [DOI] [PubMed] [Google Scholar]
- 47. Waldenstrom J., et al. 2010. Campylobacter jejuni colonization in wild birds: results from an infection experiment. PLoS One 5:e9082. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.